Discovery of GS-8374, a potent human immunodeficiency virus type 1 protease inhibitor with a superior resistance profile†
Gong-Xing
He
a,
Zheng-Yu
Yang
a,
Matthew
Williams‡
a,
Christian
Callebaut
b,
Tomas
Cihlar
b,
Bernard P.
Murray
c,
Chris
Yang
c,
Michael L.
Mitchell
a,
Hongtao
Liu
a,
Jianying
Wang‡
a,
Murty
Arimilli‡
a,
Eugene
Eisenberg
c,
Kirsten M.
Stray
b,
Luong K.
Tsai
b,
Marcos
Hatada‡
d,
Xiaowu
Chen
d,
James M.
Chen‡
d,
Yujin
Wang
c,
Melody S.
Lee‡
c,
Robert G.
Strickley
e,
Quynh
Iwata
e,
Xubin
Zheng
c,
Choung U.
Kim
a,
Swami
Swaminathan
d,
Manoj C.
Desai
a,
William A.
Lee
f and
Lianhong
Xu
*a
aDepartment of Medicinal Chemistry, Gilead Sciences Inc, 333 Lakeside Drive, Foster City, CA 94404, USA. E-mail: lianhong.xu@gilead.com.
bDepartment of Biology, Gilead Sciences Inc, 333 Lakeside Drive, Foster City, CA 94404, USA
cDepartment of Drug Metabolism, Gilead Sciences Inc, 333 Lakeside Drive, Foster City, CA 94404, USA
dDepartment of Structural Chemistry, Gilead Sciences Inc, 333 Lakeside Drive, Foster City, CA 94404, USA
eDepartment of Formulation, Gilead Sciences Inc, 333 Lakeside Drive, Foster City, CA 94404, USA
fGilead Research, Gilead Sciences Inc, 333 Lakeside Drive, Foster City, CA 94404, USA
First published on 7th September 2011
Abstract
Introduction of a unique phosphonate moiety at the P1 position of the TMC-126 (3) scaffold provided a series of novel HIV-1 protease inhibitors (PIs) with an improved resistance profile against highly resistant variants. Optimization of the linker and phosphonate moieties lead to the identification of GS-8374 (1). Compound 1 is a potent and orally bioavailable HIV-1 PI with a superior resistance profile. Synthesis and characterization of 1 are reported.
Human immunodeficiency virus type 1 (HIV-1) protease inhibitors (PIs) have continued to play a prominent role in the treatment of HIV infection since their first introduction in 1995. Highly active antiretroviral therapy (HAART) regimens employing protease inhibitors in combination with other antiretrovirals can produce a rapid and sustained decline of plasma viral RNA and a concomitant increase in CD4 T cell count in HIV-infected individuals, and prevent the progression of HIV related disease.1–3 Clinical use of PIs, together with other antiretroviral drugs, made the long term management of HIV disease possible. As a class, however, HIV-1 PIs are rapidly metabolized, primarily by cytochrome P-450 enzymes of the 3A subfamily (CYP3A) in the liver and intestine, resulting in low systemic exposure and short half-lives after oral administration. Consequently PIs, especially those of the early generation, usually require high doses and high dosing frequency, and are associated with significant side effects.4,5Ritonavir (RTV), approved as an HIV-1 PI (therapeutic dose 600 mg twice daily), was shown to inhibit CYP3A and significantly “boost” pharmacokinetic (PK) profiles of coadministered PIs at a subtherapeutic dose (often 100–200 mg administered once or twice daily). Boosting of PIs with low-dose RTV is now widely used to achieve desirable drug plasma concentrations, which lead to improved viral suppression while decreasing both the dosing frequency and pill burden.6–10 Current HIV treatment guidelines recommend ritonavir-boosted atazanavir (ATV) or darunavir (DRV) as a third agent of choice for first line antiretroviral therapy.11,12 Although there are nine PIs currently licensed for the treatment of HIV-1, and new generation PIs show better pharmacological properties than the earlier agents, profiles of most of the currently approved PIs can be further improved. The clinical benefit of this class of antiretrovirals in general is still limited by several factors including resistance and long-term safety and tolerability. Among these, the emergence of drug-resistant HIV-1 variants continues to be a major cause of treatment failure and presents a major challenge to the control of HIV infection. In addition, with the exception of ATV, other PIs require high dose and/or frequent dosing regimens, limiting their use in fixed dose combination regimens. Therefore, the design of novel PIs with more favorable resistance profiles and convenient dosing regimens remains a consistent interest in the scientific community. This communication describes the discovery and characteristics of GS-8374 (compound 1, Fig. 1), a potent and orally bioavailable PI with a superior resistance profile against a spectrum of patient-derived HIV-1 variants highly resistant to multiple PIs. Because of its favorable pharmacological profile, GS-8374 (compound 1) has been further explored as a potential candidate for clinical development.
 | ||
| Fig. 1 Structures of GS-8374, darunavir (DRV) and TMC-126. | ||
In our multi-pronged approach in searching for novel HIV-1 PIs, we observed that the incorporation of polar moieties at the P1 site of a PI can improve the resistance profile of a parent PI scaffold and thus render it to be active against several multidrug-resistant variants of HIV-1 such as the I84V/L90M mutant. Among the approved HIV-1 PIs, the peptidomimetic bis-tetrahydrofuran sulfonamide analog DRV (2), possesses arguably the most favorable resistance profile.13 We pursued further optimization of the antiviral potency, activity against PI-resistant HIV-1 variants, as well as other pharmacological properties. HIV-1 strains with high level PI resistance containing M46I/I50V or I84V/L90M mutations in protease were selected for the first tier resistance profiling of the new analogs. M46I/I50V mutant virus was selected in vitro in the presence of amprenavir, and the I84V/L90M variant is a patient-derived recombinant strain exhibiting markedly reduced susceptibility to all tested PIs, except DRV. Both HIV-1 variants also show residual resistance to the peptidomimetic bis-tetrahydrofuran sulfonamide class of PIs. In a profiling cascade, new compounds were screened in enzymatic assays, and in antiviral assays in cell cultures against wild type virus and the two resistant variants. In addition, it has been shown that PIs have the propensity for binding to human serum proteins, resulting in a reduction in their antiviral potency,14 the effect of 50% human serum (HS) on the antiviral activity of selected analogs was also evaluated. Both compound 2 and 3 (TMC-126) were considered as the potential scaffolds for the introduction of polar moieties at the P1 site. However, compound 3 was chosen as the preferred scaffold since results showed that derivatives of 3 in general have better overall biological and pharmacological profiles than the corresponding analogs of 2.
A series of moieties were introduced at the para- position of the P1 phenyl group of compound 3. Not surprisingly, in keeping with previous observations, incorporation of polar moieties into compound 3 consistently provided PIs with better resistance profiles (summarized in Table 1). For example, introduction of a polar –OH substitution at P1 (compound 4) resulted in a much improved resistance profile compared with TMC-126 (compound 3), against I84V/L90M (0.8 vs. 12 fold resistance) and M46I/I50V (5.1 vs. 72 fold resistance); the same pattern holds when comparing a polar –COOCH3 or –OH substituent with the less polar –OCH3 (compound 4 or 6vs.5). Pharmacokinetic (PK) profiling of these compounds showed that they all have metabolic liabilities. This prompted us to search for novel moieties for P1 that can maintain the desired potency and resistance profile yet overcome poor metabolic stability. We discovered that incorporation of a phosphonate moiety at the P1 site can meet these criteria. A focused survey of various P1-phosphonate linkers revealed that two-atom linkers provided better overall potency and resistance profile (compounds 1 and 9vs.7 and 8, Table 1). Investigation of different phosphonate esters in the context of the methyleneoxy linker indicated that both ethyl and isopropyl phosponates (1 and 11) offered the desired profile, while the corresponding methyl phosponate 10 showed less favorable properties, especially reduced antiviral potency. Additional profiling of compounds 1, 9 and 11 demonstrated that compound 1 (GS-8374) exhibits the best overall profile including the antiviral activity against the two key PI-resistant HIV-1 variants and protein adjusted antiviral potency; therefore it was selected for further evaluation.
|
|
||||||
|---|---|---|---|---|---|---|
| Compound | Structure R= | Ki (pM) | EC50 (nM) WT | EC50 fold shift in 50% HS | EC50 fold change | |
| I84V/L90M | M46I/I50V | |||||
| 3 (TMC-126) | –H | 2.3 | 0.35 | — | 12 | 72 |
| 4 | –OH | 5.3 | 6.9 | — | 0.8 | 5.1 |
| 5 | –OCH3 | — | 0.06 | — | 17 | 157 |
| 6 | –COOCH3 | 5.4 | 0.3 | — | 1.2 | 11 |
| 7 |
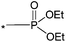
|
1.2 | 7.5 | — | 0.5 | 4.5 |
| 8 |

|
8.4 | 5.9 | 2.4 | 5.8 | 36 |
| 9 |

|
9.1 | 2.1 | 2.6 | 0.4 | 3.1 |
| 1 |
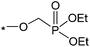
|
8.1 | 3.5 | 2.7 | 0.7 | 2.4 |
| 10 |

|
18.1 | 18.8 | — | 0.3 | — |
| 11 |

|
5.7 | 1.9 | 3.6 | 1.3 | 6.0 |
The synthesis of compound 1 is outlined in Scheme 1. Treatment of the commercial available erythro-N-Boc-O-benzyl-L-tyrosine epoxide 12 with isobutyl amine provided compound 13. Selective sulfonylation of the resulting amine with p-methoxy benzensulfonyl chloride afforded sulfonamide 14. Removal of the Boc-protecting group yielded amine 15. Chiral bis-tetrahydrofuran alcohol 16, prepared according to the literature procedure,15 was transformed to carbonate 17. Coupling of carbonate 17 with amine 15 in the presence of base provided compound 18. Removal of benzyl through hydrogenolysis followed by treatment the resulting phenol with (diethoxyphosphoryl)methyl trifluoromethanesulfonate gave compound 1.
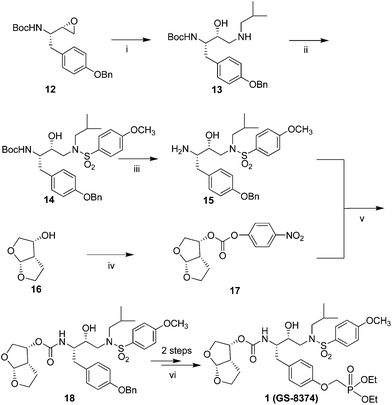 | ||
| Scheme 1 Synthesis of Compound 1 (GS-8374). Reagents and conditions: (i) isobutylamine, 2-propanol (iPrOH), reflux. (ii) 4-methoxybenzenesulfonyl chloride, diisopropylethylamine, CH2Cl2, 0 °C. (iii) TFA. (iv) bis-(4-nitrophenyl)carbonate, Et3N, CH2Cl2, rt. (v) N,N-dimethylaminopyridine, CH3CN. (vi) a. H2 (1atm), 10% Pd/C, EtOH, EtOAc. b. (diethoxyphosphoryl)methyl trifluoromethanesulfonate, Cs2CO3, THF. | ||
Compound 1 was extensively profiled in comparison with other PIs including the closely related bis-tetrahydrofuran analogs 2 and 3 (Table 2). Among the newly tested PIs, compound 1 was one of the most potent inhibitors of HIV-1 protease with a Ki value less than 10 pM. In addition, antiviral activity in the presence of HS was comparable with the most frequently prescribed PIs (DRV, lopinavir and ATV), and significantly improved relative to some the earlier generation PIs. Finally, it also showed minimal cytotoxicity.
| Compound | Enzyme inhibition Ki (pM) | EC50 (nM) WT | EC50 fold shift in 50% HS | CC50 (μM) | |
|---|---|---|---|---|---|
| MT-2 | HepG2 | ||||
| 1 (GS-8374) | 8.1 | 3.5 | 2.7 | >100 | 61.3 |
| 2 (Darunavir) | 4.6 | 2.3 | 3.6 | >100 | 95.5 |
| Lopinavir | 37.1 | 5.6 | 3.5 | 67.9 | 27.8 |
| Atazanavir | 47.9 | 2.2 | 2.4 | 79.8 | 26.5 |
| Saquinavir | 221 | 13.6 | — | 32.3 | 12.4 |
| Ritonavir | 314 | 32.2 | — | 65.0 | 18.4 |
| Amprenavir | 345 | 24 | — | >100 | 98.4 |
| Indinavir | 571 | 12.4 | — | >100 | 74.7 |
| Nelfinavir | 732 | 4.9 | 19.2 | 19.1 | 7.4 |
In addition to the two PI-resistant mutant viruses (M46I/I50V and I84V/L90M), compound 1 was further evaluated using four additional recombinant HIV-1 variants derived from PI-experienced patients that contained most of the known primary PI-resistance mutations. Activity of compound 1 against these viruses was compared directly with most approved PIs and with the close non-phosphate analog compound 3 (TMC-126). In the phenotypic assays, five of six PI-resistant strains tested showed either no significant change in susceptibility or hypersensitivity to compound 1 (Table 3). Mutant strain M46I/I50V exhibited approximately 2.5 fold reduced susceptibility relative to wild-type virus, compared with 17.8 fold reduction with DRV. The summary of resistance profiling is shown graphically in Fig. 2 and indicates a superior resistance profile for compound 1 compared to the other tested PIs, suggesting a potential for clinical activity against a broad range of PI-resistant viruses. Compound 1 was later characterized using a PhenoSense™ assay against a panel of 24 patient-derived mutant viruses with high-level phenotypic resistance to multiple clinically approved PIs. Compound 1 demonstrated superior resistance profile when compared with the other tested PIs16 including DRV that exhibits the most favorable resistance profile among all clinically approved PIs.
| Compound | Fold resistance relative to WT HIV-1 | |||||
|---|---|---|---|---|---|---|
| I84V/L90M | G48V/I54V/V82S | M46I/V82T/L90M | D30N/54V/L90M | G48V/V82A/I84V/L90M | M46I/I50V | |
| a Viruses are designated according to their primary resistance mutations present in protease. All viruses except the M46I/I50V are patient-derived recombinant strains. M46I/I50V virus was selected in vitro in the presence of amprenavir. | ||||||
| 1 (GS-8374) | 0.7 ± 0.1 | 0.1 ± 0.0 | 0.9 ± 0.1 | 0.2 ± 0.0 | 0.6 ± 0.1 | 2.4 ± 1.6 |
| 3 (TMC-126) | 12.3 ± 3.5 | 0.2 ± 0.0 | 1.8 ± 0.7 | 0.6 ± 0.5 | 4.0 ± 2.4 | 72.2 ± 37.8 |
| 2 (Darunavir) | 1.8 ± 0.6 | 0.1 ± 0.0 | 1.2 ± 0.1 | 0.2 ± 0.0 | 1.0 ± 0.5 | 17.8 ± 6.3 |
| Lopinavir | 17.9 ± 7.9 | 22.7 ± 3.2 | 7.2 ± 3.2 | 28.8 ± 9.8 | 33.9 ± 6.1 | 59.9 ± 15.8 |
| Atazanavir | >100 | 127 | 43.9 ± 17.4 | 141 ± 69 | 102 ± 54 | 1.9 ± 1.4 |
| Saquinavir | 55.4 ± 21.8 | 21.6 | 3.1 ± 1.2 | 11.9 ± 6.9 | 75.5 ± 35.3 | 1.5 ± 0.2 |
| Ritonavir | >47 | 35.9 ± 21.5 | >31 | >41 | >50 | 14.0 ± 4.9 |
| Amprenavir | 16.3 ± 6.9 | 0.9 ± 0.2 | 5.9 ± 2.8 | 0.8 ± 0.1 | 20.0 ± 3.3 | 159 ± 67 |
| Indinavir | >51 | 20.1 ± 5.5 | 24.5 ± 8.4 | 18.9 ± 1.3 | 24.6 ± 2.3 | 12.4 ± 7.1 |
| Nelfinavir | >92 | 25.4 ± 16.3 | 26.0 ± 6.6 | >68 | 30.6 ± 1.3 | 16.5 ± 8.1 |
 | ||
| Fig. 2 Comparison of GS-8374 resistance profile with that of approved and investigational PIs. | ||
The mechanism for the improved resistance profile of compound 1 through polar substitution at P1 was not immediately apparent. Thermodynamic studies, comparing the interactions of compounds 1 and 3 with wild-type and mutant proteases, showed that the reduction of the enthalpic component of binding energy due to resistance mutations were compensated by significant increases of entropic contributions.17 When binding to the mutant protease, the reduction of enthalpy was similar for both compounds 1 and 3, yet 1 had a greater increase of entropy than 3, resulting in minimal or no change of free binding energy compared to binding to wild-type protease. Crystal structures of both wild-type and I84V/L90M mutant protease in complex with compounds 1 and 3 were obtained and compared. The solvation of the phosphonate group provides an anchor point for the inhibitor, allowing more flexibility of the binding of the inhibitor to the active site of the mutant enzyme, and thus enabling it to accommodate variability in the binding pocket.17 The differences in resistance profiles of the phosphonate analogs with different linkers (compounds 7–11) indicated that the optimal solvent-anchoring effect can be obtained by adjustment of the angle and length of the polar function attachment.
With such a superior resistance profile, compound 1 was further investigated to obtain the rate of selection of mutations. Resistance selection performed with compound 1 for 11 months yielded a virus with approximately 15-fold resistance. The selected HIV-1 variant contained multiple mutations in Gag, located mainly outside the cleavage sites, in combination with one genotypic change in protease (R41K, a naturally occurring polymorphism).18 Further analysis demonstrated that the Gag mutations confer resistance to 1, indicating a high genetic barrier for the selection of resistance mutations in the protease itself. In comparison, in vitro resistance to lopinavir (LPV),19ATV20 or DRV21 emerged after 3 to 6 months of virus passaging, and in all cases the selected viruses contained resistance mutations in the protease gene.
As discussed previously, low-dose ritonavir is widely used to provide pharmacokinetic enhancement for HIV-1 PIs to maintain drug plasma levels leading to improved viral suppression while decreasing both dosing frequency and pill burden. Recently a novel pharmacoenhancer, cobicistat (COBI), which is devoid of anti-HIV activity, was discovered.22 Both preclinical and clinical studies have shown that it can reduce the metabolic clearance and thereby boost the systemic exposure of CYP3A substrates.22,23
The metabolic stability of compound 1 was studied in human hepatic microsomal fractions. Although the human microsomal stability of compound 1 was improved compared to DRV, ATV and LPV, the predicted human PK was still not compatible with once-daily dosing. However, the oxidative metabolism of compound 1 is catalyzed primarily by CYP3A and can be effectively blocked with either COBI or RTV, suggesting that it is a good candidate for pharmacokinetic enhancement (Table 4). In addition, the relative metabolic stability and boosting potential of compound 1 comparing with other marketed PIs were studied and the results are illustrated in Fig. 3.
| Compound | Microsomal CLint (μl min−1 mg−1) | ||
|---|---|---|---|
| No booster | +1 μM RTV | +1 μM COBI | |
| 1 | 9.2 ± 1.3 | 2.6 ± 1.7 | 1.0 ± 0.7 |
| Lopinavir | 544 ± 26.6 | 1.6 ± 0.7 | 2.7 ± 0.7 |
| Darunavir | 30.9 ± 2.4 | 1.6 ± 0.5 | 0.9 ± 1.2 |
| Atazanavir | 23.0 ± 2.1 | 1.4 ± 0.6 | 0.9 ± 0.2 |
 | ||
| Fig. 3 Relative human hepatic metabolic stability and boosting partner potential of PIs of HIV-1. | ||
Compound 1 has low metabolic stability with hepatic microsomal fraction from nonclinical species, such as beagle dog (predicted hepatic extraction 93%). This was confirmed in vivo, as after a dose of 2 mg kg−1 by intravenous infusion over 30 min, the clearance was 2.13 ± 0.06 L h−1 kg−1 (mean ± SD, n = 3). Such poor stability complicates interpretation of bioavailability studies in this species as it is difficult to determine whether poor bioavailability is due to low intestinal permeability or high first-pass metabolism. We thus performed oral pharmacokinetic studies in beagle dogs co-dosed with COBI to reduce the metabolic clearance. Compound 1 was dosed by oral gavage as a solution at a dose of 20 mg kg−1 in the absence or presence of codosed COBI (10 mg kg−1). Oral bioavailability in the absence of COBI was 6.9% ± 5.4% (mean ± SD, n = 3) but codosing with COBI increased exposure 15-fold to ∼100% apparent oral bioavailability (Fig. 4). This confirms the high absorption potential of compound 1.
 | ||
| Fig. 4 Pharmacokinetics of compound 1 (GS-8374) in beagle dogs. | ||
In summary, compound 1 has favorable pharmacological and PK profiles, including potent antiviral effect against both wild-type HIV-1 and many mutant variants with high level resistance to clinically approved PIs, which suggest it has potential for clinical efficacy against a broad range of PI-resistant viruses. Preclinical studies indicated that, assuming metabolism by CYP3A was the major route of elimination, compound 1 could be dosed once daily with a pharmacokinetic enhancer to achieve necessary trough concentration for effective clinical suppression of HIV replication. Compound 1 was selected as a clinical candidate for further evaluation in humans.
Acknowledgements
The authors would like to thank Holly McArthur, Haolun Jin, Maria Fardis, Hongyan Guo, Christopher Lee, Leah Tong, Edward Doerffler, James Tario, Gina Bahador, Xiaohong Liu, Martin McDermott, Azar Dastgah, Kathryn Cantrell and Carina Canizzaro for their contributions.References
- F. J. Palela, Jr, K. M. Delaney, A. C. Morrman, M. O. Loveless, J. Fuhrer and G. A. Satten, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV outpatient study investigators, N. Engl. J. Med., 1998, 338, 853–60 CrossRef.
- C. Flexner, HIV protease inhibitors, N. Engl. J. Med., 1998, 338, 1281–1292 CrossRef CAS.
- A. M. J. Wensing, N. M. van Maarseveen and M. Nijhuis, Fifteen years of HIV protease inhibitors: raising the barrier to resistance, Antiviral Res., 2010, 85, 59–74 CrossRef CAS.
- G. Moyle, Principle and practice of HIV-protease inhibitor pharmacoenhancement, AIDS Med., 2001, 2, 105–113 CAS.
- J. G. Gerber, Using pharmacokinetics to optimize antiretroviral drug-drug interactions in the treatment of human immunodeficiency virus infection, Clin. Infect. Dis., 2000, 30 (Suppl 2), S123–9 CrossRef CAS and references therein.
- C. L. Cooper, R. P. G. van Heeswijk, K. Gallicano and D. W. Cameron, A review of low-dose ritonavir in protease inhibitor combination therapy, Clin. Infect. Dis., 2003, 36, 1585–1592 CrossRef CAS.
- M. Barry, F. Mulcahy, C. Merry, S. Gibbons and D. Back, Pharmacokinetics and potential interactions amongst antiretroviral agents used to treat patients with HIV infection, Clin. Pharmacokinet., 1999, 36, 289–304 CrossRef CAS and references therein.
- M. Youle, Overview of boosted protease inhibitors in treatment-experienced HIV-infected patients, J. Antimicrob. Chemother., 2007, 60, 1195–1205 CrossRef CAS and references therein.
- K. H. Busse and S. R. Penzak, Pharmacological enhancement of protease inhibitor with ritonavir: an update, Expert Rev. Clin. Pharmacol., 2008, 1, 533–545 CrossRef CAS and references therein.
- L. Xu and M. C. Desai, Pharmacokinetic enhancers for HIV drugs, Curr. Opinion in Investig. Drugs, 2009, 10, 775–786 CAS.
- S. M. Hammer, J. J. Eron, P. Reiss, R. T. Schooley, M. A. Thompson, S. Walmsley, P. Cahn, M. A. Fischl, J. M. Gatell, M. S. Hirsh, D. M. Jacobsen, J. S. G. Montaner, D. D. Richman, P. G. Yeni and P. A. Volberding, Antiretroviral treatment of adult HIV infection: 2008 recommendations of the international AIDS society USA panel, J. Am. Med. Assoc., 2008, 300, 555–70 CrossRef CAS and references therein.
- Panel on Antiretroviral Guidelines for Adults and Adolescent, Department of Health and Human Services (DHHS). Guidelines for the use of antiretroviral agents in HIV-1 infected adults and adolescents. ( January 10, 2011) 1–174 and references therein. Available at http://aidsinfo.nih.gov/contentfiles/AdultandAdolescentGL.pdf Search PubMed.
- D. L. N. G. Surleraux, A. Tahri, W. G. Verschueren, G. M. E. Pille, H. A. de Kock, T. H. M. Jonckers, A. Peeters, S. De Meyer, A. Azijin, R. Pauwels, M.-P. de Bethune, N. M. King, M. Prabu-Jeyabalan, C. A. Schiffer and P. B. T. P. Wigerinck, Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor, J. Med. Chem., 2005, 48, 1813–1822 CrossRef CAS.
- A. Molla, S. Vasavanonda, G. Kumar, H. L. Sham, M. Johnson, B. Grabowski, J. F. Denissen, W. Kohlbrenner, J. J. Plattner, J. M. Leonard, D. W. Norbeck and D. J. Kempf, Human serum attenuates the activity of protease inhibitors toward wild-type and mutant huma immunodeficiency virus, Virology, 1998, 250, 255 CrossRef CAS.
- A. K. Ghosh and Y. Chen, Synthesis and optical resolution of high affinity P2-ligand for HIV-1 protease inhibitors, Tetrahedron Lett., 1995, 36, 505–508 CrossRef CAS.
- C. Callebaut, K. Stray, L. Tsai, M. Williams, Z.-Y. Yang, C. Cannizzaro, S. Leavitt, X. Liu, K. Wang, B. Murray, A. Mulato, M. Hatada, T. Priskich, N. Parkin, S. Swaminathan, W. A. Lee, G.-X. He, L. Xu and T. Cihlar, In vitro characterization of GS-8374, a novel phosphonate-containing inhibitor of HIV-1 protease with a favorable resistance profile, AAC, 2011, 55, 1366–1376 Search PubMed.
- T. Cihlar, G.-X. He, X. Liu, J. M. Chen, M. Hatada, S. Swaminathan, M. J. McDermott, Z.-Y. Yang, A. S. Mulato, X. Chen, S. A. Leavitt, K. M. Stray and W. A. Lee, Suppression of HIV-1 protease inhibitor resistance by phosphonate-mediated solvent anchoring, J. Mol. Biol., 2006, 363, 635–647 CrossRef CAS.
- C. Callebaut; K. M. Stray; L. K. Tsai; L. Xu; W. A. Lee; T. Cihlar. In vitro HIV resistance selection to GS-8374, a novel phosphonate protease inhibitor: comparison with atazanavir, lopinavir and darunavir. 16th International HIV Drug Resistance Workshop 2007 Search PubMed.
- A. Carrillo, K. D. Stewart, H. L. Sham, D. W. Norbeck, W. E. Kohlbrenner, J. M. Leonard, D. J. Kempt and A. Molla, In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor, J Virology, 1998, 72, 7532–7541 CAS.
- Y.-F. Gong, B. S. Robinson, R. E. Rose, C. Deminie, T. P. Spicer, D. Stock, R. J. Colonno and P.-F. Lin, In vitro resistance profile of the human immunodeficiency viruse type 1 protease inhibitor BMS-232632, AAC, 2000, 44, 2319–2326 CAS.
- S. De Meyer, H. Azijn, D. Surleraux, D. Jochmans, A. Tahri, R. Pauwels, P. Wigerinck and M.-P. de Béthune, TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses including a broad range of clinical isolates, AAC, 2005, 49, 2314–2321 CAS.
- L. Xu, H. Liu, B. P. Murray, C. Callebaut, M. S. Lee, A. Hong, R. G. Strickley, L. K. Tsai, K. M. Stray, Y. Wang, G. R. Rhodes and M. C. Desai, Cobicistat (GS-9350): a potent and selective inhibitor of human CYP3A as a novel pharmacoenhancer, ACS Med. Chem. Lett., 2010, 1, 209–213 CrossRef CAS.
- A. Mathias, P. German, B. P. Murray, L. Wei, A. Jain, S. West, D. Warren, J. Hui and B. P. Kearney, Pharmacokinetics and pharmacodynamics of GS-9350: a novel pharmacokinetic enhancer without anti-HIV activity, Clin. Pharmacol. Ther., 2010, 86, 322–329 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Biological assays, preclinical pharmacokinetics, experimental procedures and analytical data for all compounds. See DOI: 10.1039/c1md00147g |
| ‡ These authors are no longer with Gilead Sciences. |
| This journal is © The Royal Society of Chemistry 2011 |

