DOI:
10.1039/C1MD00095K
(Concise Article)
Med. Chem. Commun., 2011,
2, 735-742
New analyses of MIC90 data to aid antibacterial drug discovery†
Received
31st March 2011
, Accepted 18th May 2011
First published on 10th June 2011
Abstract
In this work we present a number of statistical and visualization methods derived from MIC90 data designed to aid decision-making in antibacterial drug discovery research. A statistical method known as bootstrapping was applied to MIC90 raw data to uncover data trends and a metric termed Net Percent Superior (NPS) was developed to capture a strain-by-strain analysis of analogs to enable rank-ordering of similar compounds. We also present novel methods of reporting the data using a variety of visualization techniques. Furthermore, the work was cross-validated using experimental results generated with siderophore-conjugated monocarbam analogs to demonstrate the effectiveness of the various parameters and visualization techniques. The methods reported herein have been incorporated in a Scitegic Pipeline Pilot protocol to enable facile, automated generation of MIC90 analyses from experimental raw data to aid prospective medicinal chemistry design as well as retrospective analyses.
Introduction
New antibiotics are desperately needed to treat the increasing number of life-threatening bacterial infections that are resistant to current therapies. In particular, the emergence and spread of drug-resistant ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) have been highlighted as areas of particular medical concern as these bacteria have the ability to “escape” the effects of current antimicrobial agents.1 Historically, antibacterial medicinal chemistry programs have relied heavily on MIC data to drive analog design.2 Typically, a panel of bacterial strains is assembled and utilized as a first-tier screen. Secondary evaluation of promising analogs typically includes MIC90 determination,3 which may be a staged process wherein initial MIC90 panels of small to moderate size are assembled (approximately 10–50 strains) for each bacterial species of interest with the most promising analogs proceeding to larger MIC90 panels containing perhaps 100 or more strains per species. In each scenario, an attempt is made to assemble panels containing clinically relevant strains which span a broad susceptibility range with respect to current therapies and include some strains with known resistance phenotypes. For example, inclusion of strains known to express the various classes of β-lactamase or aminoglycoside modifying enzymes is standard practice for programs focused on those chemotypes.4 Ideally, performance in the initial MIC panel would be predictive of performance in the follow-on MIC90 panels thus enabling efficient design cycles for the medicinal chemist. In the following discussion, we present new statistical and data visualization methods derived from MIC90 data designed to enhance learning and improve decision-making.
Results and discussion
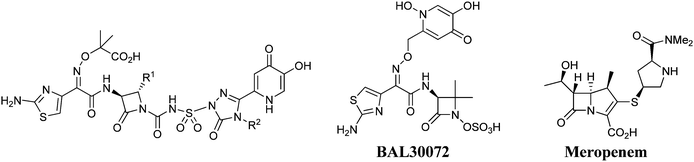
The MIC90 value is heavily utilized by antibacterial drug research teams in both decision-making around compound advancement as well as in generating predicted human doses. By convention, the MIC90 value (as well as MIC and MIC50) is a distinct number falling within a predefined drug concentration range which is expressed in two-fold increments (e.g. 0.06, 0.125, 0.25, 0.5, 1,…, 64 μg mL−1). While the MIC90 value is an important parameter, the individual MIC values which comprise the MIC90 experiment can potentially provide a wealth of information, and we have attempted to capture this knowledge in a number of heatmap views. The data described herein are derived from an MIC90 study conducted with a series of recently disclosed siderophore-conjugated monocarbam analogs (1–5)5 as well as the siderophore-conjugated monosulfactam BAL300726 and the commercial antibacterial agent Meropenemvs. 51 clinically relevant Pseudomonas aeruginosa strains (Table 1).7
Table 1 Structures and data summary
| Compound |
R
1
|
R
2
|
MIC90a (n = 51) |
BMIC90b |
BMIC90 95% CIc |
NPSd |
NPS 95% CIe |
GMf |
GM 95% CIg |
|
The drug concentration which inhibits the visible growth of ≥90% of the bacterial strains within a test population.
Bootstrap MIC90.
95% confidence interval for BMIC90.
Net Percent Superior.
95% confidence interval for BMIC90.
Geometric mean.
95% confidence interval for geometric mean.
Published MIC90 data (n = 206).6b
|
|
1 (MC-1) |
–H |
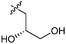
|
1 |
0.95 |
(0.5, 8) |
0 |
— |
0.27 |
(−0.45, 1.00) |
|
2
|
–Me |

|
0.5 |
0.52 |
(0.5, 0.5) |
70.6 |
(55, 86) |
0.14 |
(−0.03, 0.31) |
|
3
|
–Me |

|
0.5 |
0.50 |
(0.25, 1) |
19.6 |
(2, 37) |
0.21 |
(0.06, 0.37) |
|
4
|
–Me |

|
1 |
0.79 |
(0.5, 2) |
2 |
(−16, 20) |
0.27 |
(−0.35, 0.89) |
|
5
|
–Me |
–Me |
1 |
1.11 |
(1, 4) |
−54.9 |
(−73, −35) |
0.39 |
(−0.05, 0.82) |
| BAL30072 |
— |
— |
4 (8)h |
3.02 |
(1, 8) |
−82.4 |
(−92, −71) |
0.86 |
(0.14, 1.58) |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeropenem
|
— |
— |
>64 |
59.5 |
(32, 64) |
−72.5 |
(−90, −53) |
2.85 |
(−3.75, 9.44) |
The “Standard Heatmap” view of these data was created (Table 2) wherein the activity of each compound vs. each strain is color-coded according to a rough estimate of the activity required to kill the pathogen in a clinical setting (MIC ≤ 0.25: dark green, MIC = 0.5–1: light green, MIC = 2: yellow, MIC = 4–8: orange and MIC ≥ 16: red). The table is sorted left to right based on the MIC90 value with most potent compounds appearing on the left. The table is also sorted vertically based on the Meropenem data. From this view, one can immediately appreciate the exceptional level of potency the monocarbam analogs generally achieve vs. these clinically relevant strains. It also highlights those few strains which demonstrate a moderate level of resistance to the chemotype (1766-00, PA-1757, 1509-08, 1510-08) and the effectiveness of the monocarbam analogs vs. the competitor agent BAL30072. This view also illustrates the superior performance of the monocarbam analogs vs. a range of strains highly resistant to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeropenem. Bacteria have developed effective mechanisms of resistance to many β-lactam drugs, including expression of a wide variety of β-lactamases, expression of efflux pumps, downregulation of porin channels and active site mutations of the penicillin binding proteins, which are the biological targets of β-lactam antibacterial agents.8 One can appreciate the additional value this kind of heatmap view could provide if the bacterial strains utilized in an MIC90 study were characterized with regard to their resistance phenotypes as this would enable one to potentially correlate potency (or the lack thereof) with the presence of one or more mechanisms of resistance. This heatmap view could also be useful when conducting retrospective analyses on large datasets.
Table 2 Standard heatmap view
One limitation of the standard heatmap view is that it does not effectively highlight relative potency differences between similar compounds and, therefore, does not enhance the ability to rank-order closely related analogs, especially those with equivalent MIC90 values. One needs to appreciate that MIC90 experiments are rather labor intensive, therefore, decisions around compound advancement are often driven by learnings derived from a single experiment. Given the limited sample size of an intermediate MIC90 panel like this, coupled with the inherent error associated with individual MIC values (approximately +/−2-fold), decisions based on these data can be somewhat speculative. Oftentimes, inspection of MIC90 raw data suggests the potential for the MIC90 value to change if the experiments were repeated. This tendency towards moving upward or downward can be important to recognize and utilize in decision-making and can be visualized in Table 3. Here, the MIC data in each column have been independently sorted lowest to highest value and the row representing the MIC90 value is highlighted in purple. The orange shaded area is provided to illustrate whether the MIC90 value is “trending” toward a higher or lower value. Ideally, the MIC90 value would fall roughly in the middle of the orange shaded area as this would provide confidence that if the experiments were repeated, minor changes in activity throughout the panel would not lead to a change in the MIC90 value. The closer the MIC90 value is to the upper or lower boundary of the orange shaded area, the greater the likelihood that the MIC90 could change if the experiments were repeated. Based on this discussion, one might surmise that compounds 1, 4 and BAL30072 are leaning toward a lower MIC90 value, compound 2 is leaning toward a higher value and the MIC90s for compounds 3, 5 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeropenem appear fairly solid. Medicinal chemists conducting antibacterial research are accustomed to looking for these tendencies, and the ability to quantify them as numerical values would be useful.
Table 3 Standard MIC90 view.a
|
Each row is independently sorted, therefore, the association of individual MIC values with particular bacterial strains is not maintained in this view.
Bootstrap MIC90.
|

|
To capture this MIC90 tendency, we introduce a new parameter based on the use of a statistical methodology called the bootstrap.9 In general, the bootstrap method is a re-sampling technique that uses many alternate versions of the data, called bootstrap samples, to estimate the statistical distribution of the MIC90. A bootstrap sample is a sample taken with replacement, meaning that after a species is selected to go into the bootstrap sample, it can be selected again. The size of the bootstrap sample is the same as the original dataset (51 data points in this case), and will likely contain duplicate values of some strains due to the re-sampling process. A large number of bootstrap samples are taken (1,000,000 times for this analysis) and, for each, the MIC90 value is computed. These computed MIC90s constitute the “bootstrap distribution” for the statistic. Statistical theory allows the use of this distribution to estimate statistical summaries of the MIC90, such as the average or confidence intervals. The bootstrap estimate of the MIC90, denoted as the BMIC90 (Fig. 1), is the simple average from the bootstrap distribution. Since the MIC90 is defined to be a dilution within a series, the bootstrap method for defining confidence intervals results in highly conservative (i.e. wide) intervals and is not useful with regard to rank-ordering compounds (Table 1). However, other procedures can be used to determine if there are statistically meaningful differences in MIC90 values between compounds. A permutation test9 was used to estimate the p-value for the hypothesis that the MIC90 values are the same for two compounds. If the p-value is small, this would indicate that there is evidence that the values are not equal. However, when testing all combinations of compounds, there is a concern that multiple statistical tests will overly inflate the confidence intervals. To compensate for this, the raw p-values were adjusted to control the false positive rate (also known as the false discovery rate or FDR).10 While this procedure is more conservative than no adjustment, it is more liberal than classical p-value correction procedures that protect against any false positive results. A corrected p-value less than 0.05 suggests that a statistically meaningful difference in MIC90 value exists. Table 4 shows the FDR values for each pair-wise comparison and suggests that the MIC90 values for all compounds in this set are statistically different, with the exception of compound 1 being indistinguishable from compound 4. Strictly speaking, the FDR values alone do not enable rank-ordering of analogs, but can be used in conjunction with other parameters as will be discussed later.
 |
| | Fig. 1 Bootstrap method. | |
Table 4
FDR values
|
FDR (adjusted p-values from pair-wise analysis) for MIC90 values |
| |
1
|
2
|
3
|
4
|
5
|
BAL30072 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeropenem
|
|
1 and 4 are not different based on p-value.
|
|
1
|
— |
0.0000 |
0.0414 |
0.4917a |
0.0002 |
0.0000 |
0.0000 |
|
2
|
|
— |
0.0035 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
|
3
|
|
— |
0.0414 |
0.0000 |
0.0000 |
0.0000 |
|
4
|
|
— |
0.0004 |
0.0000 |
0.0000 |
|
5
|
|
— |
0.0000 |
0.0000 |
|
BAL30072
|
|
— |
0.0001 |
The “Finland-o-gram” is occasionally utilized to visualize the differences between similarly active compounds.11 This involves plotting the cumulative % inhibition of susceptible strains vs. the MIC values (Fig. 2). The curves for the most potent analogs are left-shifted relative to less potent analogs. To further aid in visualizing differences between closely related analogs the “Relative Heatmap” view was designed (Table 5). In this view, the activity of a set of new analogs is compared to the activity of a related reference compound in a strain-by-strain analysis. The color in each cell is determined based on how well the new compound performed vs. a particular bacterial strain relative to the activity of the reference compound (1 in this example).12 The compounds in the table are arranged left to right based on MIC90 potency and the data are sorted vertically based on the individual MIC values for the reference compound. In this view, it is readily apparent that most of the compounds are not superior to the reference compound 1 with compounds 2 and 3 being the possible exceptions. This is not surprising given that the MIC90 values for 2 and 3 are superior to 1 (0.5 μg mL−1 and 0.5 μg mL−1vs. 1 μg mL−1). This view clearly demonstrates that the superiority of 2 relative to 1 is consistently observed across the entire panel of strains, including the toughest to treat strains which are least susceptible to 1 (magenta shaded strains in Table 5). The superiority of 3 to 1 is somewhat less consistent over the entire panel, but again is demonstrated consistently vs. those strains least susceptible to reference compound 1. A more interesting comparison would be compounds 1 and 5 as these have equivalent MIC90 values (1 μg mL−1), yet the Relative Heatmap view clearly suggests that 5 is inferior to 1. Just as the BMIC90 parameter was developed to capture MIC90 data trends, a new parameter termed the “Net Percent Superior” or NPS was developed to capture trends observed in the Relative Heatmap view. This parameter is calculated by subtracting the percentage of strains with activity inferior to the reference compound from the percentage of strains with activity superior to the reference compound. The reference compound will obviously have an NPS value equal to 0; analogs with NPS values >0 are defined as potentially being superior to the reference compound and those with an NPS value <0 are potentially inferior. Since these values have higher resolution than the MIC90 values, the bootstrap procedure can be used here to generate meaningful confidence intervals (Table 1 and Fig. 3). The weakness of this parameter as calculated is that it does not account for the magnitude of activity differences between the reference compound and the analogs; a 2-fold difference in activity is treated the same as a 16-fold difference. As a result, this parameter does not effectively rank-order compounds that consistently demonstrate activity different from the reference compound on a strain-by-strain basis. For example, while compound 5 is clearly superior to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeropenem based on MIC90 values (1 μg mL−1vs. >64 μg mL−1), the NPS values (−54.9 vs. −72.5) and the corresponding confidence intervals (Fig. 3) do not reflect this superiority. That said, the goal of this work was to develop methods to identify and rank-order the most promising analogs in a series with similar activity. The NPS parameter clearly identifies compounds 1–4 as being superior to 5, BAL30072 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeropenem and can contribute to rank-ordering as discussed below.
 |
| | Fig. 2 Finland-o-gram. | |
Table 5 Relative heatmap view
 |
| | Fig. 3 95% confidence intervals for Net Percent Superior (NPS) values. | |
There are a number of examples in the literature describing statistical analyses of MIC90 raw data (arithmetic mean, geometric mean, median, mode, etc.) to enable rank-ordering compounds with the use of the geometric mean being somewhat common.13 Therefore, we have also included the geometric mean in our analysis for comparison (Table 1). As was the case with the BMIC90 parameter, confidence intervals for the geometric mean values are also quite wide and not useful with regard to rank-ordering analogs. As alluded to above, antibacterial research teams often make decisions around compound progression based on MIC90 data. Rank-ordering this set of analogs based solely on the MIC90 data would provide the following: 2 = 3 > 1 = 4 = 5 > BAL30072 > COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeropenem. By using the parameters described above, we can attempt to provide a greater degree of separation. Recall that the FDR values strongly suggest that MIC90 values for all compounds are statistically different with 1 and 4 being the exceptions. With this in mind, recognizing the ranking trends observed for the BMIC90, NPS and geometric mean values could lead to the following rank-order: 2 > 3 > 1 ≈ 4 > 5 > BAL30072 > COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeropenem. Obviously, given the wide confidence intervals associated with the BMIC90 and geometric mean values, this ranking cannot be stated with 100% confidence. Drug research teams regularly make decisions with regard to compound advancement based on a variety of information (potency, pharmacokinetics, safety, stability, ease of synthesis, etc.). The methods described above were designed to illustrate potential trends in potency data for single experiments; there is an inherent risk that the analysis could occasionally mislead. That said, the reality of antibacterial drug research involves teams making decisions based on MIC90 data, and while the methods described herein do not enable decisions with 100% confidence, we are hopeful that they may lead to better informed, higher quality decisions.
Conclusion
The MIC90 experiment provides a rich set of raw data which can be utilized in a variety of ways to enhance learnings. Here, we introduce two new parameters, the BMIC90 and NPS, to aid in understanding MIC90 tendencies and rank-ordering of analogs. Two heatmap data visualizations are introduced as well, and the entire analysis has been automated via a Pipeline Pilot protocol (SciTegic Pipeline Pilot version 7.5.2; Accelrys, San Diego, CA) to enable rapid generation of MIC90 data summaries for ongoing Antibacterial drug research programs.
Acknowledgements
The authors wish to thank Michael Huband, Lisa Mullins and Joseph Penzien for generating the MIC90 data, Jianmin Sun for preparing the new analogs disclosed herein and the following individuals for providing thoughtful input: Dr Jeremy Starr, Dr Thomas Magee, Dr Mark Noe, Dr Mark Plummer and Dr Veerabahu Shanmugasundaram. This research was sponsored by Pfizer, Inc.
Notes and references
- H. W. Boucher, G. H. Talbot and J. S. Bradley, , et al., Clin. Infect. Dis., 2009, 48, 1–12 CrossRef.
- The MIC (Minimum Inhibitory Concentration) is the lowest concentration of drug that inhibits visible growth of bacteria following overnight incubation. In order to determine the MIC value, a range of drug concentrations (e.g. 0.06 μg mL−1 to 64 μg mL−1) are incubated with a defined strain of bacteria. Typically, the drug concentration range is broken down into 2-fold increments and the various drug concentrations are all individually incubated overnight with approximately the same number of bacteria. The MIC is then determined by visually inspecting the drug effect at each concentration, and the MIC is the lowest drug concentration that inhibits bacterial growth. Typically, bacteria continue to grow at drug concentrations lower than the MIC and do not grow at concentrations at or above the MIC.
- The MIC90 is the drug concentration which inhibits the visible growth of ≥90% of the bacterial strains within a test population.
- K. Bush and G. H. Miller, Curr. Opin. Microbiol., 1998, 1, 509–515 Search PubMed.
- Compounds 1–3 were prepared as described in: M. E. Flanagan and S. J. Brickner, , et al., ACS Med. Chem. Lett., 2011, 2, 385–390 Search PubMed , for the preparation of compounds 4–5, see ESI†.
-
(a)
BAL30072 was prepared as described in: WO2008/116813;;
(b) M. G. P. Page, C. Dantier and E. Desarbre, Antimicrob. Agents Chemother., 2010, 54, 2291–2302 Search PubMed;
(c) S. Mushtaq, M. Warner and D. Livermore, J. Antimicrob. Chemother., 2010, 65, 266–270 Search PubMed.
- MIC testing was conducted according to the Clinical and Laboratory Standards Institute (CLSI, formerly NCCLS) guidelines. See: Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard-Eighth Edition. CLSI document M7–A8 [ISBN20 1-56238-689-1]. Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898, USA, 2006.
- S. B. Levy and B. Marshall, Nat. Med., 2004, 10, S122–S129 CrossRef CAS.
-
B. Efron and R. J. Tibshirani, An Introduction to the Bootstrap, Chapman and Hall, New York, 1993 Search PubMed.
- Y. Benjamini and Y. Hochberg, J. Roy. Stat. Soc. B Stat. Meth., 1995, 57, 289–300 Search PubMed.
- S. G. Sackel, S. Alpert, B. Rosner, W. M. McCormack and M. Finland, Antimicrob. Agents Chemother., 1977, 12, 31–36 Search PubMed.
- Color key: dark green: ≥4× better than reference, light green: 2× better than reference, white: equal to reference, yellow: 2× worse than reference, orange: 4× worse than reference, and red: ≥8× worse than reference.
-
(a) K. E. Aldridge and W. D. Johnson, J. Antimicrob. Chemother., 1997, 39, 319–324 Search PubMed;
(b) R. L. White, M. B. Kays, L. V. Friedrich and V. E. Del Bene, J. Antimicrob. Chemother., 1993, 31, 57–64 Search PubMed;
(c) R. P. Kowalski and K. A. Yates, , et al., Ophthamology, 2005, 112, 1987–1991 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c1md00095k |
|
| This journal is © The Royal Society of Chemistry 2011 |
Click here to see how this site uses Cookies. View our privacy policy here.