Structure–function study of gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097†‡
Tiphaine
Huet§
a,
Hubert
Maehr¶
b,
Hong Jin
Lee
b,
Milan R.
Uskokovic
b,
Nanjoo
Suh
b,
Dino
Moras
a and
Natacha
Rochel
*a
aDépartement de Biologie et de Génomique Structurales, IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Centre National de la Recherche Scientifique, Institut National de la Santé de la Recherche Médicale, Université de Strasbourg, 1 rue Laurent Fries, 67404, Illkirch, France. E-mail: rochel@igbmc.fr
bDepartment of Chemical Biology, Ernest Mario School of Pharmacy, Rutgers, The State University of New Jersey, 164 Frelinghuysen Road, Piscataway, NJ 0885, USA
First published on 17th March 2011
Abstract
Derivatives of vitamin D3 containing a second side-chain emanating at C-20 are known as gemini and act as vitamin D receptor agonists. Recently, two of these, namely Gemini-0072 and the epimeric Gemini-0097, were selected for further studies in view of their high biological activities and lack of hypercalcemic effects. We now show that the two analogs recruit coactivator SRC-1 better than the parental gemini and act as VDR superagonists. The crystal structures of complexes of zVDR with Gemini-0072 and Gemini-0097 indicate that these ligands induce an extra cavity within the ligand-binding pocket similar to gemini and that their superagonistic activity is due to an increased stabilization of helix H12.
Introduction
1α,25-dihydroxyvitamin D3 [1α,25(OH)2D3], the active form of vitamin D, regulates complex gene networks involved in bone development and metabolism, calcium homeostasis, growth-inhibitory, prodifferentiating, and immunomodulatory activities. Its genomic actions are mediated through the Vitamin D Nuclear Receptor (VDR).1–3 Therapeutic applications of 1α,25(OH)2D3, which encompass treatments for renal osteodystrophy, osteoporosis, psoriasis, cancer, autoimmune diseases and prevention of graft rejection, are limited by its intrinsic hypercalcemic effect causing hypercalcemia, increasing bone resorption, and soft tissue calcification. Therefore, VDR ligands with dissociated tissue-selective/cell-context-dependent profiles have been developed.4 Many analogs of 1α,25(OH)2D3 were synthesized with the goal to increase physiological potency and specificity. As a result of these efforts, derivatives of 1α,25(OH)2D3 were created wherein the C-21 methyl group was extended to form a second side-chain. These compounds are known as gemini (Fig. 1). The first example of this class of compounds features two identical side chains, characteristic for 1α,25(OH)2D3, and is being referred herein as the parental gemini. It binds the VDR ligand binding pocket (LBP) and activates gene transcription.5,6 In the presence of an excess of corepressor, the VDR–gemini complex shifts from an agonist to an inverse agonist conformation with the recruitment of N-CoR (Nuclear Receptor Co-Repressor) and mediates repression.6 New gemini derivatives were subsequently synthesized with chemical modifications designed to enhance their biological activity by increasing their resistance toward metabolic degradation.7–9 These modifications include a 19-nor A-ring and two different side chains, one side chain similar to the natural one wherein the geminal methyl groups are replaced by trideuteromethyls, and the second side chain with trifluoromethyl groups and C-23 unsaturation. These chemical features have been shown to prevent or delay biological degradation initiated by 24-hydroxylation.8,10 Deuteration of the geminal methyl groups also extends the half-life10,11 and was expected to stabilize the interactions within the VDR complex. The two C-20 epimeric Gemini-0072 and Gemini-0097 (Fig. 1) have been shown to be more efficient in reducing tumor growth than the non-deuterated analogs.12,13 This increased potency was also observed in their induction of human leukemia cell differentiation or human breast cancer MCF10 cell proliferation.9 Furthermore, Gemini-0072 and Gemini-0097 prevent estrogen-receptor positive and negative mammary tumorigenesis with comparable potencies in vivo without hypercalcemic toxicity13 and suppressed mammary tumor growth in the ErbB2-overexpressing transgenic mice.14 | ||
| Fig. 1 Chemical structures of 1α,25(OH)2D3, gemini, Gemini-0072 (C20S) and Gemini-0097 (C20R). | ||
We previously reported crystal structures of the VDR ligand-binding domain (LBD) in complexes with 1α,25(OH)2D3, and with synthetic agonists, and have shown that all compounds are anchored to the same residues in the LBP with the hydroxyl groups of the A-ring and of the side chain; therefore, they are locked in identical positions and form the same hydrogen bonds.15,16 Our previous structure of the gemini–VDR complex revealed a ligand-dependent structural rearrangement of the protein core, thus opening a channel that accommodates the second side chain while preserving the essential agonist features of the 1α,25(OH)2D3 bound LBD.17
The present study gains insights into the structure–activity relationships of the two epimeric Gemini-0072 and Gemini-0097. The biological assays revealed that these two ligands are more active than the parental gemini and nearly equipotent. The crystal structures of these two compounds bound to zVDR LBD explain their superagonist activity. In addition to the therapeutic interest, our study helps to clarify the functional behavior of these molecules.
Results and discussion
Gemini-0072 and Gemini-0097 act as VDR superagonists
Gemini-0072 and Gemini-0097 have been characterized as potent inhibitors of mammary tumors and inducers of leukemia cell differentiation.13 We have now investigated the transactivation potency of the VDR in the presence of these two ligands in MCF-7 cells. Previous studies with gene reporter assays have shown that vitamin D superagonists, including KH1060, have the potency to stimulate transcription at 10 to 100-fold lower than the natural ligand.18 Using reporter gene assays, we now show that Gemini-0072 or Gemini-0097 are 1α,25(OH)2D3 superagonists as they are more potent in directing transactivation than 1α,25(OH)2D3 by a factor of 36 and 22-fold higher, respectively (EC50 for 1α,25(OH)2D3 = 5.5 ± 1.5 nM, EC50 for Gemini-0072 = 0.15 ± 0.1 nM, EC50 for Gemini-0097 = 0.25 ± 0.1 nM) (Fig. 2 and Fig. S1, ESI‡). Compared to the parental gemini, Gemini-0072 and Gemini-0097 are 8 and 5-fold more active, respectively (EC50 for gemini = 1.2 ± 0.4 nM). Furthermore, the comparison between the dose-dependent profiles, obtained in the presence of Gemini-0072 or Gemini-0097, shows no difference between the two compounds in transactivation potency. We found that the C-20 configuration in gemini derivatives does not influence the VDR-directed transcription under our experimental conditions. | ||
| Fig. 2 Gemini-0072 and Gemini-0097 are highly potent VDR superagonists in vitro. Transient transfections and luciferase reporter gene assays using MCF-7 human breast cancer cells were performed as reported in the experimental procedures. Relative luciferase activities were calculated by dividing the luciferase activity by the respective β-galactosidase activity to correct for differences in transfection efficiency. For every triplicate the mean and S.D. were calculated. | ||
Gemini-0072 and Gemini-0097 are more potent inhibitors of MCF10 cell growth than 1α,25(OH)2D3
We determined the growth inhibitory effects of Gemini-0072 and Gemini-0097 on the cell proliferation of three of the MCF10 series of human breast cancer cells, namely estrogen receptor (ER)-positive mammary precancerous MCF10AT1, malignant metastatic MCF10CA1a and the MCF10DCIS.com xenograft model of ER-negative mammary tumors. Both Gemini-0072 and Gemini-0097 are superior inhibitors of cell growth when compared to the naturally occurring 1α,25(OH)2D3. Both Gemini-0072 and Gemini-0097 showed subnanomolar ranges of IC50 values in the growth inhibition of human breast cancer cells, which were 10-fold or better than that of 1α,25(OH)2D3 in the three cell lines (Table 1 and Fig. S2, ESI‡).| Compounds | IC50 (nM) MCF10AT1 | IC50 (nM) MCF10CA1a | IC50 (nM) MCF10DCIS.com |
|---|---|---|---|
| 1α,25(OH)2D3 | 3.9 ± 2.1 | 6.2 ± 2.5 | 1.75 ± 0.57 |
| Gemini-0072 | 0.23 ± 0.11 | 0.09 ± 0.07 | 0.23 ± 0.02 |
| Gemini-0097 | 0.51 ± 0.25 | 0.15 ± 0.06 | 0.36 ± 0.07 |
Gemini-0072 and Gemini-0097 increase the binding affinity of the SRC-1 peptide for zVDR compared to the parental gemini
The human transcriptional coactivator SRC-1 interacts with VDR.19 We monitored the induced recruitment of a fluorescently labeled SRC-1 peptide bearing the second LXXLL motif to the zVDR LBD in the presence of increasing concentrations of Gemini-0072, Gemini-0097 or the parental Gemini by fluorescence anisotropy. Our results show a significant increase in the binding constant (kd = 1.8 ± 0.07 μM and 1.8 ± 0.08 μM) of the SRC-1 NR2 peptide for zVDR in the presence of Gemini-0072 and Gemini-0097 compared to the parental gemini (kd = 3.3 ± 0.2 μM). In agreement with the results of the transcription assays, the modifications introduced in the two analogs are a significant parameter of stabilization for the recruitment of coactivator.Overall structures of the VDR-LBD bound to Gemini-0072 and Gemini-0097
To gain structural insights into the molecular mechanism underlying the superagonistic property of Gemini-0072 and Gemini-0097, we solved the crystal structures of these two ligands with the zebrafish zVDR LBD and a GRIP-1/TIF-2 coactivator peptide containing the second LXXLL motif recognized by the nuclear receptor. Crystals of wild-type zVDR LBD, bound to Gemini-0072 or Gemini-0097 and GRIP-1 peptide, were obtained under similar conditions as for zVDR LBD–gemini.17 We have previously demonstrated that zVDR LBD and human VDR LBD bound to the natural hormone adopt the canonical agonist conformation,17 validating zVDR as an appropriate model for VDR structural studies.Structures of zVDR LBD in complex with Gemini-0072 or Gemini-0097 were refined at a resolution of 2.4 Å and 2.5 Å, respectively (see statistics in Table S1, ESI‡). Both complexes adopt the canonical, active conformation, characteristic of all previously reported agonist-bound nuclear receptor LBDs. Like zVDR LBD–1α,25(OH)2D3 and zVDR LBD–gemini complexes, the flexible region between H2 and H3 was not visible in the electron density map, which reflects its disorder. Furthermore, the position and conformation of the activation helix 12 are strictly conserved. After refinement of the proteinzVDR LBD alone, maps revealed an unambiguous electron density to fit the ligand (Fig. 3A). In both complexes, the ligands are buried in the predominantly hydrophobic pocket and assume the same orientation as the parental gemini. The side-chain containing the trifluoromethyl groups adopts the orientation of the natural chain of 1α,25(OH)2D3 (Fig. 3); the trideuteromethyl containing side arm occupies the additional pocket, created by the reorientation of the side chain at Leu337, similar to the zVDR LBD–gemini complex.16 Compared with the structure of the zVDR LBD–gemini complex, the atomic coordinates of all Cα atoms of zVDR bound to Gemini-0072 and to Gemini-0097 show root mean square deviations of 0.21 Å and 0.38 Å, respectively. The volumes of the two epimeric Gemini-0072 and Gemini-0097 are 474 Å3 each (461 Å3 for the parental gemini). The volumes of the ligand binding cavities are 776, 757, and 729 Å3 and the ligands occupy 61, 63, and 63% of the pocket in zVDR LBD–Gemini-0072, –Gemini-0097, and –gemini complexes, respectively. Regarding the interface VDR/coactivator peptide, we observed that contacts between protein residues and the coactivator peptide are conserved in both complexes.
 | ||
| Fig. 3 Conformations and interactions of the bound ligands in the LBP of zVDR at a 4.0 Å cut-off. (A) The Gemini-0072 (left) and Gemini-0097 (right) are shown in their Fo – Fc electron density omit map contoured at 3σ. Ligands are shown in stick representation with carbon, oxygen and fluorine atoms in yellow, red and grey, respectively. (B) Superposition of Gemini-0072 (magenta), Gemini-0097 (orange) and 1α,25(OH)2D3 (grey). (C) zVDR LBD–Gemini-0072 and (D) zVDR LBD–Gemini-0097 are shown in magenta and orange, respectively. The gemini ligands are represented in stick with carbon, oxygen and fluorine atoms in yellow, red and light blue, respectively. Superimposition with 1α,25(OH)2D3 is shown with 1α,25(OH)2D3 in grey and red for the carbon and oxygen atoms, respectively. Additional interactions compared to the parental Gemini are marked in red. | ||
Additional interactions of the side chain containing the trifluoromethyl groups in Gemini-0072 and Gemini-0097 explain the increased biological activities of these agonists
In the two structures of zVDR LBD bound to Gemini-0072 or Gemini-0097, the A-, seco B-, C-, and D-rings present conformations which are similar to those observed in the presence of the natural ligand (Fig. 3). The hydroxyl groups of the A-ring of both ligands make the same hydrogen bonds as the zVDR LBD bound to 1α,25(OH)2D3 or gemini. More specifically, 1-OH interacts with Ser265 and Arg302, and 3-OH with Tyr175 and Ser306. The hydroxyl group of the trifluoromethyl-containing side chain of Gemini-0072 interacts similarly to the parental gemini but forms weaker H-bonds with His333 or His423 which may actually be weaker or altered. In the zVDR LBD–Gemini-0097 complex, we notice that His333 does not contact the hydroxyl group of the trifluoromethyl containing side chain (Fig. 3D). This loss of interaction may be the result of steric hindrance due to the vicinity of fluorine atom F-2 with the Nε2 of His333. This assertion is corroborated by the weak electron density and the high temperature B factors (>80 Å2) of the side chain of His333 in the Gemini-0097 complex. However, the interaction between the hydroxyl group of the trifluoromethyl-containing side arm and Nε2 of His423 is stronger in zVDR LBD–Gemini-0097 (distance is 2.7 Å instead of 3.0 Å in the case of Gemini-0072) and may compensate for the loss of stabilization at His333. In both complexes, His333 interacts with the two hydroxyl groups of the two side chains.The comparison between zVDR LBD–Gemini-0072 and zVDR LBD–gemini reveals that the two trideuteromethyl groups adopt a similar orientation and engage similar contacts. However, in the zVDR LBD–Gemini-0097 complex, the two trideuteromethyl groups adopt a different orientation due to the R-configuration at C-20 (Fig. 3B). The two crystal structures of Gemini-0072 and Gemini-0097 reveal additional, or stronger, contacts of the fluorine substituents compared to the hydrogens of the geminal methyl groups present in the parental gemini (Fig. 4). These interactions are manifested with the hydrophobic residues Val262 (H3), Tyr427(H11), Val444 and Phe448 (H12) for the three fluorine atoms branched at C-26 and C-25 of Gemini-0072 and -0097, respectively, and residues Leu255(H3), Leu430(H11) and Leu440(H12) for the three fluorine atoms branched at C-25 and C-26 of Gemini-0072 and -0097, respectively. Due to the 19-nor configurations of the two gemini, the contact between the A ring of the ligand and Ile299 disappears, as observed in the crystal structures of 19-nor-analogs.20 Further, the S-configuration at C-20 in Gemini-0072 induces an additional interaction between C-27 of the deuterated side chain and Cδ1 of Ile296. This contact is also observed in the zVDR LBD bound to Gemini-0097, but in this case with the C-21 atom of the trifluoromethyl-containing side chain of Gemini-0097. Synergistically, the additional contacts between fluorine atoms and hydrophobic residues of H3, H11 and H12 stabilize the agonist conformation of VDR in a significant manner (Fig. 4) and are likely to contribute to the increased interaction with SRC-1 peptide and superagonist potency of these compounds.
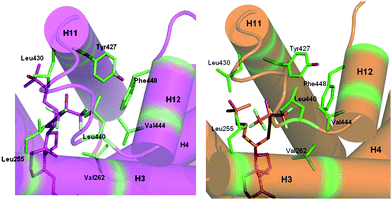 | ||
| Fig. 4 Additional contacts involving the fluorine side chain in zVDR LBD–Gemini-0072 and zVDR LBD–Gemini-0097 complexes. The zVDR LBD–Gemini-0072 and –Gemini-0097 complexes are shown in magenta and orange, respectively. The ligands are represented in stick with oxygen and fluorine atoms colored in red and light blue, respectively. Hydrophobic residues involved in additional contacts with the ligand are shown in green. | ||
Conclusion
This study shows that the novel gemini analogs, Gemini-0072 and Gemini-0097, are potent superagonist ligands. Similar to the parental gemini, the second side chains of these new compounds induce the creation of a new channel extending the original LBP. Specific additional interactions of the side-chain fluorine atoms stabilize VDR H12 and contribute to the increased transactivation properties and pro-differentiating action in cancer cells.Experimental section
Compounds
1α,25(OH)2D3 was purchased from Sigma, all gemini vitamin D analogs (>95% purity) (Fig. 1) were prepared as described,9 and all compounds were dissolved in ethanol.Steady-state fluorescence anisotropy measurements of SRC-1 peptide recruitment
Anisotropy titrations were carried out by adding increasing zVDR LBD concentrations to 1 μM fluorescent labeled SRC-1 NR2 peptide (RHKILHRLLQEGSPS). Details can be found in the ESI.‡Structure analysis of zVDR-LBD in complexes with Gemini-0072 or Gemini-0097
Details on expression, purification, crystallization, X-Ray data collection and structure determination can be found in the ESI.‡Transfection and transactivation assay
Transient transfection assays were performed with zVDR LBD. Human breast cancer MCF-7 cells were seeded into 24-well plates (105cells per well) and grown overnight in phenol red-free Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% charcoal-treated fetal bovine serum (FCS−), 5% gentamicin and 0.6 μg ml−1insulin. Liposomes were formed using the transfection reagent jetPEI™ (Polyplus Transfection), which was used according to the manufacturer's instructions. Cells were transfected with 25 ng of the expression plasmid pXJ440-Gal4DBD-zVDR (156–453), 250 ng of the reporter plasmid ptk-LUC, 50 ng of the pCH110–β-galactosidase vector (used as an internal control to normalize variation in transfection efficiency) and 675 ng of the carrier plasmid pBluescript (Stratagene). Ten hours later, cells were washed with freshly prepared phosphate-buffered saline and graded concentrations (0.1, 1, 10 and 100 nM) of 1α,25(OH)2D3 or Gemini ligands versussolvent (DMSO) were added to the cells in phenol red-free DMEM, supplemented with 10% FCS−. Twenty-four hours after the onset of stimulation, cells were rinsed in phosphate-buffered saline and lysed in 100 μl of reporter gene lysis buffer (Roche Diagnostics). Cell extracts were assayed for luciferase and β-galactosidase activities as described by Antony et al.21 and the protocol used is described in the ESI.‡Cell culture and determination of cell proliferation by [3H]thymidine uptake assay
MCF10 series of human breast cancer cell lines, MCF10AT1 (MCF10AT1k.clone2), MCF10DCIS (MCF10DCIS.com), and MCF10CA1a (MCF10CA1a.clone1) cell lines were established and provided by Dr Fred Miller at the Karmanos Cancer Institute (Detroit, MI).22–25 The cells were maintained in DMEM/F12 medium supplemented with 5% horse serum, 1% penicillin/streptomycin and 1% HEPES solution at 37 °C, 5% CO2. The culture medium for the MCF10AT1 cell line was the same as the medium for MCF10DCIS and MCF10CA1a, except that it was supplemented with 10 μg ml−1insulin, 20 ng ml−1EGF, 0.5 μg ml−1hydrocortisone, and 100 ng ml−1 cholera toxin. The cells were passaged every 3–4 days to maintain log-phase growth. 1α,25(OH)2D3 and Gemini vitamin D3 derivatives were dissolved in dimethylsulfoxide (DMSO) before addition to cell cultures; final concentrations of DMSO were 0.1% or less. Controls with DMSO alone were run in all cases.The MCF10 series of human breast cancer cells (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/well in a 24-well plate) were incubated with 1α,25(OH)2D3, Gemini-0072 and Gemini-0097 at five concentrations of 0.01, 0.1, 1, 10, 100 nM in 5% horse serum DMEM/F12 medium for 3 days. One μCi of [3H]thymidine was added to each well 3 h before the harvest. The cells were washed with PBS and precipitated with 10% trichloroacetic acid for 10 min. The cells were solubilized with the NaOH solution containing the salmon sperm DNA and the radioactivity of [3H]thymidine, incorporated into the cells, was analyzed with a liquid scintillation counter (Beckman Coulter, Fullerton, CA).
000 cells/well in a 24-well plate) were incubated with 1α,25(OH)2D3, Gemini-0072 and Gemini-0097 at five concentrations of 0.01, 0.1, 1, 10, 100 nM in 5% horse serum DMEM/F12 medium for 3 days. One μCi of [3H]thymidine was added to each well 3 h before the harvest. The cells were washed with PBS and precipitated with 10% trichloroacetic acid for 10 min. The cells were solubilized with the NaOH solution containing the salmon sperm DNA and the radioactivity of [3H]thymidine, incorporated into the cells, was analyzed with a liquid scintillation counter (Beckman Coulter, Fullerton, CA).
Protein data bank accession number
The PDB accession numbers for the coordinates of the structures of the zVDR LBD–Gemini-0072 and zVDR LBD–Gemini-0097 complexes reported in this article are 3O1D and 3O1E, respectively.Abbreviations
| NR | nuclear receptor |
| VDR | vitamin D NR |
| LBD | ligand binding domain |
| LBP | ligand binding pocket |
Acknowledgements
We thank the beam-line staff at the ESRF (Grenoble, France) for help during data collection. This work was supported from CNRS, INSERM, ULP, Association pour la Recherche sur le Cancer (ARC), l'Agence Nationale de la Recherche (ANR), the European Commission as SPINE2-complexes (contract-no LSHG-CT-2006-031220) under the RDT program ‘Quality of Life and Management of Living Resources’, in part by NIH R01 CA127645, NIEHS P30 ES005022 and the Board of Trustees Fellowship for Scholarly Excellence at Rutgers University. TH is a recipient of the Regional Council of Alsace and of ARC. We thank Yves Mély for advice on the fluorescence experiments.References
- M. J. Campbell and L. Adorini, The vitamin D receptor as a therapeutic target, Expert Opin. Ther. Targets, 2006, 10, 735–748 CrossRef CAS.
- S. Christakos, P. Dhawan, B. Benn, A. Porta, M. Hediger, G. T. Oh, E. Jeung, Y. Zhong, D. Ajibade and K. Dhawan, et al. Vitamin D: molecular mechanism of action, Ann. N. Y. Acad. Sci., 2007, 1116, 340–348 CrossRef CAS.
- G. Eelen, C. Gysemans, L. Verlinden, E. Vanoirbeek, P. De Clercq, D. Van Haver, C. Mathieu, R. Bouillon and A. Verstuyf, Mechanism and potential of the growth-inhibitory actions of vitamin D and ana-logs, Curr. Med. Chem., 2007, 14, 1893–1910 CrossRef CAS.
- S. Nagpal, S. Na and R. Rathnachalam, Noncalcemic actions of vitamin D receptor ligands, Endocr. Rev., 2005, 26, 662–687 CrossRef CAS.
- A. W. Norman, P. S. Manchand, M. R. Uskokovic, W. H. Okamura, J. A. Takeuchi, J. E. Bishop, J. I. Hisatake, H. P. Koeffler and S. Peleg, Characterization of a novel analogue of 1alpha,25(OH)(2)-vitamin D(3) with two side chains: interaction with its nuclear receptor and cellular actions, J. Med. Chem., 2000, 43, 2719–2730 CrossRef CAS.
- Y. Bury, M. Herdick, M. R. Uskokovic and C. Carlberg, Gene regulatory potential of 1alpha,25-dihydroxyvitamin D(3) analogues with two side chains, J. Cell. Biochem., 2001, 81(S36), 179–190 CrossRef.
- H. Maehr, M. R. Uskokovic, G. S. Reddy and L. Adorini, Calcitriol derivatives with two different side chains at C-20. 24-hydroxy derivatives as metabolic products and molecular probes for VDR exploration, J. Steroid Biochem. Mol. Biol., 2004, 89–90, 35–38 CrossRef CAS.
- H. Maehr, M. R. Uskokovic, L. Adorini and G. S. Reddy, Calcitriol derivatives with two different side chains at C-20. II. Diastereoselective syntheses of the metabolically produced 24(R)-hydroxygemini, J. Med. Chem., 2004, 47, 6476–6484 CrossRef CAS.
- H. Maehr, H. J. Lee, B. Perry, N. Suh and M. R. Uskokovic, Calcitriol derivatives with two different side chains at C-20. V. Potent inhibitors of mammary carcinogenesis and inducers of leukemia differentiation, J. Med. Chem., 2009, 52, 5505–5519 CrossRef CAS.
- M. Inaba, S. Okuno, Y. Nishizawa, Y. Imanishi, T. Katsumata, I. Sugata and H. Morii, Effect of substituting fluorine for hydrogen at C-26 and C-27 on the side chain of 1,25-dihydroxyvitamin D3, Biochem. Pharmacol., 1993, 45, 2331–2336 CrossRef CAS.
- C. S. Spina, L. Ton, M. Yao, H. Maehr, M. M. Wolfe, M. Uskokovic, L. Adorini and M. F. Holick, Selective vitamin D receptor modulators and their effects on colorectal tumor growth, J. Steroid Biochem. Mol. Biol., 2007, 103, 757–762 CrossRef CAS.
- H. Maehr, M. Uskokovic, L. Adorini, G. Penna, R. Mariani, P. Panina, N. Passini, E. Bono, S. Perego and M. Biffi, et al. Calcitriol derivatives with two different side chains at C-20. III. An epimeric pair of the gemini family with unprecedented antiproliferative effects on tumor cells and renin mRNA expression inhibition, J. Steroid Biochem. Mol. Biol., 2007, 103, 277–281 CrossRef CAS.
- H. J. Lee, S. Paul, N. Atalla, P. E. Thomas, X. Lin, I. Yang, B. Buckley, G. Lu, X. Zheng and Y. Lou, et al. Gemini vitamin D analogues inhibit estrogen receptor-positive and estrogen receptor-negative mammary tumorigenesis without hypercalcemic toxicity, Cancer Prev. Res., 2008, 1, 476–484 Search PubMed.
- H. J. Lee, J. Y. So, A. Decastro, A. Smolarek, S. Paul, H. Maehr, M. Uskokovic and N. Suh, Gemini vitamin D analog suppresses ErbB2-positive mammary tumor growth via inhibition of ErbB2/AKT/ERK signaling, J. Steroid Biochem. Mol. Biol., 2010, 121, 408–412 CrossRef CAS.
- N. Rochel, J. M. Wurtz, A. Mitschler, B. Klaholz and D. Moras, The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand, Mol. Cell, 2000, 5, 173–179 CrossRef CAS.
- N. Rochel and D. Moras, Ligand binding domain of vitamin D receptors, Curr. Top. Med. Chem., 2006, 6, 1229–1241 CrossRef CAS.
- F. Ciesielski, N. Rochel and D. Moras, Adaptability of the Vitamin D nuclear receptor to the synthetic ligand Gemini: remodelling the LBP with one side chain rotation, J. Steroid Biochem. Mol. Biol., 2007, 103, 235–242 CrossRef CAS.
- R. Bouillon, A. Verstuyf, J. Zhao, B. K. Tan and H. Van Baelen, Nonhypercalcemic vitamin D analogs: interactions with the vitamin D-binding protein, Horm. Res., 1996, 45, 117–121 Search PubMed.
- M. A. Arai, K. Takeyama, S. Ito, S. Kato, T. C. Chen and A. Kittaka, High-throughput system for analyzing ligand-induced cofactor recruitment by vitamin D receptor, Bioconjugate Chem., 2007, 18, 614–620 CrossRef CAS.
- G. Eelen, N. Valle, Y. Sato, N. Rochel, L. Verlinden, P. De Clercq, D. Moras, R. Bouillon, A. Muñoz and A. Verstuyf, Superagonistic fluorinated vitamin D3 analogs stabilize helix 12 of the vitamin D receptor, Chem. Biol., 2008, 15, 1029–1034 CrossRef CAS.
- P. Antony, R. Sigüeiro, T. Huet, Y. Sato, N. Ramalanjaona, L. C. Rodrigues, A. Mouriño, D. Moras and N. Rochel, Structure-function relationships and crystal structures of the vitamin D receptor bound 2 alpha-methyl-(20S,23S)- and 2 alpha-methyl-(20S,23R)-epoxymethano-1 alpha,25-dihydroxyvitamin D3, J. Med. Chem., 2010, 53, 1159–1171 CrossRef CAS.
- F. R. Miller, H. D. Soule, L. Tait, R. J. Pauley, S. R. Wolman, P. J. Dawson and G. H. Heppner, Xenograft model of progressive human proliferative breast disease, J. Natl. Cancer Inst., 1993, 85, 1725–1732 CrossRef CAS.
- F. R. Miller, S. J. Santner, L. Tait and P. J. Dawson, MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ, J. Natl. Cancer Inst., 2000, 92, 1185a–1186 CrossRef.
- L. B. Strickland, P. J. Dawson, S. J. Santner and F. R. Miller, Progression of premalignant MCF10AT generates heterogeneous malignant variants with characteristic histologic types and immunohistochemical markers, Breast Cancer Res. Treat., 2000, 64, 235–240 CrossRef CAS.
- S. J. Santner, P. J. Dawson, L. Tait, H. D. Soule, J. Eliason, A. N. Mohamed, S. R. Wolman, G. H. Heppner and F. R. Miller, Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells, Breast Cancer Res. Treat., 2001, 65, 101–110 CrossRef CAS.
Footnotes |
| † The authors declare that they have no competing financial interests. |
| ‡ Electronic supplementary information (ESI) available: Material and methods for purification, crystallization, X-Ray data collection and structure determination, protein expression vectors and transactivation. See DOI: 10.1039/c1md00059d |
| § Current address: Department of Molecular Biology, University of Geneva, Sciences III 30, quai Ernest-Ansermet, CH-1211, Geneva 4, Switzerland. |
| ¶ Current address: Department of food science and technology, Chung-Ang university, Anseng, 456-756 South Korea. |
| This journal is © The Royal Society of Chemistry 2011 |
