2-[(1,3-Dihydro-2H-isoindol-2-yl)methyl]melatonin – a novel MT2-selective melatonin receptor antagonist†
David
Heckman
a,
Mohamed I.
Attia‡
b,
Mira A. M.
Behnam
c,
Amal M. Y.
Mohsen
c,
Christian
Markl
b,
Justin
Julius
a,
Shalini
Sethi
a,
Paula A.
Witt-Enderby
a and
Darius P.
Zlotos
*c
aDivision of Pharmaceutical Sciences, School of Pharmacy, Duquesne University, 421 Mellon Hall, Pittsburgh, PA 15282, USA
bInstitute of Pharmacy and Food Chemistry, Pharmaceutical Chemistry, University of Würzburg, Am Hubland, 97074, Würzburg, Germany
cThe German University in Cairo, New Cairo City, 11835 Cairo, Egypt. E-mail: darius.zlotos@guc.edu.eg; Fax: +20 2 2758 1041; Tel: +20 2 2758 1041
First published on 25th August 2011
Abstract
A synthesis and pharmacological evaluation of new melatonin receptor ligands obtained by 2-substitution of melatonin with (indol-1-yl)methyl, (isoindolin-2-yl)methyl, and (tetrahydroiso-quinolin-2-yl)methyl groups is reported. The isoindoline analogue a displays high MT2 binding affinity (Ki = 2 nM) and high selectivity towards the MT2 subtype (Ki MT1/Ki MT2 = 124) behaving as an MT2-antagonist.
Introduction
The neurohormone melatonin (MLT, 1) exerts its diverse physiological actions mostly through activation of the two high affinity G-protein-coupled MT1 and MT2 receptors.1 | ||
| Fig. 1 Structures of melatonin 1 and of the MT2-selective reference ligand 2. | ||
The nonselective MT1/MT2 agonists ramelteon2 and agomelatine3 have been successfully launched for the treatment of primary insomnia and major depression, respectively. Other nonselective melatoninergic agonists are undergoing clinical trials as hypnotic agents.4,5Melatonin receptor antagonists have been only evaluated in preclinical studies, for instance, luzindole for its antidepressant-like effects,6 S22153 in circadian rhythm entrainment experiments,7 and ML-23 in the treatment of Parkinson's disease.8 In addition to the modulation of the sleep-wake cycle and of circadian rhythms,9MLT has been reported to possess antiinflammatory,10 pain modulatory,11retinal,12 vascular,13 antitumor,14antioxidant,15 stroke-protective,16 and neuroprotective17 properties. An accurate characterisation of melatonin receptor-mediated functions requires MT1 and MT2 selective ligands.18–20 However, pronounced MT1 selectivity is still a challenge with only few examples of selective ligands published so far,21–23 most of them possessing dimeric structures. The common structural feature of MT1-selective agents is a presence of a bulky hydrophobic substituent in a position topologically equivalent to the methoxy group of MLT. In contrast, the selectivity towards the MT2 subtype can be much easily achieved and many series of MT2-selective agents have been reported in the last decade, most of them behaving as competitive antagonists or partial agonists.24–32 Very recently, a highly potent and selective MT2 agonist bearing a methylcyclohexyl group in a position equivalent to C2 of MLT has been synthesized.33 The most MT2-selective ligand known up to date is the N-[3-(3-methoxyphenyl)propyl] amide 2 (Fig. 1). In radioligand binding assays using [3H]melatonin, 2 has been recently reported to display 1,000,000 times higher affinity for the human MT2 receptors expressed in CHO cells than for the MT1 receptors.34 A common structural feature in most of MT2-selective antagonists is the presence of a lipophilic substituent located out of the plane of their core nucleus in a position corresponding to positions 1 and 2 in melatonin.35 In accordance with this pharmacophore model, we have recently synthesized a series of (indolin-1-yl)methyl melatonin analogues exemplified by compound 3.30 (Fig. 2). In radioligand binding studies at human MT1 and MT2 receptors expressed in CHO cells using 2-[125I-]iodomelatonin, 3 displayed an excellent binding affinity and selectivity towards the MT2 subtype behaving as a competitive antagonist at both melatonin receptor subtypes (MT1: Ki = 115 nM; MT2: Ki = 1.2 nM).30
 | ||
| Fig. 2 Structures of the target melatoninergic ligands 4, 5a, and 6a. | ||
In order to examine how the ring expansion of the indoline ring system in 3 to tetrahydroisoquinoline, the introduction of a double bond to give indole, and replacing the indoline scaffold by the isoindoline skeleton affects binding affinity at MT1 and MT2 receptors, we have now synthesized and pharmacologically evaluated the corresponding analogs 4, 5a, and 6a (see Fig. 2). The ligand displaying the highest affinity and selectivity towards MT2 receptors, the isoindoline analog 6a, has been subjected to further structure modifications by replacing the methyl group of the amide moiety by ethyl, allyl, cyclopropyl, and cyclobutyl groups (6b–e). For the sake of comparison, the most MT2 selective ligand reported to date, 2, was included in our study.
Results and discussion
Chemistry
Isoindoline was prepared by reduction of phthalimide using borane-THF complex.36 The synthetic route towards the target compounds 4 and 6a–e is displayed in Scheme 1. The key intermediate, 3-(cynomethyl)-5-methoxyindole-2-carboxylic acid 7, was prepared from the commercially available methyl 5-methoxy-indole-2-carboxylate in four steps including Mannich-Eschenmoser aminomethylation, quaternisation using MeI, substitution of the trimethylamino moiety with cyanide, and ester hydrolysis, as previously reported.30 | ||
| Scheme 1 Reagents and conditions: (i) EDCI·HCl, isoindoline, CH2Cl2, rt.; (ii) EDCI·HCl, tetrahydroisoquinoline, CH2Cl2, rt.; (iii) LiAlH4, Et2O, THF, 0 °C – reflux; (iv) 6a: acetic anhydride, Et3N, CH2Cl2, 0 °C – rt; 6b–e: appropriate carboxylic acid, EDCI·HCl, CH2Cl2, −10 °C – rt; (v) acetic anhydride, Et3N, CH2Cl2, 0 °C – rt. | ||
Condensation of 7 with isoindoline and tetrahydroisoquinoline was carried out in CH2Cl2 using 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDCI·HCl) as a coupling reagent to give the cyanoamides 8, and 9 in 76% and 62% yield. Simultaneous nitrile and amide reduction using LiAlH4 in diethyl ether/THF afforded the ethylamines 10 and 11 which were converted to the desired melatoninergic ligands 6a and 4 by N-acetylation using acetic anhydride/NEt3 in CH2Cl2 (65% and 41% over two steps). The target amides 6b–e were obtained by condensation of 10 with the appropriate acid using EDCI·HCl as a coupling reagent (Scheme 1).
The indole-substituted target compounds, acetamide 5a and cyclobutanecarboxamide 5b, were prepared by dehydrogenation of the corresponding indoline analogs employing dry heating at 150 °C in the presence of Pd/C 10%.37 (Scheme 2). The MT2-selective reference ligand 2 was prepared according to the previosly published procedure.34
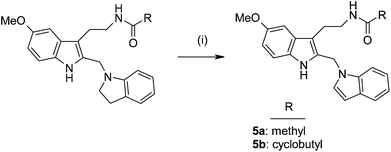 | ||
| Scheme 2 Reagents and conditions: (i) Pd/C 10%, 150 °C. | ||
Pharmacology
The affinity of the target compounds for human MT1 or MT2 melatonin receptors expressed in CHO cells was measured by competition binding analysis using the radioligand, 2-[125I]-iodomelatonin as described previously.38Melatonin competition assays were run in parallel and the affinity of melatonin for the MT1 or MT2 melatonin receptors is in the range of the reported literature. The results are compiled in Table 1. For the most MT2-selective ligand 6a, functional studies were carried out using cyclic AMP assay in CHO cells expressing human MT1 or MT2 receptors. The results are shown in Fig. 3.| pKi MT1 ± SEM | pKi MT2 ± SEM | |
|---|---|---|
| melatonin | 9.34 ± 0.10 | 9.02 ± 0.09 |
| 2 | 5.87 ± 0.32 | 8.77 ± 0.08 |
| 3 30 | 6.94 ± 0.02 | 8.93 ± 0.05 |
| 4 | 6.60 ± 0.02 | 7.67 ± 0.01 |
| 5a | 6.74 ± 0.05 | 8.24 ± 0.01 |
| 5b | 7.24 ± 0.02 | 7.09 ± 0.01 |
| 6a | 6.55 ± 0.01 | 8.65 ± 0.04 |
| 6b | 8.25 ± 0.04 | 8.43 ± 0.02 |
| 6c | 7.62 ± 0.01 | 8.03 ± 0.00 |
| 6d | 7.21 ± 0.04 | 7.47 ± 0.03 |
| 6e | 6.95 ± 0.09 | 6.70 ± 0.03 |
 | ||
| Fig. 3 Functional analysis of melatonin and 6a on forskolin-stimulated cAMP formation in CHO cells expressing either the MT1 receptor or the MT2 receptor. Each data point represents the mean ± SEM of 2–3 independent experiments performed in duplicate (see text for discussion). | ||
Discussion
The novel melatoninergic ligands described in this work are structure modifications of our previously reported MT2-selective antagonist 3. 3 displays high affinity towards the MT2 receptors (MT2: Ki = 1.2 nM), and only moderate affinity towards the MT1 subtype (MT1: Ki = 115 nM) resulting in a selectivity ratio Ki(MT1)/Ki(MT2) = 95. The structural feature responsible for its antagonistic activity at MT2 receptors is the indoline substituent in position 2 of melatonin arranged “out of plane” of the indole nucleus. In order to examine the steric tolerance of a binding pocket accomodating the “out of plane” substituent, the isoindoline scaffold in 3 was expanded by insertion of a methylene group between the indolic nitrogen and the benzene ring. The resulting tetrahydroisoquinoline analog 4 showed significantly decreased binding at both MT1 and MT2 receptors when compared to the parent agent 3. Moreover, the selectivity towards MT2 receptors was reduced to 12-fold indicating the sterically restricted binding pocket. Consequently, the size of the C2 substituent was kept virtually unchanged in the subsequent structure modifications of 3. Introduction of a double bond into the indoline ring system also worsened the pharmacological profile. The resulting ligand 5a displayed a 5-times lower affinity for the MT2 receptors (MT2: Ki = 5.7 nM) than 3 while binding to the MT1 receptors was almost unchanged (MT1: Ki = 184 nM). On the contrary, relocation of the nitrogen atom in the indoline ring of the parent compound 3 from position 1 to position 2 generated an MT2 high-affinity ligand 6a with the best selectivity profile for melatonin receptors. The isoindoline analog 6a was 124-times more selective for the MT2 than for the MT1 receptors showing low nanomolar MT2 affinity (MT2: Ki = 2.3 nM).In an attempt to further optimize the pharmacological profile of 6a, we modified the terminal acyl moiety by replacing the methyl group with ethyl (6b), allyl (6c), cyclopropyl (6d), and cyclobutyl (6e) substituents. Unfortunately, none of the ligands 6b–e displayed pronounced MT2 selectivity. A similar loss of selectivity was found in the cyclobutanecarboxamide 5b when compared to the corresponding acetamide 5a.
Previous studies on melatonin receptor ligands indicate that binding constants for the same ligand may differ depending on the cell line used in the radioligand binding asssays.21 Moreover, binding data obtained from different laboratories may vary even if the same cell lines were used.39 To allow a better direct comparison of our compounds with other melatoninergic ligands, N-[3-(3-methoxyphenyl)propyl] amide 2, the most MT2 selective ligand known up to date was included in our studies. In radioligand binding assays using [3H]melatonin, 2 has been recently reported to display 1,000,000 times higher affinity for the human MT2 receptors expressed in CHO cells than for the MT1 receptors. (MT1: Ki = 705 nM; MT2: Ki = 0.00069 nM). In our binding assay using the same cell line but a different radioligand (2-[125I]-iodomelatonin), 2 displayed a similar affinity for the MT1 receptors (MT1: Ki = 1350 nM) while binding for the MT2 receptors was more than 2,000-times reduced (MT2: Ki = 1.7 nM) resulting in a much lower 790-fold selectivity towards the MT2 receptors. Nevertheless, our data confirm that, to the best of our knowledge, the reference ligand 2 still remains one of the most MT2-selective agent known to date.
Our most MT2-selective ligand 6a was subjected to functional analysis using forskolin-stimulated cAMP formation assay in CHO cells expresing MT1 or MT2 receptors. Similar to the parent indoline analogue 3, 6a was shown to be a competitive antagonist at MT1 receptors. 6a (10 nM or 100 nM), added in combination with increasing concentrations of melatonin competitively antagonized melatonin's actions at MT1 receptors reflected by a rightward shift in the melatonin concentration-response curve in the presence of 6a (Fig. 3). When tested alone, 6a displayed no intrinsic activity against MT1 receptors (data not shown).
As for its actions at the MT2 receptor, 6a displayed no intrinsic activity against MT2 receptors when tested alone (data not shown). However, when added in combination with melatonin (0.01pM-100nM), 6a (10nM or 100nM) antagonized the effects of melatonin at all concentrations tested. These data suggest that 6a binds tightly to the MT2 receptor, perhaps at a crucial portion of the MT2 receptor affecting Gi-protein activation, thus requiring higher concentrations of melatonin (>100nM) to reverse its antagonism (Fig. 3). Another possibility could be that 6a is binding irreversibly to MT2 receptors at the MT2 receptor/Gi-protein interface thus affecting MT2 receptor-mediated activation of Gi proteins. The functional behaviour of compound 6a at MT2 receptors is similar to that observed for irreversible ligands bearing electrophilic functional groups which are able to form covalent bonds with specific receptors sites, such as isothiocynates, Michael acceptors, haloacetamides, aldol esters, and nitrogen musturds.38,40 As, for the best of our knowledge, there are no reports about the electrophilic character of isoindoline derivatives, the mechanism accounting for the possible tight (irreversible) binding behaviour of the title compound at melatonin receptors is still to be investigated.
Acknowledgements
We thank Prof. Dr Ulrike Holzgrabe, Würzburg University for her support and Anita Betz, Würzburg University for synthesizing compound 7.Notes and references
- S. M. Reppert, D. R. Weaver and C. Godson, Trends Pharmacol. Sci., 1996, 17, 100 CrossRef CAS.
- M. Miyamoto, CNS Neurosci. Ther., 2009, 15, 32 Search PubMed.
- C. de Bodinat, B. Guardiola-Lemaitre, E. Mocaër, P. Renard, C. Muñoz and M. J. Millan, Nat. Rev. Drug Discov., 2010, 9(8), 628 CAS.
- R. Hardeland, Expert Opin. Invest. Drugs, 2010, 19, 747 CrossRef CAS.
- M. Mor, S. Rivara, D. Pala, A. Bedini, G. Spadoni and G. Tarzia, Expert Opin. Ther. Pat., 2010, 20, 1059 Search PubMed.
- I. C. Sumaya, M. I. Masana and M. L. Dubocovich, J. Pineal Res., 2005, 39, 170 CrossRef CAS.
- X. M. Li, J. Beau, P. Delagrange, E. Mocaër and F. Lévi, J. Pineal Res., 2004, 37, 176 CrossRef CAS.
- G. L. Willis, Drug News Perspect., 2005, 18, 437 CrossRef CAS.
- F. A. J. L. Scheer and C. A. Czeisler, Rhythms. Sleep Med. Rev., 2005, 9, 5 Search PubMed.
- T. Genovese, E. Mazzon, C. Muia, P. Bramanti, A. De Sarro and S. Cuzzocrea, J. Pineal Res., 2005, 38, 198 CrossRef CAS.
- M. F. P. Peres, Cephalalgia, 2005, 25, 403 CrossRef CAS.
- P. M. Iuvone, G. Tosini, N. Pozdeyev, R. Haque, D. C. Klein and S. S. Chaurasia, Prog. Retinal Eye Res., 2005, 24, 433 Search PubMed.
- E. Sewerynek, Neuroendocrinol. Lett., 2002, 23(S.1), 79 Search PubMed.
- P. A. Witt-Enderby, N. M. Radio, J. S. Doctor and V. L. Davis, J. Pineal Res., 2006, 41, 297 CrossRef CAS.
- E. Sofic, Z. Rimpapa, Z. Kundurovic, A. Sapcanin, I. Tahirovic, A. Rustembegovic and G. Cao, J. Neural Transm., 2005, 112, 349 CrossRef CAS.
- M. R. Macleod, T. O'Collins, L. L. Horky, D. W. Howells and G. A. Donnan, J. Pineal Res., 2005, 38, 35 CrossRef CAS.
- V. Srinivasan, S. Pandi-Perumal, D. Cardinali, B. Poeggeler and R. Hardeland, Behav. Brain Funct., 2006, 2, 15 Search PubMed.
- D. P. Zlotos, Arch. Pharm., 2005, 338, 229 CrossRef CAS.
- G. Spadoni, A. Bedini, in Melatonin: From Molecules to Therapy, ed. S. R. Pandi-Perumal and D. P. Cardinali, Nova Science Publishers, Inc., 2007, ch. 3, pp. 33–45 Search PubMed.
- P. J. Garret and A. Tsotinis, Mini-Rev. Med. Chem., 2007, 7, 1075 CrossRef.
- V. Audinot, F. Mailliet, C. Lahaye-Brasseur, A. Bonnaud, A. Le Gall, C. Amossé, S. Dromaint, M. Rodriguez, N. Nagel, J.-P. Galizzi, B. Malpaux, G. Guillaumet, D. Lesieur, F. Lefoulon, P. Renard, P. Delagrange and J. A. Boutin, Naunyn-Schmiedebergs Arch. Pharmacol., 2003, 367, 553 CrossRef CAS.
- L. Q Sun, J. Chen, M. Bruce, J. A. Deskus, J. R. Epperson, K. Takaki, G. Johnson, L. Iben, C. D. Mahle, E. Ryan and C. Xu, Bioorg. Med. Chem. Lett., 2004, 14, 3799 CrossRef CAS.
- C. Mesangeau, B. Peres, C. Descamps-Francois, P. Chavatte, V. Audinot, S. Coumailleau, J. A. Boutin, P. Delagrange, C. Bennejean, P. Renard, D. H. Caignard, P. Berthelot and S. Yous, Bioorg. Med. Chem., 2010, 18, 3426 CrossRef CAS.
- V. Wallez, S. Durieux-Poissonnier, P. Chavatte, J. A. Boutin, V. Audinot, J.-P. Nicolas, C. Bennejean, P. Delagrange, P. Renard and D. Lesieur, J. Med. Chem., 2002, 45, 2788 CrossRef CAS.
- R. Faust, P. J. Garratt, R. Jones, L.-K. Yeh, A. Tsotinis, M. Panoussopoulou, T. Calogeropoulou, M.-T. Teh and D. Sugden, J. Med. Chem., 2000, 43, 1050 CrossRef CAS.
- G. Spadoni, C. Balsamini, G. Diamantini, A. Tontini, G. Tarzia, M. Mor, S. Rivara, P. V. Plazzi, R. Nonno, V. Lucini, M. Pannacci, F. Fraschini and B. M. Stankov, J. Med. Chem., 2001, 44, 2900 CrossRef CAS.
- V. Lucini, M. Pannacci, F. Scaglione, F. Fraschini, S. Rivara, M. Mor, F. Bordi, P. V. Plazzi, G. Spadoni, A. Bedini, G. Piersanti, G. Diamantini and G. Tarzia, J. Med. Chem., 2004, 47, 4202 CrossRef CAS.
- A. Bedini, G. Spadoni, G. Gatti, S. Lucarini, G. Tarzia, S. Rivara, S. Lorenzi, A. Lodola, M. Mor, V. Lucini, M. Pannacci and F. Scaglione, J. Med. Chem., 2006, 49, 7393 CrossRef CAS.
- S. Rivara, A. Lodola, M. Mor, A. Bedini, G. Spadoni, V. Lucini, M. Pannacci, F. Fraschini, F. Scaglione, R. O. Sanchez, G. Gobbi and G. Tarzia, J. Med. Chem., 2007, 50, 6618 CrossRef CAS.
- D. P. Zlotos, M. I. Attia, J. Julius, S. Sethi and P. A. Witt-Enderby, J. Med. Chem., 2009, 52, 826 CrossRef CAS.
- C. Mésangeau, M. Fraise, P. Delagrange, D. H. Caignard, J. A. Boutin, P. Berthelot and S. Yous, Eur. J. Med. Chem., 2011, 46, 1835 CrossRef CAS.
- S. Durieux, A. Chanu, C. Bochu, V. Audinot, S. Coumailleau, J. A. Boutin, P. Delagrange, D. H. Caignard, C. Bennejean, P. Renard, D. Lesieur, P. Berthelot and S. Yous, Bioorg. Med. Chem., 2009, 17, 2963 CrossRef CAS.
- T. Koike, Y. Hoashi, T. Takai, M. Nakayama, N. Yukuhiro, T. Ishikawa, K. Hirai and Osamu Uchikawa, J. Med. Chem., 2011, 54, 3436 CrossRef CAS.
- Y. Hu, M. K. C. Ho, K. H. Chan, D. C. New and Y. H. Wong, Bioorg. Med. Chem. Lett., 2010, 20, 2582 CrossRef CAS.
- S. Rivara, M. Mor, S. Lorenzi, A. Lodola, P. V. Plazzi, G. Spadoni, A. Bedini and G. Tarzia, Arkivoc, 2006, VIII, 8.
- R. E. Gawley, S. R. Chemburkar, A. L. Smith and T. V. Anklekar, J. Org. Chem., 1988, 53, 5381 CrossRef CAS.
- M. I. Attia, J. Julius, P. A. Witt-Enderby and D. P. Zlotos, Tetrahedron, 2007, 63, 754 CrossRef CAS.
- P. A. Witt-Enderby, G. H. Chu, M. L. Gillen and P. K. Li, J. Med. Chem., 1997, 40, 4195 CrossRef CAS.
- C. Markl, W. P. Clafshenkel, M. I. Attia, S. Sethi, P. A. Witt-Enderby and D. P. Zlotos, Arch. Pharm. Chem. Life Sci., 2010 DOI:10.1002/ardp.201100125.
- A. Hauck Newman, Ann. Rep. Med. Chem., 1990, 25, 271 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedure and analytical data for compounds 4, 5a–b and 6a–e, 8, 9. See DOI: 10.1039/c1md00149c |
| ‡ Current address: Department of Pharmaceutical Chemistry, College of Pharmacy, King Saud University, Riyadh 11451, Saudi Arabia. |
| This journal is © The Royal Society of Chemistry 2011 |
