The R-diastereomer of 6′-O-toluoyl-carba-LNA modification in the core region of siRNA leads to 24-times improved RNA silencing potency against the HIV-1 compared to its S-counterpart†
Suman
Dutta
a,
Nipa
Bhaduri
a,
Ram Shankar
Upadhayaya
bc,
Neha
Rastogi
a,
Sunita G.
Chandel
a,
Jaya kishore
Vandavasi
b,
Oleksandr
Plashkevych
d,
Ramakant A.
Kardile
b and
Jyoti
Chattopadhyaya
*d
aInstitute of Molecular Medicine, BIPL, Building-B, Ist Floor, Block-EP & GP, Salt Lake Electronics Complex, Sector-V, Kolkata, 700 091, India
bInstitute of Molecular Medicine, International Biotech Park, Genesis Campus, Phase II Opp Infosys, Tal. Mulshi Hinjewadi, Pune, 411 057, India
cInstitute of Biomedicinal Chemistry, G-16, additional MIDC Jejuri, Tal. Purandhar, Pune, 412303, India
dProgram of Chemical Biology, Institute of Cell and Molecular Biology, Biomedical Centre, Uppsala University, SE-75123, Uppsala, Sweden. E-mail: jyoti@boc.uu.se; Fax: +46-18-554495; Tel: +46-18-4714577
First published on 27th September 2011
Abstract
The modified siRNA with pure [6′(S)-O-(p-toluoyl)-7′(S)-methyl]-carba-LNA [6′(S)-O-toluoyl-jcLNA] at position T13 displayed an IC50 of 79.8 nM, which has been found to be nearly 24-times less potent as a HIV-1 RNAi silencing agent against TAR RNA than that of the corresponding pure [6′(R)-O-(p-toluoyl)-7′(S)-methyl]-jcLNA [6′(R)-O-(p-toluoyl)-jcLNA] counterpart [IC50 3.3 nM]. The later [6′(R)-O-(p-toluoyl)-jcLNA]-modified siRNAs have been found to be nearly 2-fold more efficient as a silencing agent than the corresponding 6′-deoxy-jcLNA modified siRNA [IC50 8.1 nM], and also nearly 3-fold more effective as a silencing agent than that of LNA-modified siRNA [IC50 11.7 nM], thereby showing that the 6′-carbon center in the jcLNA-modified siRNA in the core region is relatively more exposed to the Ago protein in the RISC with a clear chirality preference for the siRNA cleavage reaction. It is noteworthy that the IC50 of jcLNA-modified siRNAs are very comparable to that of the native siRNA [1.8 nM]. The jcLNA derivatized siRNAs, however, have a clear advantage of being, in general, considerably more stable in human serum. The main structural difference in duplexes of the antisense strand of the 6′(R or S)-O-(p-toluoyl)-jcLNA modified siRNA and target RNA duplex is found to be the spatial orientation of the 6′(R)-O-toluoyl group, which is exposed towards the edge of the duplex backbone, while the 6′(S) makes the minor groove relatively inaccessible for the Ago protein in the RISC. Clearly, any further C6′-modification in jcLNA-modified siRNAs with any hydrophobic group for tighter binding and cleavage or for cross-linking in the core region should preferably be done in the 6′(R)-stereochemistry.
Introduction
The potential use of siRNA as therapeutic tool has been validated in multiple animal studies.1–3 Some essential requirements of a successful siRNA have been tackled effectively with chemically modified siRNAs4–8 by improving nuclease stability, delivery into the cell with enhanced targeting ability, and with reduced off-targeting with minimal cytotoxic threat. In this regard, LNA9–15 and its carba-modifications5,16–21 (Fig. 1) have emerged as promising alternatives to native siRNAs.4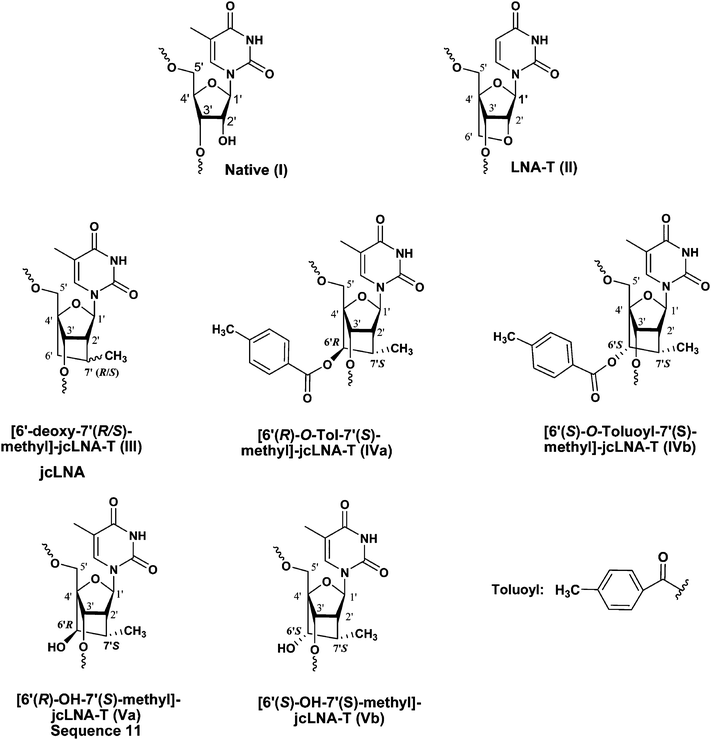 | ||
| Fig. 1 Chemical structures of native-T (I), LNA-T (II), 6′-deoxy-jcLNA-T (III), 6′(R)-O-Tol-jcLNA-T (IVa) and 6′(S)-O-Tol-jcLNA-T (IVb) nucleotides used in the siRNA modifications. Corresponding IC50 values of the native and modified siRNAs as well as their serum stability are shown in Table 1. | ||
In our previous study20 we had successfully employed locked carba-LNAs [6′-deoxy-7′(R/S)-methyl]-carba-LNA, abbreviated as ‘jcLNA’20 (III), (Fig. 1) modified siRNAs in the guiding antisense strand in the successful in vitro inhibition of HIV-1 replication. More importantly, these jcLNA modified siRNAs targeting the HIV-1 TAR region have displayed significant improvement in potency as well as stability in human serum over the LNA counterparts in a position dependent manner.20 The radical cyclisation method used in the synthesis of jcLNA22 paved the way for incorporation of chemical moieties at the C6′ position of the fused carbocyclic ring in jcLNA. As a further development, we had incorporated the C6′-OH group in R or S configuration in jcLNA 6′(R)-OH, as in Va in Fig. 1, or 6′(S)-OH-jcLNA, as in Vb, (Fig. 1) in dinucleotides which conferred enhanced stability of the corresponding dinucleoside monophosphates towards SVPDE23 in comparison with jcLNA or LNA. Interestingly, distinct stereochemical preference of the 6′-OH group (R versus S configuration at the C6′ position, as VaversusVb in Fig. 1) in jcLNA influences the stability of the corresponding dinucleotides23 when the 6′-OH (Fig. 1) was placed either at the 5′- or 3′-end. When placed at the 5′-end the Kcat/Km value of the 6′(S)-OH-jcLNA, as in Vb, containing dinucleotide (Kcat/Km = 0.0078 μM min−1) was comparable to that of the parent jcLNA, as in III (Kcat/Km = 0.0059 μM min−1), containing counterpart. However both III and Vb had shown the Kcat/Km ∼75 times lower (which means more stable duplex) than the corresponding 6′(R)-OH-jcLNA, as in Va, counterpart (Kcat/Km = 0.5865 μM min−1), as well as in the corresponding LNA, as in II (Kcat/Km = 0.625 μM min−1). This significantly lower nuclease resistance (higher Kcat/Km value) of the 6′(R)-OH isomer was explained by the steric proximity of the 6′(R)-OH to the internucleotidic phosphate resulting in stabilisation via hydrogen bonding and leading to enhanced catalysis by the nuclease. At the 3′-terminus the 6′(S)-OH-jcLNA, as in Vb in Fig. 1, counterpart showed no cleavage whereas the Kcat/Km value of the 6′(R)-OH-jcLNA, as in Va in Fig. 1, counterpart was comparable to that of jcLNA, as in III in Fig. 1.
Interesting properties of 6′(S)-OH-jcLNA and 6′(R)-OH-jcLNA oligos evaluated previously23 as antisense oligonucleotides prompted us to study further the influence of other types of 6′-substituents for their gene silencing ability against HIV-1 replication. We have focused our attention on hydrophobic toluoyl esters (IVa and IVb in Fig. 1) which have been introduced in the present work into siRNAs targeting the HIV-1 TAR1 region. Our studies reveal that 6′(S)-O-Tol-jcLNA and 6′(R)-O-Tol-jcLNA substituted siRNAs (10–17) have ca. 3-fold stronger RNA silencing ability than that of the LNA-modified (2–5) counterpart (Fig. 1).
We herein report the biological evaluation of 6′-O-Tol-jcLNA mono- and di-substituted siRNAs (with both pure 6′R and 6′S isomers) towards the TAR1 region of HIV-1 including RNAi silencing efficiency and siRNA stability in the blood serum.
Results and discussion
The 6′-O-Tol-jcLNA-T nucleotide was substituted for U at different positions within the antisense strand of siRNAs targeting the HIV-1 TAR-1 region (Table 1). The efficiency of silencing was obtained by assessing the IC50 values through a single cycle replication in vitro model24–26 of each siRNA from the dose response studies in co-transfection experiments with 100 ng of a HIV-1 molecular clone pNL4-3 and varying doses of the modified siRNAs in HEK293T cells. 48 h post-transfection p24 ELISA was done with culture supernatants to evaluate virus formation.20 We have also performed20 Western blot to estimate the IC50 through determination of residual intracellular protein (Fig. S3 in the ESI†) and RT-PCR to estimate the residual mRNA levels (Fig. S4 in the ESI†) in order to estimate the IC50 of HIV-1 TAR RNA inhibition.| a Deprotection of exocyclic amino groups using methanolic ammonia resulted in hydrolysis of 6′(R)-toluoyl ester to give 6′(R)-OH-jcLNA (as shown by MALDI for siRNA 11, and one of the O-Tol group in 13 and 17, see ESI: Fig. S17, S19 and S23†). |
|---|

|
Only substitution in the core region of 6′(S)-O-Tol-jcLNA and C6′(R)-O-Tol-jcLNA modified siRNAs (substitution at T13) led to significant diastereospecific differences in IC50 while substitution of these two stereoisomers in other positions has been found to produce no difference at all (Table 1). For the T13 modification, siRNA 14(S) displayed nearly 24-fold poorer RNA silencing potency (i.e. higher IC50) than that of the R isomer, siRNA 10(R), in that the latter displayed an IC50 of 3.3 ± 0.2 nM (inset A, Fig. S1 in the ESI†) which was nearly 2-fold more efficient than the corresponding jcLNA (siRNA 6) (Fig. 2 and Table 1) and nearly 3-fold more potent than the LNA counterpart (siRNA 2) (Fig. 2 and Table 1). 6′(R)-OH-jcLNA modified siRNA 11(R) (IC50 of 6.4 ± 1.0 nM) and 6′(S)-O-Tol-jcLNA 15(S) modifications (IC50 of 7.1 ± 1.2 nM) at T1 resulted in more than 2-fold higher silencing efficiency compared to jcLNA (siRNA 7, IC50 = 17.7 ± 1.2 nM) (Fig. 2 and Table 1). This unique observation supports that the C6′ position substituted with a hydrophobic group (toluoyl ester) or a hydrophilic group (hydroxyl) is well tolerated and modulates the siRNA inhibition efficiency successfully. Further comparison of activity suggests that the S diastereomer [siRNA 15(S), IC50 = 7.1 ± 1.2 nM] displayed 2-fold better silencing efficiency than that of jcLNA (siRNA 7, IC50 = 17.7 ± 1.2 nM) (insets C and D, Fig. S1 in the ESI†). For the T20 modification marginal improvement of RNA silencing efficiency was observed for the 6′-O-Tol-jcLNA diastereomer [siRNA 12(R), IC50 = 2.3 ± 0.4 nM and siRNA 16, IC50 = 3.6 ± 0.5 nM] (insets E and F, Fig. S1 in the ESI†) compared to jcLNA (siRNA 8, IC50 = 4.0 ± 0.9 nM) (Fig. 2 and Table 1) and more than 2-fold improvement over LNA (siRNA 4, IC50 = 7.1 ± 1.6 nM) (Fig. 2 and Table 1).
 | ||
| Fig. 2 Summary of IC50 values. IC50 values have been obtained from dose response studies of LNA-T modified siRNAs (2–5 in red in Table 1), jcLNA-T modified siRNAs (6–9 in green in Table 1) and 6′(R)-O-Tol-jcLNA-T (IVa) modified siRNAs 10, 12 and 13, (deep-blue); siRNA 11 has 6′(S)-OH-jcLNA-T (Va) and [6′(S)-O-Tol-jcLNA-T (IVb) modified siRNAs 14–17 (sky-blue) from p24 ELISA of culture supernatant. Right inset shows the zoomed scale for IC50 values for T1 + T20 modifications, which have been found to be the most active silencing agents for target HIV-1 TAR RNA. Results are average of at least three independent experiments. | ||
Interestingly, introduction of two simultaneous modifications at positions 1 and 20 (T1 + T20 in the isomeric pair of siRNAs with either all R or all S configuration, 13(R) versus17(S), Table 1) negatively affects the silencing efficiency for the 6′-O-Tol-jcLNA modifications in that a nearly 8-fold increase in IC50 was observed for these two double-substituted siRNAs [with 6′-O-Tol-jcLNAs (13(R) and 17(S))] over jcLNA (siRNA 9) (Fig. 2 and Table 1): siRNA 13 (R isomer) and siRNA 17 (S isomer), having T1 + T20 modification, displayed an IC50 of 3.2 ± 0.9 nM and 3.6 ± 0.5 nM respectively (insets G and H, Fig. S1 in the ESI†), whereas the IC50 of siRNA 9 with double substitution with jcLNA was found to be 0.5 ± 0.1 nM which represents a nearly 3-fold stronger activity compared to that of the native (siRNA 1) and 2-fold lower than the corresponding LNA (siRNA 5) (Table 1). MALDI spectra (ESI, Fig. S19 and S23†) of the double substitution at T1 and T20 siRNAs (siRNAs 13 or 17) have shown that only one of the 6′-O-Tol groups has been deprotected to 6′-OH. This suggests that the deprotection took place at the terminal T1 modification as observed in the case of siRNA 11(R) which has hydrolysed under our deprotection conditions selectively because it was more exposed to the bulk ammonia solution, while the T20 modified 6′-O-Tol group was protected from the reagent and hence found to stay protected.
These results clearly suggest that end modifications of the antisense strand do not present any isomeric bias in activity (in terms of IC50), whereas the modifications in the core region (T13), which is in the vicinity of the RISC cleavage site, markedly lead to a difference in silencing efficiency between the 6′(R)-O-Tol-jcLNA and 6′(S)-O-Tol-jcLNA stereoisomers. It is noteworthy that the RISC mediated target cleavage has been shown to follow the mechanism of dsDNA hydrolysis directed by restriction enzymes and RNase III enzyme leading to subsequent formation of 3′-OH and 5′-PO43− termini.21 Bulky 2′-O-methyl substitution at a cleavage site residue (position 9) of substrate RNA has been shown to significantly inhibit RISC mediated target hydrolysis.21 This has been attributed to steric hindrance of the conformational transitions by catalytic residues or metal cofactors or by the bulky substituent present in that region.27
Since the cleavage site residues of the 21-mer siRNA spans in the position 9–12 in the antisense strand strand,33a,b the significantly improved silencing efficiency associated with 6′(R)-O-Tol-jcLNA modification at the T13 site in siRNA 17(S) hints its direct stereochemical contribution to the RISC endonucleolytic activity at the HIV TAR1 site. The encouraging results of this study suggest a possibility of introduction of other types of chemical modifications covering hydrophobic and hydrophilic substituents at C6′ in jcLNA to control the gene silencing RNAi activity or to understand molecular interactions between the siRNA and Ago in RISC by fluorescent labelling experiments.
To compare the IC50 of virus inhibition with the suppression of intracellular viral transcripts the IC50 of p55Gag downregulation was calculated from Western blot experiments (Table 1 and Fig. S3 in the ESI†).20 As with the carba-LNAs and LNA the IC50 of p55Gag inhibition tallied well with those obtained from ELISA. Furthermore RTPCR experiments (qualitative) reflected dose dependent down regulation of Gag as well as TAR amplicons (Fig. S4 in the ESI†).20
For a potential therapeutic candidate low cytotoxicity as well as survival in body fluids like serum is of utmost importance. 48 h post-transfection cells transfected with 6′(R)-O-Tol-jcLNA containing siRNAs (10–13) and 6′(S)-O-Tol-jcLNA containing siRNAs (14–17) which showed more than 70% cell viability as revealed by MTT assay (Fig. S5 in the ESI†).20
To test the nuclease stability of the 6′(R or S)-O-Tol-jcLNA containing siRNAs, the samples were incubated in 100% human serum at a final concentration of 13 μM of ds siRNA for different time intervals and subsequently 3 μl aliquots were withdrawn and subjected to 20% non-denaturing TBE–PAGE and stained with EtBr.20 Next, t1/2 values were calculated from the % ds-siRNA remaining at different time points from the best fit first order exponential decay curve. Similar to silencing efficiency no isomeric distinction in the nuclease stability has been observed (Table 1). However, in all cases both the 6′(R) and 6′(S) isomers presented t1/2 values higher than those of the native as well as LNA-modified siRNAs (Table 1). Significant improvement of t1/2 of 6′-O-Tol-jcLNA modification over that of the jcLNA was observed for T1 position modification. siRNA 11 6′(R) and siRNA 15 6′(S) displayed t1/2 values of 8.0 h and 7.5 h respectively (insets C and D, Fig. S2 in the ESI†) which are almost 2-fold higher than that of the jcLNA (siRNA 7, 4.3 h) (Table 1), although siRNA 11 was identified as having the 6′(R)-OH. For modifications T13 and T1 + T20 the 6′(S) isomers presented t1/2 values (Table 1) marginally higher than the 6′(R) isomers; the values of 6′(S) isomers being comparable with those of jcLNA modified siRNA 6 (Table 1). siRNA 10 with 6′(R) and siRNA 14 with C6′(S), having T13 modification, displayed t1/2 values of 6.3 h and 8.1 h (insets A and B, Fig. S2 in the ESI†) as against 8.7 h of corresponding jcLNA (siRNA 6) (Table 1). For T1 + T20 bis-modifications siRNA 13(R) and siRNA 17(S) displayed t1/2 values of 8.3 h and 9.1 h (insets G and H, Fig. S2 in the ESI†) respectively as against 13.6 h of corresponding jcLNA (siRNA 9) (Table 1). Stabilities of 6′-O-Tol-jcLNAs were found to be lower in comparison to jcLNA for T20 modification. siRNA with 12(R) and siRNA with 16(S) presented t1/2 values of 8.8 h and 7.5 h respectively (insets E and F, Fig. S1 in the ESI†), compared to 11.9 h (Table 1) of the corresponding 6′-deoxy-jcLNA (siRNA 8). Although diastereomeric distinction towards SVPDE or blood serum digestions has indeed been observed23 for the diastereomeric 6′-OH-jcLNA substituted 2′-deoxy-dinucleoside monophosphates or single-stranded antisense 2′-deoxy-oligonucleotides (up to 48 h), the double-stranded siRNAs in the present study have shown consistently lower levels of the nuclease stability (compared to 2′-deoxy analogs) in human serum without any diastereomeric bias. The degradation of our ds-siRNA depends on the property of human serum which has been proven to be a reservoir of the ds-RNA degrading RNase A like enzymes. These enzymes along with other exonucleases have a profound effect on the kinetics of degradation of dsRNA versus ssRNA in the intracellular milieu, and hence a ‘thumb rule’ pattern of the stereoisomer-dependent nuclease stability may not be apparent. Nevertheless, the observed general improvement in silencing efficiency of 6′-O-Tol-jcLNA modified siRNAs [compound IVa (siRNA 10–12) and IVb (siRNA 14–16), Fig. 1 and Table 1 over the jcLNA counterpart [compound III (siRNA 6–8), Fig. 1 and Table 1] and comparative stability in human serum makes 6′-O-Tol-jcLNA attractive modifications to be studied further in the therapeutic context.
Molecular structure of modified siRNA–RNA duplexes containing 6′(S)-O-Tol- and 6′(R)-O-Tol-jcLNA stereoisomers
Preliminary structures of the chemically modified double-stranded 6′-O-Tol (R or S)-jcLNA modified siRNAs (sequences 10 and 14 in Table 1) have been calculated using molecular mechanics and Amber 94 force field as implemented in Amber 10 (ref. 28), utilizing recently modified backbone parameters by Pérez et al.,29 explicit water solvent using periodic boxes containing 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 806 TIP3P30 water molecules and 40 Na+ around the native and modified siRNA–RNA duplexes and the force field parameters for the modified residues obtained from 6–31G** ab initio geometry optimizations performed using GAUSSIAN 0331 (see Table S1 in the ESI†). The minimized structures suggest that the remarkable 24-fold difference in IC50 values (3.3 ± 0.2 nM for 6′(R)-O-Tol and 79.8 ± 17.0 nM for 6′(S)-O-Tol, Table 1) is probably a result of intramolecular interaction between the hydrophobic O-Tol group at C6′ and the Ago protein. The main structural difference between these siRNA–RNA duplexes is found to be the stereochemical position of the 6′-O-Tol group which in the case of 6′(R)-diastereoisomer is exposed towards the edge of the duplex backbone while 6′(S)-O-Tol is located hindering the access to the minor groove of the 6′(S)-O-Tol-jcLNA modified siRNA-RNA duplex (Fig. 3).
806 TIP3P30 water molecules and 40 Na+ around the native and modified siRNA–RNA duplexes and the force field parameters for the modified residues obtained from 6–31G** ab initio geometry optimizations performed using GAUSSIAN 0331 (see Table S1 in the ESI†). The minimized structures suggest that the remarkable 24-fold difference in IC50 values (3.3 ± 0.2 nM for 6′(R)-O-Tol and 79.8 ± 17.0 nM for 6′(S)-O-Tol, Table 1) is probably a result of intramolecular interaction between the hydrophobic O-Tol group at C6′ and the Ago protein. The main structural difference between these siRNA–RNA duplexes is found to be the stereochemical position of the 6′-O-Tol group which in the case of 6′(R)-diastereoisomer is exposed towards the edge of the duplex backbone while 6′(S)-O-Tol is located hindering the access to the minor groove of the 6′(S)-O-Tol-jcLNA modified siRNA-RNA duplex (Fig. 3).
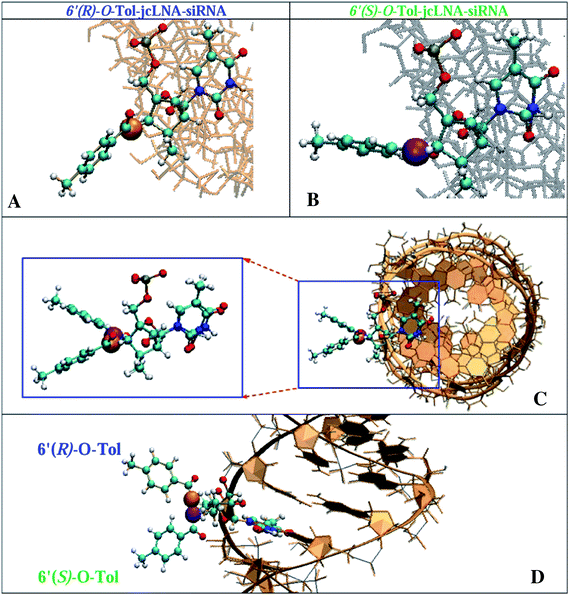 | ||
Fig. 3 Molecular structure of the central region of C6′(R)-O-Tol (panel A, O-atom of the O-Tol group is marked in orange) and C6′(S)-O-Tol (panel B, O-atom of O-Tol group is marked in magenta)—C6′-achiral-cLNA T13 modified-siRNA duplexes (10 and 14 in Table 1) with the target RNA as well as the overlap of these structure, in panels C and D. The C6′(S)-O-Tol is located at the entry of the minor grove of the duplex while C6′(R)-O-Tol is pointing out (see also Fig. S6 in the ESI†). The structures are minimized using the molecular mechanics technique and Amber force field. Amber 94 force field as implemented in Amber 10 (ref. 28), recently modified backbone parameters by Pérez et al.29 have been employed for the simulations. Periodic boxes containing 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 806 TIP3P30 water molecules and 40 Na+ have been placed around the native and modified siRNA–RNA duplexes to mimic ionic and aqueous environment. The force field parameters for the modified residues have been obtained from 6–31G** ab initio geometry optimizations performed using GAUSSIAN 03 (ref. 31) and are provided in Table S1 in the ESI†. The molecular structures are visualized using VMD.32 The full duplex structure is shown in Fig. S6 of the ESI†. 806 TIP3P30 water molecules and 40 Na+ have been placed around the native and modified siRNA–RNA duplexes to mimic ionic and aqueous environment. The force field parameters for the modified residues have been obtained from 6–31G** ab initio geometry optimizations performed using GAUSSIAN 03 (ref. 31) and are provided in Table S1 in the ESI†. The molecular structures are visualized using VMD.32 The full duplex structure is shown in Fig. S6 of the ESI†. | ||
Conclusions
(1) In this study we have employed 6′-O-Tol (R or S)-jcLNA (10–17) modifications at various positions of the antisense (guide) strand of siRNAs targeting HIV TAR1 RNA. With the exception of the core region modification (region encompassing the RISC cleavage site, which is putatively A30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U13 base-pair in our case, although we have not done any experiment that rules out in anyway the alternative cleavage site at A10
U13 base-pair in our case, although we have not done any experiment that rules out in anyway the alternative cleavage site at A10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U33 and/or A8
U33 and/or A8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U35 and/or A6
U35 and/or A6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U37) both the 6′(R)-O-Tol and 6′(S)-O-Tol diastereomers are very similar in their siRNA activity, as evident from similar IC50 values.
U37) both the 6′(R)-O-Tol and 6′(S)-O-Tol diastereomers are very similar in their siRNA activity, as evident from similar IC50 values.
(2) This study represents the significance of chiral versus achiral nucleotide modification at the putative T13 cleavage site. The distances from the 6′(R), 6′(S) and 7′-CH(Me) to the deep minor groove point, O2 of the pyrimidine base are as follows: d[6′(R)-O-Tol ↔ O2(py) = 6.66 Å], d[6′(S)-O-Tol ↔O2(py) = 5.76 Å] and d[7′C ↔ O2(py) = 4.24 Å, whereas the distance between O2′ ↔ N3′ = 3.98 Å]. Thus this simple distance estimation from 6′-O-Tol to the interior of minor groove shows that 6′(R)-O-Tol is relatively located in the exterior part of the minor groove. Hence it is more available for intramolecular interactions, compared to that of 6′(S)-O-Tol.
(3) Our results show that silencing efficiency (IC50) increases for the single modification in a characteristic manner for LNA modification versus jcLNA modification, in which 3′-end modification gives the strongest inhibition of HIV-specific TAR RNA in terms of IC50. This is followed by modification at the putative cleavage site (T13), and then for the 5′-end modification at T1. For LNA modification, on the other hand, as in siRNAs 2–4; IC50 decreases in the following order: 13.7 nM for T1, 11.7 nM for T13 and 7.1 nM for T20, whereas IC50s for corresponding single jcLNA modification, as in siRNAs 6–8; at the identical sites are 17.7 nM for T1, 8.1 nM for T13 and 4.0 nM for T20.
The unusual diastereomeric specific silencing efficiency of 6′-O-Tol (R and S)-jcLNA at T13 located near the putative cleavage site is especially interesting in light of results of the earlier attempts to modify the guide strand at or near the cleavage site in the core region (9–11 nt region) which have shown reduced silencing efficiency for 2′-OH, 2′-OMe, and 2′-O-MOE substituted core regions of the guide strand.33a,b
(4) Hence, we have compared these IC50s with those of the modified siRNAs with single chiral 6′-O-Tol (R or S)-jcLNA (as in siRNAs 10–17): the IC50 for 6′(R)-O-Tol-jcLNA (as in siRNA 10 and 12) modification is 3.3 nM (T13) and 2.3 nM (T20), at the identical sites, which are all better silencing than those jcLNA and LNA counterparts. One exception which has been discussed earlier is that sequence 11 containing 6′-OH modification shows IC50 of 6.4 nM (T1). On the other hand, the IC50 values for 6′(S)-O-Tol-jcLNA (as in siRNA 14–16) for the corresponding site (T1, T13 and T20) are 7.1 nM (T1), 79.8 nM (T13) and 3.6 nM (T20) respectively, which are all higher than the corresponding 6′(R)-O-Tol-jcLNA modification (as in siRNA 10–12). This shows that the R-stereochemistry of 6′(R)-O-Tol-jcLNA in the modified siRNA (as in siRNA 10–13) does not depend on the position of modification (tested at sites T1, T13 or T20) and was consistently found to be more potent compared to those of the 6′(S)-O-Tol-jcLNA (as in siRNA 14–17) modified counterpart.
(5) The most interesting results were found for the T13 modified site with 6′(R)-O-Tol-jcLNA (siRNA 10) and 6′(S)-O-Tol-jcLNA (siRNA 14) and in fact the latter is ∼24 times less effective as silencing agent compared to the former in terms of IC50. Since the internucleotidyl phosphates in the core region are known to form hydrogen bonds it is likely that while 6′(R)-O-Tol of T13 modification can form a competing electrostatic interaction with ago2, which the 6′(S)-O-Tol-jcLNA diastereomer at T13 site cannot, as reflected by their respective IC50 values. Clearly, this observation opens up a new approach to modulate the silencing efficiency by controlling the stereochemistry of the nucleotide at C6′ at the core region. It also gives an effective handle to cross-link catalytic Asp residues (or any residue of ago2 in the proximity of the core region of the guide strand) to understand the interactions involved in the siRNA promoted cleavage reaction, in general.
(6) Dramatic difference in siRNA activities of 6′(S)- and 6′(R)- substituents at position 13 can be associated with specific interactions of 6′-O-Tol groups with Ago proteins34,35 leading to influence the target RNA cleavage (which consequently may influence the target RNA cleavage).
(7) It is notable that double modifications (T1 + T20, sequences 9, 13 and 17), where parent jcLNA (siRNA 9) have displayed the best IC50 value (0.5 nM), lower than that of the native counterpart (siRNA 1, 1.8 nM). Nevertheless, the 6′(R)-O-Tol modification (siRNA 13, 3.2 nM) and 6′(S)-O-Tol (siRNA 17, 3.6 nM) isomers for the T1 + T20 Bis-modification displayed near native (1, 1.8 nM) like IC50s, with 2–3 fold enhanced serum stability compared to native siRNA.
(8) Unlike for the parent jcLNA modified siRNAs (siRNAs 6–9) the silencing efficiency of 6′(R/S)-O-Tol-jcLNA (siRNA 10,12–17) modified siRNAs did not go hand-in-hand with their increased stability in serum:Whereas, parent jcLNA modified siRNAs (siRNA 6–9) displayed t1/2 values ranging from 4.3 h to 13.6 h (Table 1), the 6′(R)-O-Tol diastereomer (siRNAs 10–13) displayed a range of 6.3 h to 8.8 h and the values for the 6′(S)-O-Tol diastereomer (siRNAs 14–17) varied in the range of 7.5 h–9.1 h, whereas the native siRNA showed a serum stability under identical conditions for 3 h and LNA modified counterpart of 3–11 h. Hence, we see very little difference in the serum stability of 21 mer siRNA modified or unmodified because both of them are protected from exo and endonucleases to a very comparable level compared to ssRNA.36 Despite this, what is more important is that jcLNA and 6′-O-Tol modified siRNA have shown ∼3-fold improved stability than that of the native counterpart hence, it is entirely the property of the modified nucleotides and the position where this modification has been included in these modified siRNAs vis-a-vis native that is reflected in serum digestion experiments (Table 1). It is noteworthy that stability of native or modified ssRNA or ssDNA in the SVPDE or blood serum extract does not provide such inter- and intracellular milieu related protection.37,38
(9) Recently, jcLNA incorporated antisense oligonucleotides (ASO) and siRNA have been studied against various biological targets to evaluate the true potential of jcLNAs in various disease models such as single cycle replication model for HIV-120 and allele selective inhibition17 of mutant Huntingtin expression involving cell culture in vitro and in vivo experiments.39 These studies clearly showed that jcLNA modified oligos are considerably better than any other modification, including LNA, in terms of IC50, and human blood serum stability. Recently in vitro and in vivo studies employing LNA, jcLNA and its exocyclic methylene derivative21 were presented to down regulate the PTEN mRNA expression. Results of these animal experiments suggest that jcLNA was slightly less potent with IC50 (7.9 μM) whereas LNA and exocyclic methylene LNA have shown 2.8 μM and 3.9 μM respectively. This study also emphasizes the importance of chemical modification and optimal chemical diversification within the jcLNA scaffold for future in vivo applications.21 However, this is the first report where we show that the silencing efficiency of jcLNA type molecules can be enhanced by controlling the stereochemistry at C6′.
(10) To be a potential therapeutic candidate low cytotoxicity as well as stability in blood serum are of utmost importance. 48 h post-transfection cells transfected with 6′-O-Tol-jcLNA (both R and S) showed more than 70% cell viability as revealed by MTT assay (Fig. S5 in the ESI†),20 which is comparable to that of the native counterpart. Hence the enhanced serum stability of jcLNA modified siRNAs, in general, and native-like RNA silencing efficiency strongly finger-points the therapeutic potential of these group of molecules.
Implication
In continuation of our previous work20 with jcLNA modified siRNAs (6–9) targeting the TAR1 region of HIV-1 we have incorporated toluoyl substitution at the C6′ centre in the alkane bridge, 4′-CH2–CH(Me)-2′, of jcLNA nucleotides. The general improvement in the silencing potency for 6′-O-Tol-jcLNA modified diastereomers (siRNAs 10–17) takes place in a position dependent manner, whereas the serum stability is very comparable and is position independent. The serum stabilities of these jcLNA-modified siRNAs are found to be significantly higher than the native counterpart, thereby making them very credible candidates against other targets. Two key findings in this study, first, significant diastereomeric bias (S versus R) in potency with 6′-O-Tol-jcLNA modified siRNAs 10–17 in the core region, and second, uniform display of high t1/2 values (6.3–9.1 hours, Table 1) in serum independent of the position of modification, can add new mechanistic details to RNAi pathway and siRNA stability. High potency of the silencing activity to the presence of the 6′-O-toluoyl substituent has prompted us to initiate further studies involving various chemical modifications (hydrophobic versus hydrophilic groups with different alkane chain lengths) of the 6′(R)-position in the jcLNA building block (IVa in Fig. 1) to fine-tune the RISC complex formation and possibly to use it as a molecular probe to reveal the biochemical mechanism of the silencing by siRNAs. The observed stereoselectivity of the RISC protein to the 6′-O-Tol-modified jcLNA also suggests that any further 6′-modification in jcLNA-modified siRNAs with any hydrophilic or hydrophobic group with an alkyl chain for tighter binding in the core region should preferably be done in R-stereochemistry, not in S.The incorporation of 6′-toluoyl and hydroxyl groups in the jcLNA nucleotides provides an opportunity to design relatively easy chemical diversification and functionalization of the C6′ site, which may have a significant advantage over other LNA-type building blocks in that the former are well tolerated in the siRNA silencing approach. Such functionalization is also a valuable approach to improve bio-viability and delivery properties of the drug candidates in RNA interference therapy.40
Experimental section
Synthesis, deprotection and purification of oligonucleotides
All oligonucleotides were synthesized using an automated DNA/RNA synthesizer by Applied Biosystems, model 394. The stepwise coupling yields of the modified phosphoramidites i.e. LNA-T (Link Technologies), 6′(R)-O-Tol-jcLNA and 6′(S)-O-Tol-jcLNA were 97%, 98% and 94% respectively. 5-(3,5-Bis(trifluoromethyl)phenyl)-1H-tetrazole (Activator 42, Proligo) as the activating reagent with 10 min coupling time for modified phosphoramidites, followed by deprotection of all base-labile protecting groups with 33% methanolic ammonia to give oligonucleotides. All oligonucleotides were purified by (20% polyacrylamide/7 M urea) PAGE, extracted with 0.3 M NaOAc, desalted with C18-reverse phase cartridges (Waters) and their purity (>95%) was confirmed by PAGE. Concentrations were determined by diluting the stock solutions in DEPC-treated water followed by analysis using a UV spectrophotometer (Shimadzu UV2550).Duplex formation
RNA duplexes were prepared by the annealing of complementary oligonucleotides in thin-walled PCR tubes containing same concentrations of sense and antisense (equimolar) of each in DEPC treated water and annealing buffer (5 ×; 50 mM Tris; pH 7.5–8.0; 100 mM NaCl in DEPC-treated water). Annealing of RNA duplexes was performed in a thermal cycler by incubating the solution for 1 min at 90 °C followed by slowly cooling to room temperature over a period of about 60 min. After annealing, the RNA duplexes were stored at −20 °C. A list of siRNAs synthesized is given in Table 1 with site of modifications.Cell culture and transfection
HEK293T cells were cultured in DMEM (Sigma) supplemented with 10% bovine serum (PAN Biotech) and 1% Penicillin–Streptomycin solution (Sigma). Transfection was carried out in 24-well plates following a standard protocol.20 For transfection in 6-well plates the calculations were stepped up as recommended by the manufacturer.p24 ELISA of culture supernatant
p24 ELISA of culture supernatant was performed using a Perkin Elmer Alliance HIV-1 p24 antigen ELISA kit as per the manufacturer's recommendation. For further details of calculation of mean percentage inhibitions using Origin® 6.1 software see ref. 20.MTT assay
The culture media were removed 48 h post-transfection and fresh media added. MTT was added to a final concentration of 0.1 mg ml−1. After incubation for 2.5 h at 37 °C in a CO2 incubator the reactions were stopped by addition of stop solution (99% isopropanol and 1% HCl) and OD was measured at 595 nm. Percentage viability was calculated over that of the mock transfected sample and results are a cumulative of at least three independent experiments.Stability assay of siRNAs in human serum
1.5 μl of native as well as jcLNA and LNA modified siRNAs from 100 μM stock were added to 10 μl of 100% human serum isolated from a healthy B+ donor.20 After incubation of the samples for required time points 3 μl aliquots were withdrawn and immediately suspended in 9 μl of 1× TBE gel loading buffer (18 mM EDTA, 5% glycerol 0.025% bromophenol blue) followed by snap freezing in liquid nitrogen and stored at −80 °C. The samples were subjected to 20% non-denaturing TBE–PAGE, following which the run gels were stained with ethidium bromide (EtBr) and visualized and documented in a BIORAD Geldoc. ImageJ software was utilized to determine the band intensities of the double strand (ds) form and the percentage of ds form remaining at each time point was calculated over that at zero time point. The mean percentages of ds form remaining for a particular time point along with standard deviation were calculated from at least three independent experiments. The plot and t1/2 calculation were done as in our previous work.20Western blot
48 h post-transfection cells were harvested and lysed in RIPA buffer 50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40 (v/v), 1% Na-deoxycholate (w/v), 0.1% SDS (w/v). After centrifugation the protein concentration in the supernatant fluid was quantitated and 40 μg of the protein samples were subjected to 12% SDS–PAGE in a Biorad Mini PROTEAN Tetra Cell electrophoresis apparatus. The proteins were electroblotted onto the nitrocellulose membrane in a Biorad Western blot unit. Blocking (with 5% BSA), subsequent incubation in primary antibody (mouse anti-HIV-1 p24 and mouse antiactin, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Santa Cruz Biotechnology), followed by secondary antibody incubation and developing was carried out using routine lab protocols as described in our previous work.16 Densitometric analyses were done as in our previous work.20
200; Santa Cruz Biotechnology), followed by secondary antibody incubation and developing was carried out using routine lab protocols as described in our previous work.16 Densitometric analyses were done as in our previous work.20
RNA isolation and RTPCR
Total cellular RNA was isolated from transfected cells using TRI (Sigma) according to the manufacturer's protocol. 1 μg of total RNA was subjected to DNAse1 treatment as per the manufacturer's instruction and then subjected to reverse transcription in a 25 μl mix containing 300 ng of random hexamer (Invitrogen), dNTP (0.2 mM) (Sigma), DTT (5 mM) (Invitrogen), Porcine RNAse inhibitor (20 U) (New England Biolabs) and reverse transcriptase (100 U) (Invitrogen). Following reverse transcription 2.5 μl of cDNA was subjected to PCR in a 25 μl mix containing each forward and reverse primers (0.4 μM) (Sigma), MgCl2 (1.5 mM), dNTP (200 μM) and Taq (1.5U) (Genei). Primer sequence details and cycle parameters are same as in our previous work.20Abbreviations
| LNA | Locked nucleic acid ((Fig. 1, compound II) |
| jcLNA | Carbocyclic locked nucleic acid (carba-LNA) or [C6′-deoxy, C7′(R/S)-methyl]-carba-LNA (Fig. 1, compound III) |
| [6′(R)-O-(p-Toluoyl)-7′(S)-methyl]-jcLNA | [6′(R)-O-(p-Toluoyl)-jcLNA] (Fig. 1, compound IVa) |
| [6′(S)-O-(p-Toluoyl)-7′(S)-methyl]-jcLNA | [6′(S)-O-(p-Toluoyl)-jcLNA] (Fig. 1, compound IVb) |
| TAR | Trans activation response |
Acknowledgements
We thank Uppsala University, Sweden for valuable scientific support through Prof. Jyoti Chattopadhyaya. Swedish Natural Science Funding Agencies VR-N and VR-M, European Framework Program (EU-FP7) to JC, Swedish National Supercomputer Center (NSC), and Department of Science and Technology, Government of India, are greatly acknowledged for their generous support of this project (Grant no. VII-PRDSF/109/05-06/TDT dated 31.03.2006 to JC and IMM). We also thank TCG Life Sciences, Kolkata and Uppsala RNA Research Center (URRC, Uppsala University) for contributing to the funding of this research. We thank Dr Sekhar Chakrabarti (National Institute of Cholera & Enteric Kolkata, India) for allowing us to use his lab for initial experiments.References
- A. P. McCaffrey, H. Nakai, K. Pandey, Z. Huang, F. H. Salazar, H. Xu, S. F. Wieland, P. L. Marion and M. A. Kay, Nat. Biotechnol., 2003, 21(6), 639–644 CrossRef CAS.
- G. S. Ralph, P. A. Radcliffe, D. M. Day, J. M. Carthy, M. A. Leroux, D. C. P. Lee, L.-F. Wong, L. G. Bilsland, L. Greensmith, S. M. Kingsman, K. A. Mitrophanous, N. D. Mazarakis and M. Azzouz, Nat. Med., 2005, 11(43), 429–433 CrossRef CAS.
- J. Soutschek, et al. , Nature, 2004, 432(7014), 173–178 CrossRef CAS.
- S. Shukla, C. S. Sumaria and P. I. Pradeepkumar, ChemMedChem, 2010, 5(3), 328–349 CrossRef CAS.
- J. B. Bramsen, et al. , Nucleic Acids Res., 2009, 37(9), 2867 CrossRef CAS.
- D. V. Morrissey, et al. , Nat. Biotechnol., 2005, 23(8), 1002–1007 CrossRef CAS.
- M. Gaglione and A. Messere, Mini-Rev. Med. Chem., 2010, 10(7), 578–595 CrossRef CAS.
- J. Hong, Y. Huang, J. Li, F. Yi, J. Zheng, H. Huang, N. Wei, Y. Shan, M. An, H. Zhang, J. Ji, P. Zhang, Z. Xi, Q. Du and Z. Liang, FASEB J., 2010, 24(12), 4844 CrossRef CAS.
- M. Manoharan, A. Akinc, R. K. Pandey, J. Qin, P. Hadwiger, M. John, K. Mills, K. Charisse, M. A. Maier, L. Nechev, E. M. Greene, P. S. Pallan, E. Rozners, K. G. Rajeev and M. Egli, Angew. Chem., Int. Ed., 2011, 50, 2284–2288 CAS.
- S. K. Singh, A. A. Koshkin, J. Wengel and P. Nielsen, Chem. Commun., 1998, 455–456 RSC.
- A. A. Koshkin, P. Nielsen, M. Meldgaard, V. K. Rajwanshi, S. K. Singh and J. Wengel, J. Am. Chem. Soc., 1998, 120, 13252 CrossRef CAS.
- J. S. Jepsen, M. D. Sørensen and J. Wengel, Oligonucleotides, 2004, 14, 130 CrossRef CAS.
- F. Darfeuille, J. B. Hansen, H. Orum, C. D. Primo and J. J. Toulme, Nucleic Acids Res., 2004, 32, 3101 CrossRef CAS.
- F. Darfeuille, S. Reigadas, J. B. Hansen, H. Orum, C. D. Primo and J. J. Toulmé, Biochemistry, 2006, 45, 12076 CrossRef CAS.
- C. D. Primo, I. Rudloff, S. Reigadas, A. Arzumanov, M. Gait and J. J. Toulmé, FEBS Lett., 2007, 581(4), 771–774 CrossRef.
- D. Corey; J. Hu; K. Gagnon; J. Liu; J. Watts; F. Bennett; E. Swayze; J. Randolph; J. Chattopadhyaya. Biol. Chem., 2011392(4), 315–325 Search PubMed.
- K. Gagnon, H. Pendergraff, G. Deleavey, E. Swayze, P. Potier, J. Randolph, E. Roesch, J. Chattopadhyaya, M. Damha, F. C. Bennett, C. Montailler, M. Lemaitre and D. Corey, Biochemistry, 2010, 9(47), 10166–10178 CrossRef.
- J. B. Bramsen, M. M. Pakula, T. B. Hansen, C. Bus, N. Langkjær, J. Chattopadhyaya, J. W. Engels, P. Herdewijn, J. Wengel and J. Kjems, Nucleic Acids Res., 2010, 38(17), 5761–5773 CrossRef CAS.
- O. R. F. Mook, J. Vreijling, S. Lena-Wengel, J. Wengel, C. Zhou, J. Chattopadhyaya, F. Baas and K. Fluiter, Artif. DNA: PNA XNA, 2010, 1, 36–44 CrossRef.
- S. Dutta, N. Bhaduri, N. Rastogi, S. G. Chandel, J. Vandavasi, R. S. Upadhayaya and J. Chattopadhyaya, Med. Chem. Commun., 2011, 2, 206–216 RSC.
- P. P. Seth, C. R. Allerson, A. Berdeja, A. Siwkowski, P. S. Pallan, H. Gaus, T. P. Prakash, A. T. Watt, M. Egli and E. E. Swayze, J. Am. Chem. Soc., 2010, 132(42), 14942–14950 CrossRef CAS.
- P. Srivastava, J. Barman, W. Pathmasiri, O. Plashkevych, M.g. Wenska and J. Chattopadhyaya, J. Am. Chem. Soc., 2007, 129(26), 8362–8379 CrossRef CAS.
- C. Zhou and J. Chattopadhyaya, J. Org. Chem., 2010, 75(7), 2341–2349 CrossRef CAS.
- Z. Ao, X. Yao and E. A. Cohen, J. Virol., 2004, 78(6), 3170–3177 CrossRef CAS.
- M. L. Yeung, Y. Bennasser, K. Watashi, S.-Y. Le, L. Houzet and K.-T. Jeang, Nucleic Acids Res., 2009, 37(19), 6575–6586 CrossRef CAS.
- J. Ahluwalia, S. Khan, K. Soni, P. Rawat, A. Gupta, M. Hariharan, V. Scaria, M. Lalwani, B. Pillai, D. Mitra and S. Brahmachari, Retrovirology, 2008, 5(1), 117, DOI:10.1186/1742-4690-5-117.
- J. Martinez and T. Tuschl, Genes Dev., 2004, 18, 975–980 CrossRef CAS.
- D. A. Case, et al., AMBER 7, University of California, San Francisco, 2002 Search PubMed.
- A. Pérez, I. Marchán, D. Svozil, J. Sponer, T. E. Cheatham Iii, C. A. Laughton and M. Orozco, Biophys. J., 2007, 92(11), 3817–3829 CrossRef.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- M. J. Frisch, et al., Gaussian 98 (Revision A.6), Gaussian, Inc, Pittsburgh PA, 1998 Search PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS.
- (a) W. F. Lima, H. Wu, J. G. Nichols, H. Sun, H. M. Murray and S. T. Crooke, J. Biol. Chem., 2009, 284(38), 26017 CrossRef CAS; (b) A number of studies have shown (Angew. Chem., Int. Ed., 2011, 50, 2284–2288; Chem. Biodiversity, 2007, 4, 858–873; J. Med. Chem., 2005, 48, 4247–4253) that the mRNA silencing efficiency by modified siRNA with 2′-F or/and 2′-OMe positioned either in the sense or antisense strand in the core region varies depending upon the target RNA sequence composition as well as on the position of the modification. These studies have shown that the 2′-F and 2′-OMe modifications are generally tolerated giving siRNA promoted cleavage even at the cleavage site (J. Med. Chem., 2005, 48, 4247–4253) with however a few exceptions (RNA, 2003, 1034–1048). For example, the 2′-OMe modification at position 11 has led to a greatly diminished RNA silencing (16% or 25%) compared to that of the native (RNA, 2003, 1034–1048). whereas the 2′-OMe modification (site of modification not specified) on both strands of the duplex siRNA has prevented RNAi activity completely. The 2′-OMe modification at the cleavage sites 9 and 12 has led to >80% and >60% inhibition of human PTEN protein production (J. Med. Chem., 2005, 48, 4247–4253). Best gene silencing of human PTEN has been achieved however by combination of alternate modifications of 2′-OMe and 2′-F at positions 8 and 10 leading to IC50 of 0.068 nM and 0.002 nM (75% target mRNA reduction), respectively (J. Med. Chem., 2005, 48, 901–904). In the same study 2′-O-MOE (2′-O-(2-methoxyethyl) was also studied at endonuclease cleavage site position 12 and found less potent than 2′-F and 2′-OMe modified siRNA. The following potency order of the RNAi cleavage was observed with modification at position 12: 2′-F > 2′ O-Me > 2′-O-MOE. Furthermore to this potency of the 2′-O-OME modified siRNA was relatively limited compared to the unmodified siRNA. 2′-O-OME modification was not tolerated for RNAi cleavage either at 5′-end or 3′-end of the antisense strand. However; it is interesting to note that 2′-O-MOE modifications were better tolerated at the middle of sequences than 2′-F and 2′-OMe. A recent study involving siRNA modified with 2′-F, 2′-O-Me, 2′-O-MOE, or locked nucleic acid (LNA), all pyrimidine residues and few of them are mixed modified sequences. The results of in vitro dose response evaluation suggest that neither the 2′-OMe nor LNA modification was tolerated (position 14 with various modifications) on the antisense strand whereas 2′-O-MOE was not tolerated on either strand. These collective studies suggest that larger groups are not well tolerated and it may (i.e., 2′-OMe and 2′-O-MOE) compromise binding by proteins involved in the RNAi pathway because steric clashes severely limit the interaction between siRNAs and target mRNA. (RNA, 2003, 1034–1048; Angew. Chem., Int. Ed., 2011, 50, 2284)The main seemingly advantage of chemical modification of siRNA is increased nuclease stability of the siRNA which should lead to enhanced inhibitory activity in vivo. There is however a few contrasting examples which clearly suggest that it is not the case at least for the 2′-F modified siRNAs which have shown to be not more potent than the unmodified siRNA (RNA, 2004, 10, 766–771). Remarkable study has shown that ago2 mediated target DNA cleavage could be activated by transforming the 2′- endo to 3′- endo conformation by choosing 2′-F and 2′-OMe modifications at the cleavage site of the target DNA strand (Nature, 2009, 461, 754). Our work suggests that 6′-O-Tol-jc-LNA modifications are well tolerated to give the desired siRNA activity irrespective of site of modification and displayed a native like silencing efficiency. Furthermore, our study shows that the jcLNA modification in the core region of TAR-1 sequence is at least as good as the native, with the additional bonus of enhanced nuclease stability, which, taking into account of readily available synthetic approaches to modify the 6′-hydroxyl group of the jcLNAs presents a new dimension of possible stereospecific modulation with efficient silencing effect.
- Y. Wang, S. Juranek, H. Li, G. Sheng, T. Tuschl and D. J. Patel, Nature, 2008, 456(7224), 921–926 CrossRef CAS.
- Y. Wang, S. Juranek, H. Li, G. Sheng, G. S. Wardle, T. Tuschl and D. J. Patel, Nature, 2009, 461(7265), 754–761 CrossRef CAS.
- J. A. H. Hoerter, V. Krishnan, T. A. Lionberger and N. G. Walter, PLoS One, 2011, 6(5), e20359 CAS.
- K. Raemdonck, K. Remaut, B. Lucas, N. N. Sanders, J. Demeester and S. C. De Smedt, Biochemistry, 2006, 45(35), 10614–10623 CrossRef CAS.
- S. A. Uhler, D. Cai, Y. Man, C. Figge and N. G. Walter, J. Am. Chem. Soc., 2003, 125(47), 14230–14231 CrossRef CAS.
- P. P. Seth, C. R. Allerson, A. Berdeja, A. Siwkowski, P. S. Pallan, H. Gaus, T. P. Prakash, A. T. Watt, M. Egli and E. E. Swayze, J. Am. Chem. Soc., 2010, 132(42), 14942–14950 CrossRef CAS.
- M. Monaghan and A. Pandit, Adv. Drug Delivery Rev., 2011, 43(4–5), 197–208 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Dose response studies using p24 ELISA for [6′(R), 7′(S); 6′(S), 7′(S)]-jcLNA modified siRNA (as in IVa or IVb in Fig. 1) showing % inhibition of viral replication in comparison to the virus control at various doses (Fig. S1); serum stability of chemically modified double-stranded 6′(R)-O-Tol-jcLNA (10–13) and 6′(S)-O-Tol-jcLNA (14–17) containing siRNAs targeting HIV1 TAR1 at different time points (Fig. S2); dose response study obtained from Western blot of 6′-(R)-OH/7′S-jcLNA modified (siRNA 10–13) and 6′-(S)-O-Tol/7′S modified counterpart siRNAs (14–17) (Fig. S3); dose response studies using RTPCR for C6′-(R/S)-O-Tol/7′S modified siRNAs (Fig. S4); cell viability assay (MTT assay) for C6′-(R/S)-O-Tol/7′S modified siRNAs (Fig. S5); atomic charges, names and types of the 6′R-O-Tol-jcLNA and 6′S-O-Tol-jcLNA nucleotides used as parameters of the MD simulations (Table S1); overlap of the molecular structure of the 6′(R)-O-Tol and 6′(S)-O-Tol-jcLNA T13 modified-siRNA duplexes (10 and 14 in Table 1) with the target RNA (Fig. S6); MALDI-MS spectra of all sequences of this study (Fig. S7–S23). See DOI: 10.1039/c1md00167a |
| This journal is © The Royal Society of Chemistry 2011 |
