A highly selective agonist for the metabotropic glutamate receptor mGluR2†
Simon D.
Nielsen‡
a,
Marica
Fulco‡
ab,
Michaela
Serpi‡
ab,
Birgitte
Nielsen
a,
Maria B.
Hansen
a,
Kasper L.
Hansen
a,
Christian
Thomsen
d,
Robb
Brodbeck
d,
Hans
Bräuner-Osborne
a,
Roberto
Pellicciari
b,
Per-Ola
Norrby
c,
Jeremy R.
Greenwood
e and
Rasmus P.
Clausen
*a
aDepartment of Medicinal Chemistry, Faculty of Pharmaceutical Sciences, University of Copenhagen, Universitetsparken 2, 2100, Copenhagen, Denmark. E-mail: rac@farma.ku.dk; Fax: +45 35 33 60 41; Tel: +45 35 33 65 66
bDipartimento di Chimica e Tecnologia del Farmaco, Università di Perugia, Italy
cDepartment of Chemistry, Göteborg University, Sweden
dLundbeck Research USA, Paramus, NJ, USA
eSchrodinger Inc., 120 W 45th St, New York, NY 10036, USA
First published on 23rd September 2011
Abstract
The three conformationally restricted cyclopropyl glutamate analogues (3, 4, 5) were synthesised and their affinity for ionotropic and activity at metabotropic glutamate receptors were probed. Compound 4 turned out to be a highly selective agonist at the metabotropic glutamate receptor mGluR2 with at least two orders of magnitude selectivity in potency compared to the very homologous mGluR3 as well as mGluR1, 4, 5, 7. We also tried to synthesise the two epimers of 6, but the two compounds underwent fast epimerisation in H2O. Furthermore, two cyclopropyl arginine analogues (7, 8) were synthesised and characterised pharmacologically at GPRC6A.
Introduction
The major excitatory neurotransmitter in the central nervous system (CNS), (S)-glutamic acid (Glu, Fig. 1), interacts with a large number of proteins including transporters and receptors. The continuous development of compounds selectively affecting the individual components of the glutamatergic system continues to play a central role in the characterisation of this system. Glu exerts its functions via two classes of receptors: the ionotropic Glu receptors (iGluRs) and the metabotropic receptors (mGluRs). Whereas the iGluRs, which are ligand-gated ion channels, are subdivided into N-methyl-D-aspartic acid (NMDA), 2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA), and kainic acid receptors, the mGluRs belong to the family of G-protein coupled receptors and comprise mGluR1–8. Group I and group II mGluRs include mGluR1, 5 and mGluR2, 3 respectively, whereas mGluR4, 6, 7, 8 constitute group III mGluRs.1,2 All subtypes of iGluRs and mGluRs are considered potential therapeutic targets for a number of neurologic and psychiatric diseases.3,4 In particular, cyclopropyl analogues of Glu have led to highly potent and selective GluR ligands.5,6 | ||
| Fig. 1 Glutamic acid and its conformationally restricted analogues (1–8). | ||
Conformational restriction of the carbon skeleton of Glu analogues has been important for achieving selectivity among Glu receptor subtypes. However, the 1,2-cyclopropyl analogues of Glu were never characterised pharmacologically. This spurred our interest in developing a new synthesis route for the Glu analogues 3 and 4 (Fig. 1).
These analogues are interesting not only as receptor ligands, but also as unnatural amino acid analogues for incorporation into peptides and proteins.
The synthesis and pharmacology of the enantiomers of TDPA (1)7 and homo-TDPA (2)8 (Fig. 1) were previously reported from our department and all four compounds affect glutamate receptors and transporters with various degrees of selectivity. Unexpectedly, both enantiomers of 1 displayed similar affinity at AMPA binding sites and both enantiomers of 2 were equipotent in the activation of mGluR2 receptors. Such selectivity for (R)-enantiomers is quite unusual at these two receptor groups since they normally prefer the S-configuration of the amino acid.9
Restricting conformational freedom and introducing an extra stereocenter via a cyclopropane ring could potentially narrow the pharmacological profile of TDPA derivatives, and lead to more selective ligands. Since the synthetic route of Glu analogues 3 and 4 could enable access to restricted TDPA analogues 5 and 6, and restricted arginine analogues 7 and 8, we also decided to explore these as target molecules. We here report the syntheses of the five conformationally restricted amino acids 3, 4, 5, 7, 8 and our attempts to make the cyclopropane epimers of TDPA trans- and cis-6 (Fig. 1). Furthermore, the pharmacological characterisation of the compounds 3, 4 and 5 on iGluRs and mGluRs and of compounds 7 and 8 on the recently discovered L-arginine activated receptor GPRC6A was established. The GPCR6A receptor is homologous to mGluRs.3
Chemistry
The synthetic strategy is based on the previous synthesis of homo-TDPA,8 where the heterocycle is built up from an aldehyde bearing an amino acid moiety as the di-Boc amide and tert-butyl ester. The first step in the route to 3, 4, 5, 7, and 8 is cyclopropanation of 9 with di-tert-butyl gluconate using sodium hydride as base. The starting material 9 was synthesised as previously described.10 The isomers 10 and 11 were separated by flash chromatography and transformed in parallel in the subsequent steps. Oxidative cleavage of the double bond and oxidation of benzyl to benzoyl were carried out in a one-pot procedure using NaIO4 and RuCl3·H2O. The products were obtained in moderate to good yields. Ethyl chloroformate was used to convert the carboxylic acids 12 and 13 to activated ester carbonates. These carbonates were converted to acyl azides which underwent the Curtius rearrangement to give isocyanate intermediates that reacted with tert-butanol to give 14 and 15. The benzoyl protecting group was hydrolysed using potassium carbonate in methanol to give the unprotected alcohols in good yields (16 71% yield and 17 60% yield) (Scheme 1).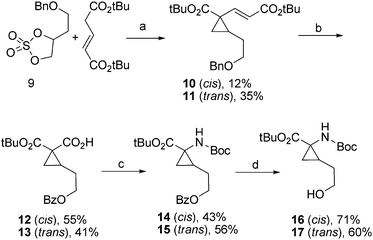 | ||
| Scheme 1 (a) NaH/DME, rt, 22 h. (b) NaIO4, RuCl3·H2O, CCl4, CH3CN, H2O, rt, 17 h. (c) (i) ClCO2Et, Et3N, THF, 0 °C, 35 min; (ii) NaN3, H2O, 0 °C to rt, 50 min; (iii) t-BuOH, reflux, 15 h. (d) K2CO3, MeOH. | ||
The two isomeric alcohols served as building blocks in the synthesis of 3, 4, 5, 7, and 8. Treatment of 16 and 17 with NaIO4 and RuCl3·H2O gave the carboxylic acids 18 and 19 in 57% yield and 82% yield respectively. Removal of the protection groups was carried out using TFA in CH2Cl2 and gave 3 and 4 in 65% and 75% yields respectively (Scheme 2). The synthesis of a single stereoisomer of compound 4 has previously been reported by Jiménez and Ortuño in 1996.11
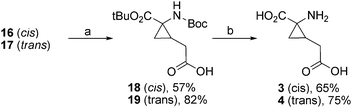 | ||
| Scheme 2 (a) NaIO4, RuCl3·H2O, CCl4, CH3CN, H2O, rt, 3 h. (b) CF3COOH, CH2Cl2, rt, 3 h. | ||
In the synthesis of 7 and 8, the Mitsunobu reaction was used to convert alcohols 16 and 17 to the guanidine-derivatives 20 and 21. Removal of the protection groups in 20 and 21 was carried out using TFA in CH2Cl2 and gave 7 and 8 in good yields (Scheme 3).
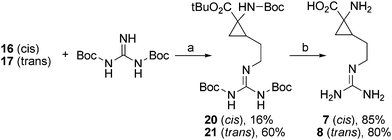 | ||
| Scheme 3 (a) Ph3P, DIAD, THF, 0 °C to rt, 20 h. (b) CF3COOH, CH2Cl2, rt, 3 h. | ||
Oxidation of 16 with DMP followed by the Strecker reaction yielded α-amino nitrile 22 in 66% yield after the two steps. The α-amino nitrile 22 was converted crude to α-amino amide 23 in 63% yield after aqueous workup. Thionylaniline in pyridine converted 23 to thiadiazole 24 in 16% yield. Removal of the protection groups was done with TFA in CH2Cl2 to give the final product 5 in 64% yield (Scheme 4).
 | ||
| Scheme 4 (a) (i) DMP, CH2Cl2, rt, 1 h; (ii) KCN, NH4OH, MeOH, rt, 16 h. (b) 1 M NaOH, H2O2, THF–EtOH, 0 °C, 2 h. (c) N-Thionylaniline, pyridine, 0 °C to rt, 2 h. (d) CF3COOH, CH2Cl2, rt, 3 h, evaporation from 1 M HCl. | ||
The first step in the route to 6 is a cyclopropanation of the protected dehydroalanine 25 (Scheme 5). Reaction with ethyl dimethylsulfuranylidene acetate gave only 26, whereas reaction with ethyl diazoacetate gave an epimeric mixture of 26 and 27. The latter contrasts with the reaction of ethyl diazoacetate with the methyl ester of 25, which only gives the trans-product. Although it enabled access to 27, even meticulous column chromatography could not remove 10% contamination with 26 which remained in the rest of the synthesis of 36. Attempts to enrich 27 by epimerisation of 26 with various bases (LiHMDS, LDA, NaH) gave only the starting material.
 | ||
| Scheme 5 Cyclopropanation of protected dehydroalanine 25. | ||
The aldehydes 29 and 33 (Scheme 6) could not be obtained by selective reduction with DIBAl-H. The reaction was hampered by removal of one Boc group and complete reduction to the alcohol leading to complex mixtures depending on the temperature and solvent. LiBH4 did not react and LiAlH4 led to degradation, but reduction of the ester with 2 eq. of DIBAl-H in diethyl ether at −40 °C for 30 min and 3 eq. of DIBAl-H in diethyl ether at −40 °C for 4 h gave satisfactory yields of the respective alcohols 28 and 32.
 | ||
| Scheme 6 (a) DIBAl-H, Et2O. (b) DMP, CH2Cl2. (c) NH4Cl, KCN, H2O, MeOH. (d) NaOH, H2O2, H2O, THF, EtOH. | ||
Oxidation with DMP gave the aldehydes trans- and 29 and 33 in good yields. Under Strecker conditions, the aldehydes 29 and 33 yielded α-amino nitriles 30 and 34, both products as a diastereomeric mixture (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). These products were converted in crude form to α-amino amides 31 and 35 which could be purified by column chromatography yielding 31 and 35 as a diastereomeric mixture (2
4). These products were converted in crude form to α-amino amides 31 and 35 which could be purified by column chromatography yielding 31 and 35 as a diastereomeric mixture (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) in 55% and 45% yields respectively, for the two steps. The diastereomers of 30 could be separated, and although one diastereomer (30a) converted well to 31 (67%) the other (30b) gave low yield (21%) due to base catalysed cyclopropane ring opening giving 36 (38%) most likely by the mechanism shown in Scheme 7. The proposed mechanism enables an establishment of the relative stereochemistry of the introduced stereocenter in 31, and emphasises the α-anion stabilising character of the di-Boc amino protecting group employed, which also led to partial racemisation in the synthesis of homo-TDPA.8
5) in 55% and 45% yields respectively, for the two steps. The diastereomers of 30 could be separated, and although one diastereomer (30a) converted well to 31 (67%) the other (30b) gave low yield (21%) due to base catalysed cyclopropane ring opening giving 36 (38%) most likely by the mechanism shown in Scheme 7. The proposed mechanism enables an establishment of the relative stereochemistry of the introduced stereocenter in 31, and emphasises the α-anion stabilising character of the di-Boc amino protecting group employed, which also led to partial racemisation in the synthesis of homo-TDPA.8
 | ||
| Scheme 7 Proposed elimination reaction leading to 36. | ||
Cyclising 31 with thionyl chloride and triethylamine at −78 °C gave 37 in 13% yield. When instead thionylaniline was employed the yield was improved to 74%. Attempts to cyclise 35 were not successful.
Final deprotection of 37 with TFA (Scheme 8) gave at first an almost clean 1H-NMR spectrum in D2O of the crude product, but surprisingly, the product epimerised after a while resulting in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 cis-6/trans-6 equilibrium mixture. The epimeric mixture was therefore subjected to preparative HPLC and elution with 0.1% TFA or 1 mM AcOH confirmed the 1
4 cis-6/trans-6 equilibrium mixture. The epimeric mixture was therefore subjected to preparative HPLC and elution with 0.1% TFA or 1 mM AcOH confirmed the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratio. However, the isolated product epimerised completely upon evaporation. HPLC analysis (Fig. 2) of the first minor peak revealed that epimerisation completes in ca. 15 min. Analysis of the latter peak confirmed this. Thus, epimerisation was apparently slower in the first more acidic, crude NMR solution. The proton on the cyclopropane ring does not exchange, and the mechanism therefore seems to involve opening of the cyclopropane ring.
4 ratio. However, the isolated product epimerised completely upon evaporation. HPLC analysis (Fig. 2) of the first minor peak revealed that epimerisation completes in ca. 15 min. Analysis of the latter peak confirmed this. Thus, epimerisation was apparently slower in the first more acidic, crude NMR solution. The proton on the cyclopropane ring does not exchange, and the mechanism therefore seems to involve opening of the cyclopropane ring.
 | ||
| Scheme 8 (a) Thionylaniline, pyridine. (b) TFA, CH2Cl2. | ||
 | ||
| Fig. 2 HPLC diagrams of: (A) a preparative separation of the epimeric mixture of 6, (B) immediate HPLC analysis of the 2.57 peak and (C) after 10 min (note the enhanced baseline between the peaks due to the constant epimerisation). Eluent: 1 mM AcOH. | ||
The unexpected epimerisation prompted us to investigate plausible paths for inversion of the ring carbons using DFT methods. The increase of rate with pH indicates that epimerisation occurs in a neutral or anionic protonation state of 6. It was found that the amine lone pair can assist in heterolytic fragmentation of the ring, leading to a tri-ionised intermediate with higher energy but only low barriers to rotation (Scheme 9).
 | ||
| Scheme 9 Epimerisation of 6. | ||
The transition state with a low bond rotation barrier (Fig. 3) is ca. 100 kJ mol−1 higher in energy than 6, in good agreement with the observed rapid reaction.
 | ||
| Fig. 3 Transition state for epimerisation of the anion of 6. | ||
Pharmacology
The affinity of compounds 3, 4 and 5 for native AMPA, KA, and NMDA receptors was initially determined in [3H]AMPA, [3H]KA, and [3H]CGP 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 653 competition radioligand displacement assays, respectively, using membranes prepared from rat cortical brain tissue (Table 1).12–14 In this assay all three compounds display very weak activity at NMDA sites and no activity at other sites. Furthermore, the compounds were characterised pharmacologically at human metabotropic glutamate receptors expressed in HEK cells.15,16 Compounds 3 and 5 displayed potent agonist activity at both group I and group II mGluRs with potencies somewhat lower (mGluR2) or comparable (mGluR1) to Glu. Interestingly, compound 4 turned out to be a highly selective agonist towards mGluR2 with no significant activity at any of the other receptor subtypes. It is notable how the conformational restriction of 5 changes the pharmacological profile from group II to group I metabotropic agonist.
653 competition radioligand displacement assays, respectively, using membranes prepared from rat cortical brain tissue (Table 1).12–14 In this assay all three compounds display very weak activity at NMDA sites and no activity at other sites. Furthermore, the compounds were characterised pharmacologically at human metabotropic glutamate receptors expressed in HEK cells.15,16 Compounds 3 and 5 displayed potent agonist activity at both group I and group II mGluRs with potencies somewhat lower (mGluR2) or comparable (mGluR1) to Glu. Interestingly, compound 4 turned out to be a highly selective agonist towards mGluR2 with no significant activity at any of the other receptor subtypes. It is notable how the conformational restriction of 5 changes the pharmacological profile from group II to group I metabotropic agonist.
| AMPA IC50/μM | KAIN IC50/μM | NMDA Ki/μM | mGluR1 EC50/μM | mGluR2 EC50/μM | mGluR3 EC50/μM | mGluR4 EC50/μM | mGluR5 EC50/μM | mGluR7 EC50/μM | |
|---|---|---|---|---|---|---|---|---|---|
| a The data are mean of 3 individual experiments performed in triplicate. In brackets, pKi values with SEM are listed. Not determined is abbreviated “nd” in the table. | |||||||||
| Glu | 0.34 | 0.38 | 0.20 | 1.6 | 1.8 | 0.06 | 3.2 | 4.7 | 1180 |
| [5.80 ± 0.07] | [5.74 ± 0.04] | [7.24 ± 0.08] | [5.49 ± 0.03] | [5.33 ± 0.02] | [2.93 ± 0.10] | ||||
| (S)-homo-TDPA8 | >100 | >100 | >100 | >1000 | 40 ± 3 | nd | >1000 | 190 ± 80 | nd |
| (R)-homo-TDPA8 | >100 | >100 | >100 | >1000 | 37 ± 3 | nd | >1000 | >1000 | nd |
| 3 | >100 | >100 | 77 | 6.1 | 23 | 200 | >1000 | 19 | >1000 |
| [4.11 ± 0.037] | [5.21 ± 0.04] | [4.65 ± 0.02] | [3.70 ± 0.05] | [4.72 ± 0.04] | |||||
| 4 | >100 | >100 | 72 | >1000 | 10 | >1000 | >1000 | >1000 | >1000 |
| [4.14 ± 0.01] | [4.98 ± 0.04] | ||||||||
| 5 | >100 | >100 | 19 | 3.0 | 251 | nd | >1000 | nd | nd |
| [4.73 ± 0.01] | [5.52 ± 0.03] | [3.60 ± 0.03] |
The guanidine derivatives 7 and 8 were characterised for their activity on the promiscuous amino acid receptor mouse GPRC6A (see Table 2). The compounds showed improved potency for 7 and comparable potency for 8 when both were compared to L-arginine.
Molecular modelling
The possible binding mode, stereoselectivity, and subtype selectivity of 4 at human group II metabotropic glutamate receptors were probed by a combination of homology modelling based on the crystal structure of rat mGluR3, followed by docking (Fig. 4). Only the (S,S)-form of trans_1 was found to be able to adopt a Glu-like binding mode, with the α-amino acid portion and distal acid making the same hydrogen bonds as seen in all agonist–mGluR (group II) crystal complexes to date. The cyclopropyl ring of the (R,R)-form would clash with the receptor and is predicted to be much less active than its enantiomer. While hmGluR2 and hmGluR3 have no residue differences in direct contact with the ligand, there are some notable differences among the second shell residues, two of which are highlighted in Fig. 4. The environment beyond an arginine close to the distal acid is quite different between the two subtypes, and small changes in the geometry and conformational energetics of the ligand could affect how well situated this group is upon domain closure (agonism). Testing this hypothesis, that selectivity arises from the precise distal environment of the receptor interacting with conformationally constrained ligands, could be facilitated by the solving of a crystal structure of an mGluR2 homologue, or by carrying out mutagenesis experiments upon key non-conserved residues such as H56, L300 (mGluR2) to D63, Q306 (mGluR3) or vice versa.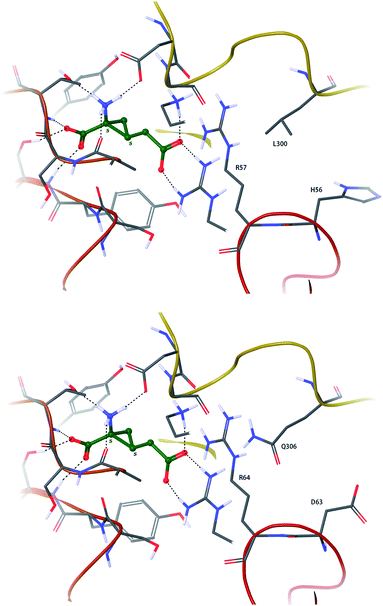 | ||
| Fig. 4 (S,S)-trans_1 docked to homology models of hmGluR2 (upper) and hmGluR3 (lower). Non-conserved second shell residues in contact with an arginine lining the distal end of the ligand binding pocket are noted. | ||
Conclusions
We have reported the synthesis and pharmacological characterisation of conformationally restricted cyclopropyl analogues of Glu (3 and 4) and homo-TPDA (5). Conformational restriction alters the pharmacological activity compared to the unrestricted compounds. In particular, compound 4 is a highly selective mGluR2 agonist and the first example of a compound displaying a significant difference in agonist potency at mGluR2 and mGluR3. Molecular modelling studies suggest a binding mode for 4 but cannot explain the high selectivity for mGluR2. Furthermore, we have reported the synthesis of compound 6 and shown that it undergoes rapid epimerisation. Finally, the conformationally restricted cyclopropyl analogues of arginine 7 and 8 were synthesised and characterised at GPRC6A.Acknowledgements
Dr Bolette Christiansen is gratefully acknowledged for technical assistance. This work was supported by the Danish Medical Research Council (HBO, RPC), GluTarget and the Lundbeck Foundation.Notes and references
- S. F. Traynelis, L. P. Wollmuth, C. J. McBain, F. S. Menniti, K. M. Vance, K. K. Ogden, K. B. Hansen, H. Yuan, S. J. Myers and R. Dingledine, Pharmacol. Rev., 2010, 62, 405–496 CrossRef CAS.
- C. M. Niswender and P. J. Conn, Annu. Rev. Pharmacol. Toxicol., 2010, 50, 295–322 CrossRef CAS.
- H. Bräuner-Osborne, P. Wellendorph and A. A. Jensen, Curr. Drug Targets, 2007, 8, 169–184 CrossRef.
- H. Bräuner-Osborne, J. Egebjerg, E.Ø. Nielsen, U. Madsen and P. Krogsgaard-Larsen, J. Med. Chem., 2000, 43, 2609–2645 CrossRef.
- J. B. Monohan, W. F. Hood, R. P. Compton, A. A. Cordi, J. P. Snyder, R. Pellicciari and B. Natalini, Neurosci. Lett., 1990, 112, 328–332 CrossRef.
- M. Kawai, Y. Horikawa, T. Ishihara, K. Shimamoto and Y. Ohfune, Eur. J. Pharmacol., 1993, 211, 195–202 CrossRef.
- T. N. Johansen, Y. L. Janin, B. Nielsen, K. Frydenvang, H. Bräuner-Osborne, T. B. Stensbøl, S. B. Vogensen, U. Madsen and P. Krogsgaard-Larsen, Bioorg. Med. Chem., 2002, 10, 2259–2266 CrossRef CAS.
- The pharmacological characterisation of (S)- and (R)-homo-TDPA was described by: R. P. Clausen, H. Bräuner-Osborne, J. R. Greenwood, M. B. Hermit, T. B. Stensbøl, B. Nielsen and P. Krogsgaard-Larsen, J. Med. Chem., 2002, 45, 4240–4245 CrossRef CAS.
- S. B. Vogensen, J. R. Greenwood, L. Bunch and R. P. Clausen, Curr. Top. Med. Chem., 2011, 11, 887–906 CrossRef CAS.
- Y. Gao and K. B. Sharpless, J. Am. Chem. Soc., 1988, 110, 7538–7539 CrossRef CAS.
- J. M. Jiménez and R. M. Ortuño, Tetrahedron: Asymmetry, 1996, 7, 3203–3208 CrossRef.
- T. Honore and M. Nielsen, Neurosci. Lett., 1985, 54, 27–32 CrossRef CAS.
- D. J. Braitman and J. T. Coyle, Neuropharmacology, 1987, 26, 1247–1251 CrossRef CAS.
- M. A. Sills, G. Fagg, M. Pozza, C. Angst, D. E. Brundish, S. D. Hurt, E. J. Wilusz and M. Williams, Eur. J. Pharmacol., 1991, 192, 19–24 CrossRef CAS.
- R. Filosa, M. Marinozzi, G. Costantino, M. B. Hermit, C. Thomsen and R. Pellicciari, Bioorg. Med. Chem., 2006, 14, 3811–3817 CrossRef CAS.
- R. Pellicciari, M. Marinozzi, A. Macchiarulo, M. C. Fulco, J. Gafarova, M. Serpi, G. Giorgi, S. Nielsen and C. Thomsen, J. Med. Chem., 2007, 50, 4630–4641 CrossRef CAS.
- B. Christiansen, K. B. Hansen, P. Wellendorph and H. Bräuner-Osborne, Br. J. Pharmacol., 2007, 150, 798–807 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures and spectroscopic characterisation. See DOI: 10.1039/c1md00186h |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2011 |
