Ester prodrugs of ciprofloxacin as DNA-gyrase inhibitors: synthesis, antiparasitic evaluation and docking studies†
Faustine
Dubar
a,
René
Wintjens
b,
Érica S.
Martins-Duarte
c,
Rossiane C.
Vommaro
c,
Wanderley
de Souza
c,
Daniel
Dive
d,
Christine
Pierrot
d,
Bruno
Pradines
e,
Alexandre
Wohlkonig
f,
Jamal
Khalife‡
d and
Christophe
Biot‡
*ag
aUniversité Lille Nord de France, Université de Lille1, Unité de Catalyse et Chimie du Solide - UMR CNRS 8181, ENSCL, Bâtiment C7, B.P. 90108, 59652, Villeneuve d'Ascq Cedex, France
bLaboratoire de Chimie Générale, Facultés de Pharmacie, Université Libre de Bruxelles (ULB), Campus Plaine (CP 206/4), boulevard du Triomphe, B-1050, Brussels, Belgium
cLaboratório de Ultraestrutura Celular Hertha Meyer, Instituto de Biofísica Carlos Chagas Filho, Universidade Federal do Rio de Janeiro, 21941-902, Rio de Janeiro, RJ, Brazil
dCIIL, Inserm U 1019, UMR CNRS 8024 Université Lille Nord de France, Institut Pasteur de Lille, 1 rue du Pr Calmette, 59019, Lille Cedex, France
eInstitut de Recherche Biomédicale des Armées, Antenne de Marseille, Unité de Parasitologie, URMITE-UMR 6236, Allée du Médecin Colonel Jamot, Parc le Pharo, BP 60109, 13262, Marseille Cedex 07, France
fStructural Biology Brussels & Molecular and Cellular Interactions, VIB, Pleinlaan 2, B-1050, Brussels, Belgium
gUniversité Lille Nord de France, Université de Lille1, Unité de Glycobiologie Structurale et Fonctionnelle, CNRS UMR 8576, IFR 147, 59650, Villeneuve d'Ascq Cédex, France. E-mail: christophe.biot@univ-lille1.fr; Fax: +33 320436555; Tel: +33 320436941
First published on 17th March 2011
Abstract
Novel esterprodrugs of ciprofloxacin were synthesized, and tested for their antimalarial and antitoxoplasma activity. These new compounds proved to be extremely efficient against these parasites. Molecular modeling and computational calculations were used to understand the mechanisms of action of these drugs.
Introduction
The Plasmodium plastid is currently considered as a potential target to develop chemotherapy.1–3Quinolones and fluoroquinolones, which target bacterial topoisomerases II and IV4,5 were found efficient on P. falciparum6 prior to the discovery of apicoplast in this parasite. A plausible explanation of the activity of this class of antibiotics on P. falciparum came from the sequencing of its entire genome,7,8 and from the recent biochemical characterization of gyrases and topoisomerase IV which could be the main targets of (fluoro)quinolones.9,10 Using a dual strategy combining a prodrug/bioorganometallic approach, we recently improved the antimalarial activity of ciprofloxacin (CIPRO 1).11 This strategy revealed that the prodrug approach resulted in a dramatic increase of the antimalarial activity. In an interesting manner, grafting a ferrocenic moiety led to an additional enhancement of the activity which could be at least in part attributed to two mode of action of the metallocene. First, an oxidative stress which could be induced in situ due to the redox properties of the ferrocene (iron II)/ferricenium (iron III).12 Second, the high lipophilicity of the ferrocene which may help transport of drugs across membranes.13 It is important to note that these two mechanisms may operate together, and are not mutually exclusive.In order to further explore these potential mechanisms of action, we synthesized new derivatives 4 and 5 (Fig. 1) carrying a phenyl or an adamantanyl substituent (instead of the ferrocenyl, 3). These substituents allow mimicking the bulky ferrocene core but prevent the ability to catalyze a Fenton-like reaction resulting in the production of hydroxyl radicals. The new molecules were compared to CIPRO 1, its ethyl ester prodrug2 and the dual metallocenic/prodrug CIPRO 3 for antimalarial and antitoxoplasma activity. To provide information on the possible role of the DNA gyrase in the mechanism of action of these drugs, we performed a docking study using a modelled structure.
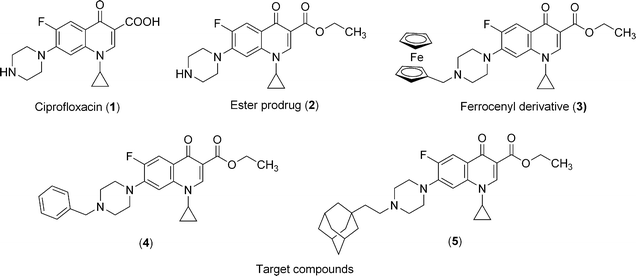 | ||
| Fig. 1 Chemical structures of ciprofloxacin and compounds investigated in this study 2–5. | ||
Our results show that compounds 4 and 5 are more active against P. falciparum and T. gondii than CIPRO or its ferrocenyl derivative, 3. In addition, a high therapeutic index was observed with product 5 when spleen primary cells were used as targets. With respect to the mode-of-action, the replacement of the ferrocene core of product 3 excluded the role of the radicals in the enhancement of activity of product 4 and 5. Based on modelled ternary complexes of DNA gyrase structures, docking studies revealed the absence of relationship between the binding free energy values and the in vitro activities of the novel compounds.
Results and discussion
Chemistry
The novel fluoroquinolones 4 and 5 were synthesized as previously reported (Scheme 1).11,14–16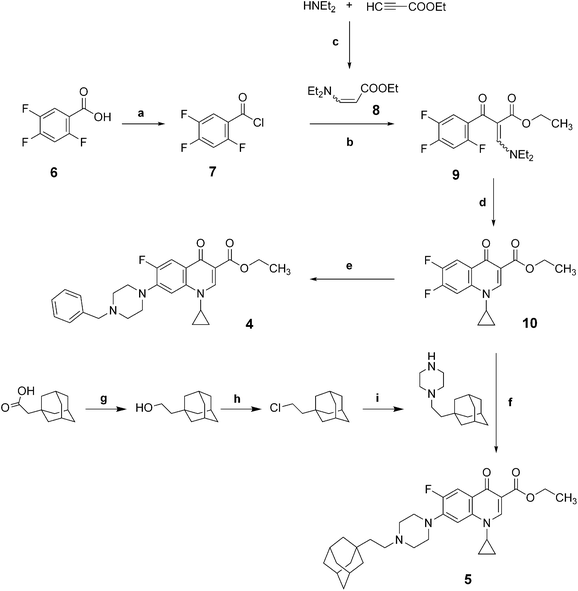 | ||
| Scheme 1 Synthetic routes to compounds 4 and 5. (a) (COCl)2, DMF, dichloromethane, N2 atm, r.t., 24h; (b) toluene, Et3N, N2 atm, reflux, 24h, (c) CH3CN, N2 atm, r.t., 24h (d) EtOH–Et2O (1/2), r.t., 3h, then DMF, K2CO3, reflux, 12h; (e) benzylpiperazine, N2 atm, CH3CN, reflux; (f) N2 atm, CH3CN, reflux; (g) LiAlH4, N2 atm, anh. THF, r.t.; (h) SOCl2, pyridine; (i) piperazine, K2CO3, CH3CN. | ||
Commercially available 2,4,5-trifluorobenzoic acid 6 reacts with Vilsmeier–Haack reagent (oxalyl chloride and dimethylformamide (DMF)) in dichloromethane to afford 7 in quantitative yield (Scheme 2). The acyl chloride7 is then condensed with ethyl 3-(diethylamino)acrylate 8 (freshly obtained from ethyl propiolate and N,N-diethylamine) in a mixture of toluene/triethylamine to provide the cetoester 9 in 73% yield. Transaminolysis of 9 with cyclopropylamine and successive cyclization with potassium carbonate in DMF gave the intermediate 10 in 79% yield. The fluoroquinolone 4 was then obtained by coupling the compound 10 in acetonitrile with 1-benzylpiperazine in 64% yield. Similarly, the fluoroquinolone 5 was obtained by coupling the compound 10 in acetonitrile with 1-(2-((adamantan-1-yl)ethyl)piperazine in 30% yield. The 1-(2-((adamantan-1-yl)ethyl)piperazine was prepared from 2-(adamantan-1-yl)acetic acid in three steps: (i) reduction of the carboxylic acid in the primary alcohol, (ii) conversion of the alcohol into the corresponding chloride, and (iii) condensation of the chloride with piperazine.
 | ||
| Scheme 2 | ||
Studies on the stability of the esters
The stability of the esterprodrugs2–5 toward chemical hydrolysis in aqueous solutions to produce the carboxylic acids6–8 was evaluated under quasi-physiological conditions by determining the half-life (t1/2) of esterhydrolysis (Table 1).17 It is well known that the pH of the parasitic cytosol is 7.4. Hence, the rates of enzymatic hydrolysis of the esterprodrugs were determined in phosphate buffer at pH 7.4 and in RPMI medium supplemented with 10% human serum. The esters2–5 were found to be highly stable toward both chemical and extracellular hydrolysis with half-lives longer than 76 h. With all compounds, the rate of cleavage is not affected by the presence of serum, suggesting that they are poor serum esterase substrastes. These results are in accordance with those obtained previously for biphosphonated fluoroquinolone esters.18| Compound | t 1/2 (h) | |
|---|---|---|
| pH 7.4 phosphate buffer | RPMI 10% human serum | |
| a The t1/2 was determined by HPLC analysis (in triplicate). | ||
| 2 | >76 | >76 |
| 3 | >76 | >76 |
| 4 | >76 | >76 |
| 5 | >76 | >76 |
Antimalarial activity
Antimalarial activities were determined against the CQ-susceptible (3D7), the CQ-resistant (W2), and the atovaquone (ATV)-resistant (Tm90C2b) P. falciparum strains (Table 2). Results showed that the novel fluoroquinolones 4 and 5 were more active than CIPRO 1 or fluoroquinolones 2 and 3 after 48 or 96 h of drug exposure. For instance, the adamantanyl derivative 5 is 29 to 60-fold more potent than CIPRO 1 against the CQ-resistant strains Tm90C2b and W2, 47-fold more potent toward the CQ-susceptible strain 3D7 after 48 h of drug exposure and 7 to 31-fold more potent after 96 h. Slightly higher IC50 values against the ATV-resistant strain were observed for the compounds 4 and 5. Antimalarial activities of all fluoroquinolones were increased in vitro after prolonged exposure. Nevertheless, this effect was less marked for the novel compounds 2–5 than for CIPRO 1. It is known that antibiotics such as CIPRO 1 or Doxicycline (DOX) exerted a “delayed death” effect in P. falciparum clones or isolates.11,19,20 In this work, synthesized fluoroquinolones 2–5 were 1.6 to 4.4-fold more potent after 96 h of drug exposure than 48 h, while CIPRO 1 was 3.7 to 7.3-fold more potent under the same conditions. Pyrimethamine (PY) and atovaquone (ATV) exert also a “delayed death” effect in P. falciparum parasites.| Cpd | P. falciparum strains | T. gondii RH strain | Cytotoxicity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 ± SD (μM) | IC50 ± SD (μM) | IC50 ± SD (μM) | LD50 ± SD (μM) | |||||||
| after 48 ha | after 96 ha | after 24 ha | after 48 ha | LLC-MK2 | murine | |||||
| 3D7 | W2 | Tm90C2b | 3D7 | W2 | Tm90C2b | cells | splenocytes | |||
| a Results are expressed as mean ± standard deviation of four to eight different experiments. b N.D., not determined. | ||||||||||
| 2 | 2.33 ± 0.25 | 3.67 ± 0.41 | 3.56 ± 0.67 | 0.89 ± 0.18 | 0.84 ± 0.28 | 2.27 ± 0.20 | 0.96 ± 0.18 | 0.42 ± 0.06 | >30 | >40 |
| 3 | 2.63 ± 0.27 | 4.82 ± 0.58 | 6.78 ± 0.72 | 1.59 ± 0.33 | 1.72 ± 0.23 | 1.82 ± 0.40 | 2.95 ± 0.92 | 1.28 ± 0.20 | >30 | >40 |
| 4 | 1.08 ± 0.19 | 1.61 ± 0.23 | 2.56 ± 0.21 | 0.32 ± 0.14 | 0.49 ± 0.23 | 2.35 ± 0.48 | 1.70 ± 0.68 | 1.24 ± 0.14 | >30 | 57 ± 13 |
| 5 | 1.00 ± 0.40 | 2.06 ± 0.18 | 2.46 ± 0.23 | 0.41 ± 0.10 | 0.71 ± 0.23 | 2.23 ± 0.37 | 0.92 ± 0.39 | 0.46 ± 0.06 | 27.8 ± 3.7 | >100 |
| 1 | 47.0 ± 5.9 | 122.3 ± 19.0 | 71.8 ± 13.0 | 12.8 ± 4.7 | 16.8 ± 2.6 | 15.3 ± 3.5 | >20 | >20 | N.D. | >40 |
| IC50 ± SD (nM) | IC50 ± SD (nM) | T. gondii RH strain | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| after 48 ha | after 96 ha | IC50 ± SD (μM) | ||||||||
| 3D7 | W2 | Tm90C2b | 3D7 | W2 | Tm90C2b | after 24 ha | after 48 ha | |||
| CQ | 18 ± 3 | 519 ± 32 | 475 ± 59 | 18 ± 2.4 | 455 ± 36 | 393 ± 58 | — | — | — | — |
| ATV | 2.3 ± 0.2 | 3.9 ± 1.4 | >10 000 | 1.9 ± 0.2 | 2.0 ± 0.3 | 388 ± 124 | — | — | — | |
| QN | 178 ± 23 | 917 ± 213 | 1068 ± 266 | 112 ± 21 | 583 ± 112 | 608 ± 124 | — | — | — | — |
| PY | 13.3 ± 3.5 | 10.4 ± 2.9 | 14.6 ± 2.4 | 2.1 ± 0.4 | 2.3 ± 0.4 | 1.4 ± 0.3 | 0.24 ± 0.04 | 0.12 ± 0.04 | N.D. | N.D. |
Antitoxoplasma activity
Next, we tested the activity of the novel fluoroquinolones against Toxoplasma gondii, another apicoplasma parasite. Results presented in Table 2 showed that these compounds exhibited strong activity against tachyzoites of T. gondii at very low concentrations. Most importantly, all tested compounds were found to be dramatically more active than CIPRO 1, in particular, fluoroquinolones 2 and 5 which inhibited parasite growth with IC50 at nanomolar range. Kinetic studies revealed that the novel fluoroquinolones blocked tachyzoites proliferation within 24 h of exposure, while CIPRO 1 inhibited parasite growth only after the second cycle of infection of new host cell, confirming previous results.2 After 48 h of exposure the IC50 were found to be lower from those obsered at 24 h, suggesting that fluoroquinolones 2–5 exert also a slight “delayed death” effect in T. gondii parasites.Cytotoxicity
The in vitro toxicity of 1–5 was tested using mouse spleen cells and LLC-MK2 cell line. (Detailed procedures are given in the ESI†). All compounds exhibited relatively weak cytotoxicity (Table 2). The adamantanyl derivative 5 exhibited the lowest cytotoxic effect. Consequently, this compound showed the highest therapeutic index (TI). For example, the TI (LD50/IC50) is >60 for T. gondii RH strain and >100 for P. falciparum3D7 strain.Lipophilicity and basicity
Lipophilicity (logP value) and basicity (pKa value) which influence the uptake and the distribution of fluoroquinolones derivatives have been predicted using Marvin Calculator Plugin from ChemAxon 5.2 (Table SI1 in the ESI†). Due to the high hydrophilic behavior of the carboxylic acids (1, 7 and 8), it is reasonable to propose that these forms are less efficient to cross the multiple membranes of the parasites including the apicoplast one than the esterprodrugs do. The critical concentration of the drugs that affect the apicoplast functions may be reached faster leading to a less pronounced “delayed effect”.Homology modeling
The amino acid sequences of GyrA and GyrB deduced from P. falciparumgenome (accession numbers from Tr-EMBL Q8I0X3 and Q8I528, respectively), were compared with those expressed by Staphylococcus aureus. Although alignments of sequences revealed the presence of large gaps and regions with weak similarities P. falciparum GyrA and GyrB showed 32.3% and 29.7% identity (56.2 and 37.2% similarity) with S. aureus GyrA and GyrB respectively. The catalytic residues are well conserved in Pf gyrases, while the Quinolone Resistance Determining Regions (QRDR) of GyrA showed one amino-acid change and one amino-acid substitution. The critical Ser84 that interacts with CIPRO 1 is replaced by Lys or Gln in P. falciparum and T. gondii, respectively, whereas Glu88 of S. aureus is substituted by Asp, which has similar properties. Due to the presence of large gaps, some regions appeared as loops in the modelled structure of P. falciparum gyrase, but as they are not involved in the CIPRO 1binding site their structures were not solved. The tri-dimensional structures of the two subunits in complex with DNA were modelled using the ternary complex of S. aureus gyrase in complex with DNA and CIPRO 1 (PDB code: 2XCT, resolution 3.35 Å).Similar observations were made when we applied the same strategy the T. gondii gyrase (accession numbers B9PYK0 and B9Q192). To the best of our knowledge, this is the first report of a modelled ternary complex (protein-ligand-DNA) based on an available structure of S. aureus gyrase ternary complex.
Docking studies
We used the zwitterionic active forms of the fluoroquinolones in the docking calculations as a noncatalytic magnesium ion was found to be important to mediate the interaction between quinolone derivatives and the protein,21,22 the presence of this Mg2+ ion was thus maintained in the docking processes. The predicted binding affinities (Table 3) of the fluoroquinolones with DNA gyrase duplex were evaluated using the binding free energies (Ebind, kcal mol−1). We considered the binding free energy of the best ranked conformations as the main parameter for analysis of Autodock4.2 results. Docking of CIPRO 1 to the P. falciparum and T. gondiiDNA gyrase models indicated that the binding is very tight with estimated binding energies less than −12 kcal mol−1.| Compound | P.falciparum E bind kcal mol−1 | T. gondii E bind kcal mol−1 |
|---|---|---|
| 1 | −12.33 | −15.14 |
| 7 | −13.94 | −16.12 |
| 8 | −13.03 | −15.28 |
The binding is largely made through van der Waals and π–π stacking interactions. As the compound was found intercalated between a DNA base pair, only few direct interactions with the protein were detected. The corresponding residues of Ser84, i.e.Lys248 or Gln348, are major interacting residues. The Mg2+ ion chelated by CIPRO 1 greatly contributed to the binding energy through interactions with the protein, as well as with DNA.
Similar docking results were obtained for compounds 7 and 8 (Fig. 2). The phenyl and the adamantanyl substituents gave additional van der Waals contacts, but the resulting estimated free energies of binding were in the same order of magnitude as CIPRO 1. As a difference of 2.0–2.5 kcal mol−1 is considered significant,23 our docking calculations were not able to discriminate between the different synthetic compounds. Thereby we can not correlate here the binding free energy values and the in vitro activities of the drugs.
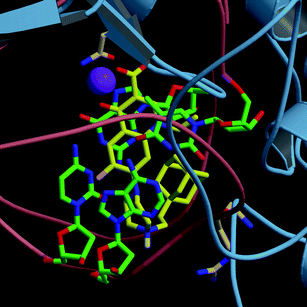 | ||
| Fig. 2 Best ranked docked conformation of compound 8 in complex with T. gondiiDNA gyrase. The protein is showed in light blue ribbon and the DNA backbone in pink coils. Residues implicated in binding are displayed (Arg938 of GyrA and Gln348 of GyrB). Residue numbering used throughout the paper is based on the corresponding gene sequence. Carbon atoms of the ligand are coloured in yellow, those of DNA and protein in green and light pink, respectively. Oxygen, nitrogen and hydrogen are in red, blue and white, respectively. The Mg2+ ion is depicted in purple. | ||
Note that the crystal structure of S. aureus gyrase does not contain water molecules. However, previous studies have shown the importance of water molecules for the coordination of the Mg2+ ion, as well as their role to mediate interactions between quinolone derivatives and the protein.22 New interacting residues could be added by performing docking calculations in presence of water molecules.
Factors that can explain the higher activities of our novel compounds
Here, we evaluated the contribution of the redox properties of the ferrocene/ferricenium, and of the hydrophobicity of fluoroquinolones to their activity against apicomplexa parasites such as malaria and T. gondii. Regarding the redox reaction promoted by ferrocene core, the main question was the potential production of hydrogen peroxide, in parasite apicoplast, required to promote the Fenton-like reaction.12,24 However, there are two possible explanations that support our data which showed that the antiparasite activity of fluoroquinolones is not due to the Fenton-like reaction: 1) the replacement of ferrocene core did not cause, even partially, any loss-of-function and 2) the unfavorable conditions in the apicoplast compartment to accumulate critical H2O2 concentrations, despite the presence of a SOD,25 as this organelle requires reductive conditions for the function of its enzymes involved in gluthation and lipoate metabolism.26 Our data support the concept that the main activity of quinolones derivatives is rather assigned to their hydrophobic properties by increasing the penetration capacity of the novel compounds 4 and 5 to pass through the multiple membranes and to target more efficiently the gyrase.27 Even if the docking studies failed to show any relation between the bulkiness of the compounds and the global activity (4 in compraison with 5), the presence of the adamantanyl group greatly reduced the toxicity of the compound 5.Conclusions
Chemical modifications of CIPRO 1 allowed us to design novel compounds which are dramatically more efficient against P. falciparum and T. gondii than the initial compound, with a promising therapeutic index. By similarity with the antibacterial action of CIPRO 1, these compounds were assumed to target the gyrase enzyme of the parasites. Nevertheless, given the sequence plasticity of the quinolone binding site, the exact mode of interaction of these compounds with gyrase protein needs further experimental investigations, including a high resolution structure of the ternary complex.Acknowledgements
The authors thank S. Lafitte, R. Amalvict, E. Baret, M. Mokrane and A. Pascual for technical support. S. Habib is acknowldedged for helpul discussion. This work was supported by the CNRS, by the Université de Lille 1, and by the Ministère de l'Enseignement Supérieur (grant attributed to F.D.) and the Ministère de la Défense. R.W. is Research Associate at the FNRS-FRC (Belgium).Notes and references
- E. L. Dahl and P. J. Rosenthal, Trends Parasitol., 2008, 24, 279 CrossRef CAS.
- M. E. Fichera and D. S. Roos, Nature, 1997, 390, 407 CrossRef CAS.
- G. I. McFadden, Protoplasma, 2010 Search PubMed.
- K. Drlica and M. Malik, Curr. Top. Med. Chem., 2003, 3, 249 CrossRef CAS.
- D. C. Hooper, Clin. Infect. Dis., 2001, 32(Suppl. 1), 9.
- A. A. Divo, A. C. Sartorelli, C. L. Patton and F. J. Bia, Antimicrob Agents Chemother, 1988, 32, 1182 CAS.
- M. A. Dar, A. Sharma, N. Mondal and S. K. Dhar, Eukaryotic Cell, 2007, 6, 398 CrossRef CAS.
- C. Garcia-Estrada, C. F. Prada, C. Fernandez-Rubio, F. Rojo-Vazquez and R. Balana-Fouce, Proc. Biol. Sci., 2010, 277 Search PubMed.
- G. Anquetin, J. Greiner and P. Vierling, Curr. Drug Targets: Infect. Disord., 2005, 5, 227 Search PubMed.
- E. L. Dahl and P. J. Rosenthal, Antimicrob. Agents Chemother., 2007, 51, 3485 CrossRef CAS.
- F. Dubar, G. Anquetin, B. Pradines, D. Dive, J. Khalife and C. Biot, J. Med. Chem., 2009, 52, 7954 CrossRef CAS.
- N. Chavain, H. Vezin, D. Dive, N. Touati, J. F. Paul, E. Buisine and C. Biot, Mol. Pharmaceutics, 2008, 5, 710 CrossRef CAS.
- C. Biot, D. Taramelli, I. Forfar-Bares, L. A. Maciejewski, M. Boyce, G. Nowogrocki, J. S. Brocard, N. Basilico, P. Olliaro and T. J. Egan, Mol. Pharmaceutics, 2005, 2, 185 CrossRef CAS.
- V. Cecchetti, A. Fravolini, M. C. Lorenzini, O. Tabarrini, P. Terni and T. Xin, J. Med. Chem., 1996, 39, 436 CrossRef CAS.
- V. Cecchetti, A. Fravolini, M. Palumbo, C. Sissi, O. Tabarrini, P. Terni and T. Xin, J. Med. Chem., 1996, 39, 4952 CrossRef CAS.
- V. Cecchetti, O. Tabarrini, S. Sabatini, H. Miao, E. Filipponi and A. Fravolini, Bioorg. Med. Chem., 1999, 7, 2465 CrossRef CAS.
- E. Davioud-Charvet, S. Delarue, C. Biot, B. Schwobel, C. C. Boehme, A. Mussigbrodt, L. Maes, C. Sergheraert, P. Grellier, R. H. Schirmer and K. Becker, J. Med. Chem., 2001, 44, 4268 CrossRef CAS.
- K. S. Tanaka, T. J. Houghton, T. Kang, E. Dietrich, D. Delorme, S. S. Ferreira, L. Caron, F. Viens, F. F. Arhin, I. Sarmiento, D. Lehoux, I. Fadhil, K. Laquerre, J. Liu, V. Ostiguy, H. Poirier, G. Moeck, T. R. Parr, Jr. and A. Rafai Far, Bioorg. Med. Chem., 2008, 16, 9217 CrossRef CAS.
- B. Pradines, C. Rogier, T. Fusai, J. Mosnier, W. Daries, E. Barret and D. Parzy, Antimicrob. Agents Chemother., 2001, 45, 1746 CrossRef CAS.
- B. Pradines, A. Spiegel, C. Rogier, A. Tall, J. Mosnier, T. Fusai, J. F. Trape and D. Parzy, Am J Trop Med Hyg, 2000, 62, 82 Search PubMed.
- B. D. Bax, P. F. Chan, D. S. Eggleston, A. Fosberry, D. R. Gentry, F. Gorrec, I. Giordano, M. M. Hann, A. Hennessy, M. Hibbs, J. Huang, E. Jones, J. Jones, K. K. Brown, C. J. Lewis, E. W. May, M. R. Saunders, O. Singh, C. E. Spitzfaden, C. Shen, A. Shillings, A. J. Theobald, A. Wohlkonig, N. D. Pearson and M. N. Gwynn, Nature, 2010, 466, 935 CrossRef.
- A. Wohlkonig, P. F. Chan, A. P. Fosberry, P. Homes, J. Huang, M. Kranz, V. R. Leydon, T. J. Miles, N. D. Pearson, R. L. Perera, A. J. Shillings, M. N. Gwynn and B. D. Bax, Nat. Struct. Mol. Biol., 2010, 17, 1152 CrossRef CAS.
- R. Huey, G. M. Morris, A. J. Olson and D. S. Goodsell, J. Comput. Chem., 2007, 28, 1145 CrossRef CAS.
- F. Dubar, T. J. Egan, B. Pradines, D. Kuter, K. K. Ncokazi, D. Forge, J. F. Paul, C. Pierrot, H. Kalamou, J. Khalife, E. Buisine, C. Rogier, H. Vezin, I. Forfar, C. Slomianny, X. Trivelli, S. Kapishnikov, L. Leiserowitz, D. Dive and C. Biot, ACS Chem Biol, 2011 Search PubMed , in press.
- P. Pino, B. J. Foth, L. Y. Kwok, L. Sheiner, R. Schepers, T. Soldati and D. Soldati-Favre, PLoS Pathog., 2007, 3, 115 Search PubMed.
- S. Kehr, N. Sturm, S. Rahlfs, J. M. Przyborski and K. Becker, PLoS Pathog., 2010, 6, e1001242 Search PubMed.
- T. Fleige, J. Limenitakis and D. Soldati-Favre, Microbes Infect., 2010, 12, 253 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed experimental and theoretical procedures and spectroscopic data of novel compounds. See DOI: 10.1039/c1md00022e/ |
| ‡ C. Biot and J. Khalife contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2011 |
