(Carboxydiamine)Pt(II) complexes of a combretastatin A-4 analogous chalcone: the influence of the diamine ligand on DNA binding and anticancer effects†
Miroslava
Zoldakova
a,
Bernhard
Biersack
a,
Hana
Kostrhunova
b,
Aamir
Ahmad
c,
Subhash
Padhye
c,
Fazlul H.
Sarkar
c,
Rainer
Schobert
*a and
Viktor
Brabec
b
aOrganic Chemistry, University of Bayreuth, 95447, Bayreuth, Germany. E-mail: rainer.schobert@uni-bayreuth.de; Fax: +49921552671
bInstitute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, CZ-61265, Brno, Czech Republic
cBarbara Ann Karmanos Cancer Institute, Detroit, Michigan 48201, USA
First published on 30th March 2011
Abstract
The combretastatin A-4 analogue m-hydroxychalcone 2 was esterified with a (1-carboxyethane-2,3-diamine)dichloridoplatinum(II) fragment to give complex 6 which was more active against various cancer cell lines (IC50 < 1 µM) than its analogue 3 bearing a 6-aminomethylnicotinate ligand. Complex 6 bound to the same sites of DNA as cisplatin but caused a larger DNA unwinding angle and ten times more interstrand cross-links. Also, DNA lesions due to binding of 6 were only half as efficiently repaired as cisplatin–DNA adducts. Complex 6 also showed a much lower affinity to the platinum detoxifier glutathione.
Introduction
The anticancer agent combretastatin A-4 (1, Fig. 1), a cis-stilbene occurring in the bark of the South African bush-willow Combretum caffrum,1 binds to the colchicine binding site of tubulin and inhibits its polymerisation.2 Compound 1 is highly active against multi-drug resistant tumours and vascular-disrupting by way of Rho activation.2,3Chalcone 2, an analogue of 1 which also inhibits tubulin polymerisation, proved strongly cytotoxic in various cancer cells.4,5 Further chalcones were reported with high anticancer activities originating from other modes of action such as the inhibition of resistance-relevant ABC-transporters.6,7 Recently, fluorinated 2-hydroxychalcones of the garcinol type were shown to owe their activity in a pancreas tumour to an up-regulated COX-2 expression,8 and a chlorambucil conjugate of 1 was found far more efficacious against neuroblastoma cells than 1 alone.9 Our group presented a dichlorido-Pt(II)-(6-aminomethyl)nicotinate 3 of chalcone 2 that combines DNA and tubulin binding properties and which inhibits the growth of certain cisplatin-resistant tumour cell lines.10 Conjugate 3 is accumulated in cancer cells mainly viacell-controlled transporters and it induces apoptosis by triggering multiple targets.11 Herein we report on a close analogue of 3 bearing a different stationary 1-carboxyethane-2,3-diamine ligand and how it differs from 2 and 3 in terms of cytotoxicity, DNA binding and robustness against deactivation by glutathione.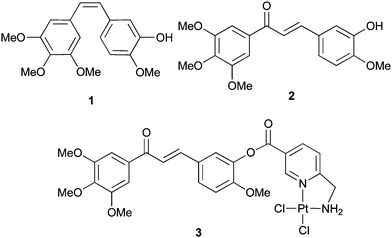 | ||
| Fig. 1 Combretastatin A-4 (1), a chalcone analogue 2, and its Pt(II)-complex 3. | ||
Results and discussion
Chemistry
Chalcone 2 was synthesised by a Claisen–Schmidt condensation of isovanillin with 3,4,5-trimethoxyacetophenone following a literature procedure.4Boc-protected (D,L)-2,3-diaminopropionic acid 4 was esterified with hydroxychalcone 2 under Yamaguchi conditions affording ester 5. Deprotection with TFA and reaction of the bis-ammonium intermediate with K2PtCl4 in aqueous THF under slightly acidic conditions gave the platinum complex 6 (Scheme 1).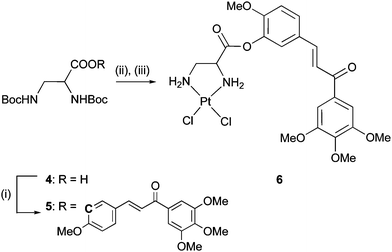 | ||
| Scheme 1 Reagents and conditions: (i) Et3N, C6H2Cl3COCl, 2, DMAP, DMF/toluene, rt, 16 h (52%); (ii) TFA, DCM, rt, 1 h; (iii) K2PtCl4, H2O/THF, rt, 24 h (40%). | ||
Biological evaluation
The antiproliferative activity of the new complex 6 was evaluated in MTT-assays against cells of leukaemia HL-60, melanoma 518A2, and the colon HCT-116, pancreas BxPC-3 and breast MDA-MB-231 carcinomas, and compared with the activities of chalcone 2 and dichlorido(6-aminomethylnicotinate)Pt(II) 3 (Table 1, ESI†). Complex 6 was most active against HL-60 cells and distinctly more active than compounds 2 and 3 in all other tested cancer cell lines. The high activity of all three test compounds in the leukaemia cells is not surprising since these are generally sensitive to both combretastatins and platinum compounds.4,12 The finding that complex 6 was superior to the analogue 3 in efficacy against the melanoma 518A2 cells is at variance with recent test results of a series of terpene–Pt complexes which inhibited their growth most when bearing a 6-aminomethylnicotinate rather than a 1-carboxyethane-2,3-diamine ligand.13,14 It becomes plausible, though, when assuming different effects by the terpene and the chalcone moieties such as lipophilic membrane shuttling vs.tubulin binding. The small IC50 values of the complexes 3 and 6 in the glutathione-rich HCT-116 cells15 when compared with cisplatin are a hint at the robustness of chalcone Pt-complex conjugates against the glutathione-based detoxification mechanism of resistant cancer cells. Complex 6 was equally active against the aggressive, hormone-refractory MDA-MB-231 cells which lack estrogen and progesterone receptors as well as ErbB2 receptor tyrosine kinases, and the BxPC-3 cells that respond well to the standard therapy with gemcitabine. In addition, complex 6 led to significant apoptosis, especially in the more aggressive MDA-MB-231 cells, at concentrations as low as 100 nM according to DNA–histone ELISA assays (Fig. 2, ESI†), thus surpassing the apoptotic impact of analogue 3 by far.| Compound/cell line | 2 | 3 | 6 | Cisplatin |
|---|---|---|---|---|
| a Values are derived from dose–response curves obtained by measuring the percentage of viable cells relative to untreated controls after 48 h exposure to test compounds using an MTT assay. Values represent means of four independent experiments. The organic solvent never exceeded 1% (DMF) during the treatment. b Values are derived from dose–response curves obtained by measuring the percentage of viable cells relative to untreated controls after 72 h exposure to test compounds using an MTT assay. Values represent means of three independent experiments. The organic solvent never exceeded 0.1% (DMSO) during the treatment. | ||||
| 518A2 | 2.0 ± 0.5 | 1.1 ± 0.3 | 0.4 ± 0.01 | 9.0 ± 2.3 |
| HL-60 | <0.001 | 0.3 ± 0.01 | <0.001 | 0.5 ± 0.3 |
| HCT-116 | — | 1.0 ± 0.1 | 0.8 ± 0.1 | 17 ± 2 |
| BxPC-3 b | 0.26 ± 0.01 | 0.31 ± 0.01 | 0.09 ± 0.01 | — |
| MDA-MB-231 b | 0.41 ± 0.01 | 0.38 ± 0.02 | 0.08 ± 0.01 | — |
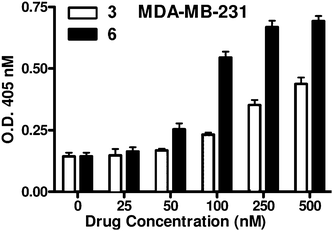 | ||
| Fig. 2 The rate of apoptosis in MDA-MB-231 breast carcinoma cells upon treatment for 72 h with various doses of platinum complex 3 or 6. | ||
The chalcones 2 and 3 inhibit tubulin polymerisationin vitro and affect the microtubular network of neural cells.10,11 Now we could show them by light microscopy to also cause inter-nucleosomal DNA cleavage in 518A2 melanoma cells (Fig. 3).
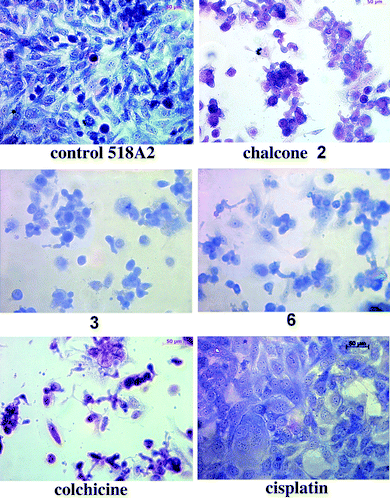 | ||
| Fig. 3 Light microscopic images of 518A2 cells treated with IC50 concentrations of test and reference compounds for 24 h, fixed and stained with Giemsa dye. | ||
While control cells treated only with Giemsa dye appeared typically shaped and regularly spread with their nuclei well defined by envelopes and with condensed chromatin visible as dark blue dots, treatment with the chalcone 2, 3 or 6 led to a decrease in the number of cells which were also packed more densely in insular aggregates. Some of them even formed connective evaginations. Treatment with the complex 3 or 6 gave rise to cells with bluish cytoplasm because of DNA metallation and to a rounded morphology typical of exposure to chalcones. Late apoptotic or dead cells lay flat with a purple or altogether missing cytoplasm. In contrast, 518A2 cells treated with cisplatin appeared swollen but remained equally distributed. Exposure to colchicine, a known inhibitor of tubulin polymerisation, rapidly reduced the number of 518A2 cells which no longer displayed nuclear envelopes, took on a pink-purple hue and an either flat or rounded shape. Thus, the morphological changes of 518A2 cells caused by the tubulin binders colchicine, chalcone 2, and by chalcone–platinum complexes 3 and 6 differed distinctly which is an indication of them acting by different modes.
In order to pinpoint the role of platinum in the anticancer effect of complexes 3 and 6 as opposed to cisplatin, we investigated their DNA binding behaviour. First, the degree of metallation of calf thymus DNA by 3 (55% rel. to the concentration applied in vitro) and 6 (75%) was determined with atomic absorption spectroscopy (AAS). Next, electromobility shift assays (EMSA) were carried out with pUC19 plasmid DNA in order to quantify the unwinding capacity of the complexes 3 and 6. The unwinding of supercoiled pUC19 plasmid DNA induced upon their binding was determined by incubating the plasmid with them for 24 h at 37 °C at various stages of platination rb (platinum per nucleotide). A typical native agarose gel of DNA modified by 6 is shown in Fig. 4. Since unwinding of the DNA reduces the number of supercoils it results in a slowdown of migration. The mean unwinding angle can be calculated from the equation Φ = −18σ/rb(c), where σ is the superhelical density and rb(c) is the rb at which the supercoiled and nicked forms comigrate.16 Under the experimental conditions σ was calculated to be −0.051 on the basis of the data for cisplatin for which the rb(c) was determined in this study and Φ = 13° was assumed.16Fig. 4 shows that 6 causes a significant DNA unwinding of Φ = 21–23°. The co-migration point rb(c) of modified supercoiled and nicked DNA was reached at rb = 0.058. This conspicuously high level of unwinding induced by 6 is even greater than that associated with the binding of cisplatin. It can be explained by assuming 6 to form many DNA cross-links (CLs). In contrast, increasing concentrations of complex 3 or chalcone 2 when added to pUC19 DNA had no effect on the electrophoretic mobility of the sc and nicked forms (not shown).
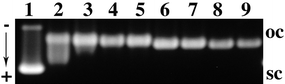 | ||
| Fig. 4 Unwinding of the supercoiled pUC19 plasmid DNA modified by 6: rb = 0.022, 0.032, 0.044, 0.048, 0.096, 0.108, 0.150, and 0.152 (lanes 2–9). Lane (1): untreated DNA; oc = nicked plasmid; sc = closed negatively supercoiled plasmid. | ||
Cisplatin also forms various types of inter- and intrastrand CLs.17 We now compared the capacities of cisplatin and the complex 6 to form interstrand cross-links (ICLs) in a 213-bp DNA fragment obtained by linearisation of the pSP73KB plasmid DNA with EcoRI. The samples were analysed for ICLs by agarose gel electrophoresis under denaturing conditions. The non-cross-linked strands of 213-bp DNA labelled with 32P at the 3′-end migrated as a 213-mer single strand, whereas the high molecular ICLs migrated more slowly. The intensity of the more slowly migrating band increased with the number of CLs induced by 6 (Fig. 5). The radioactivity of the bands in each lane was measured to determine the fractions of non-cross-linked vs. cross-linked DNA. The frequency of ICLs was calculated by the Poisson distribution from the fraction of non-cross-linked DNA in combination with the rb values and the fragment size. While the ICL frequency induced by cisplatin was thus calculated to be ca. 6 ± 0.5%, which is in line with previous results,18 the corresponding value for complex 6 was 58 ± 2.3%, i.e., ten times that of cisplatin.
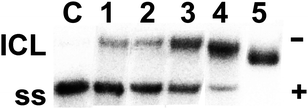 | ||
| Fig. 5 Electrophoretic autoradiograms of 3′-end 32P-labelled 213-bp fragments of linear pSP73KB plasmid DNA (2455 bp) on denaturing 1% agarose gel after incubation for 24 h with cisplatin (lane 1) at rb = 0.001, or with 6 (lanes 2–5) at rb = 0.00001, 0.00003, 0.0002, and 0.001. Lane C = control; ICL = interstrand cross-linked DNA; ss = single-strand DNA. | ||
The preference of complex 6 for binding to particular nucleotide sequences of DNA was then investigated by a transcription mapping assay. Cutting of the pSP73KB DNA by NdeI and HpaI restriction endonucleases gave a 212-bp fragment of a known sequence that contained a T7 RNA polymerase promotor. In vitroRNA synthesis by RNA polymerases on this DNA template modified by 6 at rb = 0.02 or 0.03 can be prematurely terminated at or in the proximity of sites where platinum fragments are coordinated to DNA bases (Fig. 6A). Interestingly, DNA lesions originating from bisdentate coordination of Pt fragments can effectively terminate RNA synthesis at the position of the defect whereas monodentate coordination of Pt fragments such as [PtCl(dien)]+ (dien = diethylenetriamine) is insufficient for the termination of RNA synthesis. The major stop sites, primarily guanine and adenine residues, were roughly identical for complex 6 and cisplatin (Fig. 6B). Thus, the major sites in DNA at which complex 6 preferentially binds are GG and AG sites.
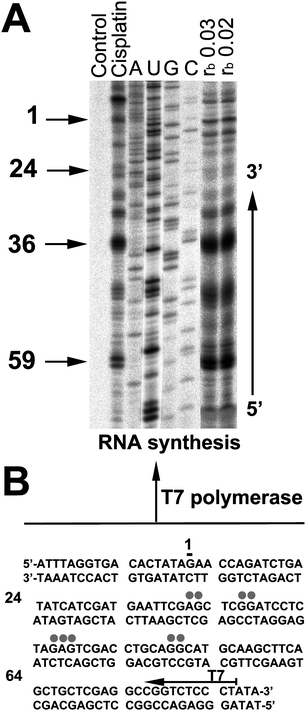 | ||
| Fig. 6 Inhibition of RNA synthesis by T7 RNA polymerase on the NdeI/HpaI fragment of pSP73KB plasmid DNA containing adducts of 6 or cisplatin. (A) The autoradiograph of 8% PAA/8 M urea sequencing gel; lanes: non-platinated control; cisplatin at rb 0.01; A, U, G, C, chain-terminated marker RNAs; 6 at rb 0.03 and 0.02 (as to AAS). Numbers correspond to the nucleotide sequence numbering of (B). (B) The sequence of the NdeI/HpaI fragment; the arrow indicates the promoter for T7 RNA polymerase, grey dots indicate sites of stop signals in (A), numbers correspond to the nucleotide numbering in the sequence map of the pSP73KB plasmid. | ||
Mammalian cells damaged by chemotherapeutics such as cisplatin may remove the bulky helix-distorting DNA adducts and modified nucleotides by DNA repair mechanisms and so evade cell death.19 Obviously, this is not desired in the case of cancer cells. In order to determine how readily DNA damage induced by platinum complex 6 is repaired we used an assay that quantifies the extent of DNA repair synthesis by cell-free tumour cell extracts (CFE).
The efficiency of CFE of repair-proficient HeLa or melanoma 518A2 cells to repair defects in pUC19 plasmid (2686 bp) globally modified by 6 at rb = 0.005–0.015 or cisplatin at rb = 0.01 was ascertained by measuring the amount of incorporated radiolabelled nucleotide, corrected for the relative DNA content in each band. Fig. 7 shows that DNA repair synthesis in the plasmid modified by 6 was considerably lower than that found for cisplatin. This effect was more pronounced when employing CFE of 518A2 cells.
![DNA repair of pUC19 pre-treated with cisplatin or 6. Top row: untreated pBR322 as internal control. Repair efficiency is given by the radioactivity of incorporated [α-32P]-dATP. Samples repaired by repair-proficient extracts of HeLa cells (lanes 1–4) and 518A2 cells (lanes 5–8). Cisplatin: rb ≈ 0.01; 6: rb ≈ 0.005, 0.01, and 0.015.](/image/article/2011/MD/c1md00042j/c1md00042j-f7.gif) | ||
| Fig. 7 DNA repair of pUC19 pre-treated with cisplatin or 6. Top row: untreated pBR322 as internal control. Repair efficiency is given by the radioactivity of incorporated [α-32P]-dATP. Samples repaired by repair-proficient extracts of HeLa cells (lanes 1–4) and 518A2 cells (lanes 5–8). Cisplatin: rb ≈ 0.01; 6: rb ≈ 0.005, 0.01, and 0.015. | ||
Glutathione (GSH) plays a key role in the metabolism and mechanism of action of platinum drugs in cancer cells.20DNA damaging agents with thiophilic atoms such as platinum get frequently deactivated by the high concentrations of GSH and GSH-S-transferase in resistant tumour cells,21–23 and so depletion of GSH levels usually leads to an enhanced cytotoxicity of cisplatin24 and maybe of the complexes 3 and 6, too. The reaction of GSH with compounds 2, 3, 6, cisplatin, and dichlorido(6-aminomethylnicotinic acid)Pt(II) (=nic-Pt), the putative hydrolysis product of 3,25 was monitored by measuring the absorbance at 280 nm.26 The absorption band at this wavelength is indicative of the binding interaction of platinum complexes with the thiolate group of GSH. The progress of the reactions was slow and they were finished only after 24 hours (Fig. 8). The reaction between GSH and nic-Pt was the fastest whereas 6 reacted most slowly. The affinity of 6 to GSH was even lower than that of cisplatin, which bodes well for future tests against cisplatin-resistant tumours. Although complex 3 showed a high affinity to GSH, its cytotoxicity in GSH-rich HCT-116 cells was comparable to that observed in 518A2 cells (1 µM). However, the poor GSH-binder 6 is more active in both cell lines than the good GSH-binder 3.
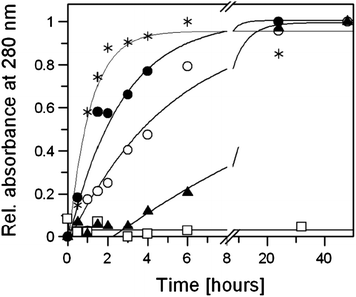 | ||
| Fig. 8 Time-dependent relative absorbance at 280 nm of the reaction of two equiv. GSH (3.33 mM) with 2 (open squares), 6 (filled triangles), cisplatin (open circles), 3 (filled circles), and nic-Pt (asterisks) in 1 mL of PBS (pH 7.4) at 37 °C. Curves obtained from mono-exponential fitting; apparent rate constants were 0.058, 0.2, 0.38 and 0.94 h−1 for 6, cisplatin, 3 and nic-Pt. | ||
In principle, GSH can bind to the metal centre or the β-carbon atom of the propenone linker of 3 and 6. In order to identify the preferred site of attack by S-nucleophiles, we monitored the reaction of 3 with an excess of N-acetyl-L-cysteine in DMF-d7 at room temperature over several days by 1H NMR spectroscopy. Under water-free conditions the reaction proceeded slowly with the peaks of 2-H (δ = 10.06 ppm) and 4-H (δ = 8.88 ppm) on the pyridine ring of 3 gradually vanishing and new signals for the corresponding protons in the N-acetylcysteine adduct cropping up at higher field at δ = 9.37 ppm for 2-H and δ = 8.61 ppm for 4-H. These results indicate an S-coordination of N-acetylcysteine to the Pt atom trans to the pyridine ring rather than an attack at the Michael system of the chalcone fragment. A similar experiment was carried out in the presence of 50% D2O which accelerated the formation of the mono-adduct (cf. ESI†).
Conclusions
The new chalconyl (1-carboxyethane-2,3-diamine)dichloridoplatinum(II) complex 6 differs advantageously, not only from cisplatin and the parent chalcone 2, but also from its close congener 3 bearing the same chalcone residue but a different stationary diamine ligand. Its superior in vitro efficacy against various cancer cell lines, its capacity to platinate DNA to a larger extent and in a way inflicting much greater unwinding and a greater number of interstrand cross-links which are less amenable to DNA repair, and its robustness against deactivation by S-nucleophiles make complex 6 a promising candidate for further anticancer tests. The decisive role of the diamino ligand leaves scope for drug optimisation. Despite the differences between 3 and 6 it becomes apparent, from the similarity of the morphological changes they inflicted in 518A2 melanoma cells and which were quite different from those caused by cisplatin, chalcone 2 or colchicine, that their mode of action synergistically combines the traits of DNA-binding platinum drugs and tubulin-binding chalcones.Materials and methods
(E)-2-Methoxy-5-[3′-oxo-3′-(3,4,5-trimethoxyphenyl)prop-1′-enyl]phenyl (D,L)-2,3-di-(tert-butoxycarbonylamino)propionate (5)
4 27 (193 mg, 0.63 mmol), DMF (2 mL), NEt3 (100 µL, 0.72 mmol), and 2,4,6-trichlorobenzoyl chloride (111 µL, 0.72 mmol) were stirred at rt for 20 min. 2 (247 mg, 0.72 mmol) and DMAP (155 mg, 1.26 mmol) in DMF/toluene (10 mL, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) were added, the mixture was stirred at rt (16 h), diluted with EtOAc, and the organic phase was washed with water, dried and concentrated. Chromatography (silica gel) left 5 (200 mg, 52%), brown solid of mp 175 °C; Rf = 0.17 (EtOAc/n-hexane 1
5) were added, the mixture was stirred at rt (16 h), diluted with EtOAc, and the organic phase was washed with water, dried and concentrated. Chromatography (silica gel) left 5 (200 mg, 52%), brown solid of mp 175 °C; Rf = 0.17 (EtOAc/n-hexane 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2); ν/cm−1 3357, 2977, 2938, 2841, 1770, 1680, 1656, 1579, 1503, 1457, 1413; 1H NMR (300 MHz, CDCl3) δ 1.3–1.4 (m, 18H), 3.6–3.8 (m, 2H), 3.8–4.0 (m, 9H), 4.5–4.6 (m, 1H), 5.3–5.4 (m, 1H), 5.6–5.7 (m, 1H), 6.90 (d, J 8.6 Hz, 1H), 7.1–7.4 (m, 5H), 7.65 ppm (d, J 15.5 Hz, 1H); 13C NMR (75 MHz, CDCl3) δ 28.1, 42.1, 54.7, 55.8, 56.1, 56.2, 60.7, 79.8, 80.1, 105.8, 112.1, 120.2, 121.4, 128.2, 129.2, 133.3, 139.4, 142.2, 143.2, 152.3, 152.9, 155.4, 156.4, 168.8, 188.6 ppm; m/z (EI) 554 (17), 344 (58), 329 (39), 207 (86), 195 (100).
2); ν/cm−1 3357, 2977, 2938, 2841, 1770, 1680, 1656, 1579, 1503, 1457, 1413; 1H NMR (300 MHz, CDCl3) δ 1.3–1.4 (m, 18H), 3.6–3.8 (m, 2H), 3.8–4.0 (m, 9H), 4.5–4.6 (m, 1H), 5.3–5.4 (m, 1H), 5.6–5.7 (m, 1H), 6.90 (d, J 8.6 Hz, 1H), 7.1–7.4 (m, 5H), 7.65 ppm (d, J 15.5 Hz, 1H); 13C NMR (75 MHz, CDCl3) δ 28.1, 42.1, 54.7, 55.8, 56.1, 56.2, 60.7, 79.8, 80.1, 105.8, 112.1, 120.2, 121.4, 128.2, 129.2, 133.3, 139.4, 142.2, 143.2, 152.3, 152.9, 155.4, 156.4, 168.8, 188.6 ppm; m/z (EI) 554 (17), 344 (58), 329 (39), 207 (86), 195 (100).
cis-{(E)-2-Methoxy-5-[3′-oxo-3′-(3,4,5-trimethoxyphenyl)prop-1′-enyl]phenyloxy-3-oxopropane-(D,L)-1,2-diamine}dichloridoplatinum(II) (6)
A mixture of 5 (190 mg, 0.30 mmol), CH2Cl2 (3 mL), and TFA (2 mL) was stirred at rt for 1 h, concentrated, and the oily residue was dissolved in THF/water. K2PtCl4 (130 mg, 0.31 mmol) dissolved in water was added, the pH was adjusted to 5 and the mixture was stirred at rt for 4 h. The product was washed with water, THF, and diethyl ether and dried in vacuum to leave 80 mg (40%) of 6; light brown solid of mp 240 °C (dec.). Anal. calcd for C22H26Cl2N2O7Pt: C, 37.94; H, 3.76; N, 4.02%. Found: C, 38.12; H, 3.81; N, 3.96%. ν/cm−1 3264, 3192, 2940, 2838, 1770, 1656, 1576, 1504, 1457, 1412, 1342, 1274, 1159, 1122, 999, 814, 766; 1H NMR (300 MHz, DMF-d7) δ 3.1–3.3 (m, 2H), 3.85 (s, 3H), 3.9–4.0 (m, 9H), 4.1–4.3 (m, 1H), 5.6–6.2 (m, 4H), 7.2–7.3 (m, 1H), 7.55 (s, 2H), 7.7–7.9 (m, 3H), 7.9–8.0 ppm (m, 1H); 13C NMR (75 MHz, DMF-d7) δ 51.0, 56.4, 60.3, 63.1, 106.6, 113.4, 121.2, 122.8, 128.6, 129.5, 133.9, 140.0, 142.8, 143.1, 153.1, 153.7, 166.8, 188.2 ppm; 195Pt NMR (64 MHz, DMF-d7) δ 2259 ppm [relative to Ξ(195Pt) = 21.4 MHz].DNA–histone ELISA apoptosis assay
The Cell Death Detection Kit (Roche, Palo Alto, CA) was used to detect apoptosis in BxPC-3 and MDA-Mb-231 cells. Briefly,28cells were treated with test compounds for 72 h, the cytoplasmic histone/DNA cell fragments were extracted and incubated in microtiter plates coated with anti-histone antibodies. Peroxidase-conjugated anti-DNA antibody was used for the detection of immobilised histone/DNA fragments followed by colour development with ABTS substrate for peroxidase. The absorbance at 405 nm was determined with a TECAN Ultra Multifunctional Microplate Reader.DNA unwinding
Unwinding of closed circular supercoiled pSP73KB plasmid DNA was assayed by an agarose gel mobility shift assay. The unwinding angle Φ per platinum–DNA adduct was calculated from the rb value at which the supercoiled to relaxed transformation of the plasmid was complete. Plasmid DNA samples were incubated with the complexes (37 °C, 24 h, dark) then precipitated with ethanol and redissolved in Tris–acetate/EDTA buffer. An aliquot of the precipitate was subjected to electrophoresis on 1% agarose gel at 25 °C with TAE buffer at 2.5 V cm−1. The gels were stained with EtdBr and photographed. Another aliquot was used to determine the rb values by AAS.Preparation of the 213-bp dsDNA fragment
The pSP73 plasmid was digested with NdeI and EcoRI. The resulting two fragments, 2473-bp and 213-bp long, were separated on 1% agarose gel in buffer containing 40 mM Tris–acetate (pH 8), 1 mM EDTA and 0.5 mM EtdBr. The 213-bp fragment (1 mg) was incubated (37 °C, 1 h) in buffer containing 50 mM NaCl, 10 mM Tris–HCl (pH 7.9), 10 mM MgCl2, 1 mM DTT, 10 mCi of [α-32P]-dATP (3000 Ci mmol−1) and 5 U of Klenow fragment (KF). Another 10 mCi of [α-32P]-dATP and 5 U of KF were added and incubated for 1 h. Unbound label molecules were removed on a Sephadex G50, the DNA was extracted with phenol/CHCl3 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), precipitated with ethanol and dissolved in 0.1 M NaClO4. It was treated with the complexes and incubated (24 h) to a specific rb.
1), precipitated with ethanol and dissolved in 0.1 M NaClO4. It was treated with the complexes and incubated (24 h) to a specific rb.
Interstrand cross-link assay
The complexes were incubated at various concentrations with 2 mg of pSP73 DNA linearised by EcoRI. The platinated samples were precipitated with ethanol and analysed for ICLs in the same way as previously described.18 The linear duplexes were first 3′-end labelled with KF of DNA polymerase I and [α-32P]-dATP. The samples were deproteinised with phenol, precipitated with ethanol, and the pellet was dissolved in 18 µL of a solution containing 30 mM NaOH, 1 mM EDTA, 6.6% sucrose, and 0.04% bromophenol blue. The intensities of the bands resulting from electrophoresis on alkaline agarose gel (1%) corresponding to single strands of DNA and ICL duplex were quantified with Molecular Dynamics PhosphorImager (Storm 860 System with Image Quantsoftware; Sunnyvale, CA).DNA repair
Repair DNA synthesis by cell-free tumour cell extracts (CFE) was assayed using the pUC19 plasmid. Each reaction (50 mL) contained 600 ng of unmodified pBR322 and 600 ng of unmodified or platinated pUC19, 2 mM ATP, 30 mM KCl, 0.05 mg mL−1creatine phosphokinase (rabbit muscle), 20 mM each of dGTP, dATP, and dTTP, 8 mM dCTP, 74 kBq of [α-32P]dCTP in a buffer composed of 40 mM HEPES–KOH, pH 7.5, 5 mM MgCl2, 0.5 mM dithiothreitol, 22 mM creatine phosphate, 1.4 mg BSA mL−1, and 20 mg of CFE from HeLa S3 or 518A2 cells. Incubation (30 °C, 3 h), termination by adding Na2H2EDTA to a final concentration of 20 mM, SDS to 0.6%, and proteinase K to 250 mg mL−1 and further incubation (20 min) left products that were extracted with phenol/CHCl3 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The DNA was precipitated from the aq. layer with 3 M NaOAc and ethanol. Incubation (−20 °C, 30 min) and centrifugation (12
1). The DNA was precipitated from the aq. layer with 3 M NaOAc and ethanol. Incubation (−20 °C, 30 min) and centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C, 0.5 h) left a pellet that was washed with 80% ethanol (0.2 mL) and dried. The DNA was linearised, submitted to electrophoresis on 1% agarose gel and stained by EtBr. The experiments were carried out in quadruplicate.
000g, 4 °C, 0.5 h) left a pellet that was washed with 80% ethanol (0.2 mL) and dried. The DNA was linearised, submitted to electrophoresis on 1% agarose gel and stained by EtBr. The experiments were carried out in quadruplicate.
Transcription mapping
A Promega kit Riboprobe® in vitroTranscription System was used. Briefly, a 212-bp DNA fragment obtained by cutting of pSP73KB DNA with NdeI and HpaI restriction endonucleases and containing a T7 RNA polymerase promotor in the upper strand close to its 3′-end was randomly modified by cisplatin at rb 0.01 at 37 °C for 24 h. The RNA synthesis was carried out at 37 °C using T7 RNA polymerase.Glutathione interaction
GSH (3.33 mM) was incubated with 1.67 mM test compound in 1 mL of PBS (8.0 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4·12H2O, 0.24 g KH2PO4 per 1 L, pH 7.4) at 37 °C. At different times over 48 h, 20 µL aliquots were withdrawn and diluted with 980 µL of PBS. The absorption spectra of the diluted samples were recorded over a range of 200–400 nm using a Beckman DU-65 spectrophotometer.Acknowledgements
This work was funded by the Deutsche Forschungsgemeinschaft (Grant Scho 402/8-3) and a scholarship from the University Bayreuth (MZ). We thank Dr Olga Novakova for AAS analyses.References
- G. R. Pettit, S. B. Singh, E. Hamel, C. M. Lin, D. S. Alberts and D. Garcia-Kendall, Experientia, 1989, 45, 209–211 CAS.
- G. C. Tron, T. Pirali, G. Sorba, F. Pagliai, S. Busacca and A. Genazzani, J. Med. Chem., 2006, 49, 3033–3044 CrossRef CAS.
- C. Kanthou and G. M. Tozer, Blood, 2002, 99, 2060–2069 CrossRef CAS.
- S. Ducki, R. Forrest, J. A. Hadfield, A. Kendall, N. J. Lawrence, A. T. McGown and D. Rennison, Bioorg. Med. Chem. Lett., 1998, 8, 1051–1056 CrossRef CAS.
- N. J. Lawrence, R. P. Patterson, L.-L. Ooi, D. Cook and S. Ducki, Bioorg. Med. Chem. Lett., 2006, 16, 5844–5848 CrossRef CAS.
- X.-L. Liu, H.-W. Tee and M.-L. Goo, Bioorg. Med. Chem., 2008, 16, 171–180 CrossRef CAS.
- C. Hirschman-Jax, A. E. Foster, G. G. Wulf, J. G. Nuchtern, T. W. Jax, U. Gobel, M. A. Goddel and M. K. Brenner, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14228–14233 CrossRef.
- S. Padhye, A. Ahmad, N. Oswal, P. Dandawate, R. A. Rub, J. Deshpande, K. V. Swamy and F. H. Sarkar, Bioorg. Med. Chem. Lett., 2010, 20, 5818–5821 CrossRef CAS.
- B. Coggiola, F. Pagliai, G. Allegrone, A. A. Genazzani and G. C. Tron, Bioorg. Med. Chem. Lett., 2005, 15, 3551–3554 CrossRef CAS.
- R. Schobert, B. Biersack, A. Dietrich, S. Knauer, M. Zoldakova, A. Fruehauf and T. Mueller, J. Med. Chem., 2009, 52, 241–246 CrossRef CAS.
- M. Zoldakova, Z. Kornyei, A. Brown, B. Biersack, E. Madarász and R. Schobert, Biochem. Pharmacol., 2010, 80, 1487–1496 CrossRef CAS.
- M. Ochoa, Jr, Ann. N. Y. Acad. Sci., 1969, 163, 921–930 CAS.
- R. Schobert, B. Biersack, A. Dietrich, A. Grotemeier, T. Müller, B. Kalinowski, S. Knauer, W. Voigt and R. Paschke, J. Med. Chem., 2007, 50, 1288–1293 CrossRef CAS.
- G. Bernhardt, B. Biersack, S. Bollwein, R. Schobert and M. Zoldakova, Chem. Biodiversity, 2008, 5, 1645–1659 CrossRef CAS.
- S. Arnould, I. Hennebelle, P. Canal, R. Bugat and S. Guichard, Eur. J. Cancer, 2003, 39, 112–119 CrossRef CAS.
- M. V. Keck and S. J. Lippard, J. Am. Chem. Soc., 1992, 114, 3386–3390 CrossRef CAS.
- A. Halamikova, O. Vrana, J. Kasparkova and V. Brabec, ChemBioChem, 2007, 8, 2008–2015 CrossRef CAS.
- V. Brabec and M. Leng, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 5345–5349 CAS.
- V. Brabec, Prog. Nucleic Acid Res. Mol. Biol., 2002, 71, 1–68 Search PubMed.
- D. M. Townsend and K. D. Tew, Oncogene, 2003, 22, 7369–7375 CrossRef CAS.
- C. C. McIlwain, D. M. Townsend and K. D. Tew, Oncogene, 2006, 25, 1639–1648 CrossRef CAS.
- A. Pompella, A. Corti, A. Paolicchi, C. Giommarelli and F. Zunino, Curr. Opin. Pharmacol., 2007, 7, 360–366 CrossRef CAS.
- S. Tsuchida and K. Sato, Crit. Rev. Biochem. Mol. Biol., 1992, 27, 337–384 CrossRef CAS.
- M. A. Fuertes, C. Alonso and J. M. Pérez, Chem. Rev., 2003, 103, 643–662.
- R. Schobert and B. Biersack, Inorg. Chim. Acta, 2005, 358, 3369–3376 CrossRef CAS.
- T. Ishikawa and F. Ali-Osman, J. Biol. Chem., 1993, 268, 20116–20125 CAS.
- S. Moradell, J. Lorenzo, A. Rovira, S. van Zutphen, F. X. Aviles, V. Moreno, R. de Llorens, M. A. Martinez, J. Reedijk and A. Llobet, J. Inorg. Biochem., 2004, 98, 1933–1946 CrossRef CAS.
- A. Ahmad, Z. Wang, R. Ali, M. Y. Maitah, D. Kong, S. Banerjee, S. Padhye and F. H. Sarkar, J. Cell. Biochem., 2010, 109, 1134–1141 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Details of microscopy, NMR kinetics, and bioassays. See DOI: 10.1039/c1md00042j |
| This journal is © The Royal Society of Chemistry 2011 |
