DOI:
10.1039/C0MD00263A
(Concise Article)
Med. Chem. Commun., 2011,
2, 486-492
Synthesis and biological evaluation of tetrazole containing compounds as possible anticancer agents
Received
15th December 2010
, Accepted 1st March 2011
First published on 29th March 2011
Abstract
A series of new tetrazole derivatives are synthesized from Baylis–Hillman allyl amines in a clean, efficient and straightforward manner. The stereochemistry of the double bond is confirmed by X-ray diffraction data. These compounds are evaluated for in vitro anticancer activity against five cancer cell lines. Most of the compounds exhibited good anticancer activity in micro molar concentration out of 16 compounds synthesized and screened. Furthermore, the compound 5o has good binding affinity to calf thymus DNA (ct DNA), as assayed by UV-vis absorption and fluorescence spectroscopic methods.
Introduction
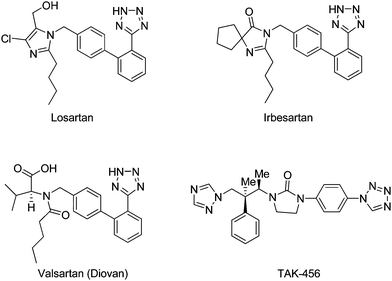
Small molecules that bind to DNA have found application in medicine, notably as cancer chemotherapeutic agents.1 These agents are also highly cytotoxic and this is attributed to their tendency to bind rather indiscriminately to DNA and any beneficial effect arises from the more rapid death rates for faster replicating cancer cells.2 Thus, it seems a reasonable proposition that agents which are able to selectively recognize specific sequences of DNA could potentially have higher therapeutic indices.3 Cancer chemotherapy has entered a new era of molecularly targeted therapeutics, which is highly selective and not associated with the serious toxicities of conventional cytotoxic drugs.4 Recently, Yang et al. and others5 invented tetrazole derivatives as promising candidates for anticancer activity. Tetrazole derivatives are well known compounds with a high level of biological activity.6 Derivatives of tetrazoles are screened for various biological activities such as antiviral, antibacterial, antifungal, antiallergic, anticonvulsant and anti-inflammatory properties.7 In drug design, tetrazoles are regarded as an isostere for the carboxylate group, and extensive work on tetrazoles has been carried out in the field of medicinal chemistry.8 Among them, 1-substituted tetrazoles have received much attention because of their wide utility.9Tetrazoles are medicinally important heterocycles that are incorporated in a large number of drugs approved by the FDA.10Losartan, irbesartan, and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundvalsartan are famous antihypertensive drugs belonging to the class of nonpeptide angiotensin-II inhibitors, and have a biphenyl tetrazolyl moiety in their structure (Fig. 1). One of the antifungal agents viz., TAK-456 also carries a tetrazole ring (Fig. 1).
Tetrazoles have also found a wide range of applications as speciality explosives, in photography, and information recording systems.11 They are important ligands for many useful transformations and also precursors for a variety of nitrogen-containing heterocycles.12 It was also noticed that toxic properties of a drug can decrease through the introduction of a tetrazole ring into the molecule.13 The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtetrazole moiety is also generally accepted to exhibit stronger resistance to in vivo metabolization than the carboxylate group, thus conferring to the corresponding drug longer lifetimes (bioavailability) in blood.14 In the field of organocatalysis,15 it is used as an attractive substitute for the carboxylic acid group to increase solubility in organic solvents. Last but not least, the tetrazole group has been investigated for its coordination properties in metal complexes.16 This initiated us constructing a system containing a tetrazole ring in the Baylis–Hillman matrix to serve as a new scaffold for the synthesis of anticancer agents. Motivated by these findings, and in continuation of our ongoing efforts endowed with the discovery of nitrogenated heterocycles17 with potential chemotherapeutic activities, it was planned to synthesize and investigate the anticancer activity of a new series of substituted tetrazoles.
Results and discussion
Chemistry
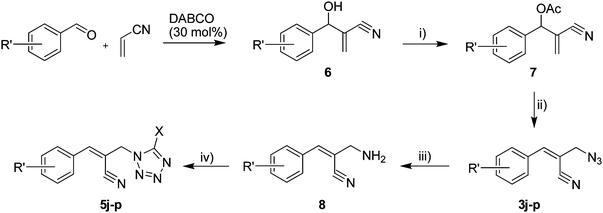
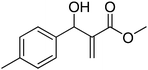
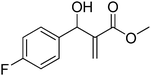
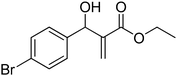
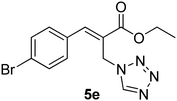
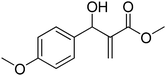
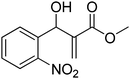
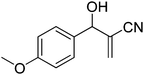
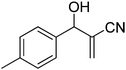
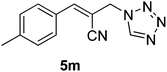
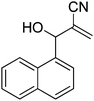
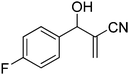
The Baylis–Hillman reaction is now a standard synthetic method to make several synthons, heterocycles and natural products.18 In connection with our project on the chemical transformations using Baylis–Hillman chemistry,17a–d we are interested in the generation of new heterocyclic entities from the Baylis–Hillman allyl amines in a synthetically practical fashion. There are only a few applications reported in the literature using allyl amines of Baylis–Hillman adducts as active substrates.19 Herein, we describe the preparation of new 1-substituted tetrazoles from allyl amines of the Baylis–Hillman adducts in an efficient manner. Synthesis of allyl amines is achieved in a convenient manner from respective azides20 derived from acetates21 of Baylis–Hillman adducts using Zn/NH4Cl22 (Scheme 1 & 2). The allyl amines obtained are directly used for the protocol of targeted tetrazoles.
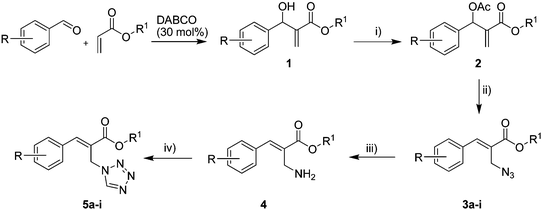 |
| | Scheme 1 Reagents and conditions. (i) CH3COCl, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundPyridine, DCM, 0 °C–rt, (ii) NaN3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMSO, rt, (iii) Zn, NH4Cl, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundEtOH/COMPOUND LINKS
Read more about this on ChemSpider
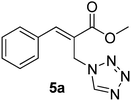
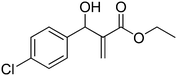
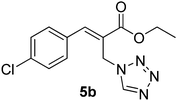
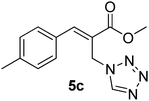
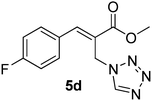
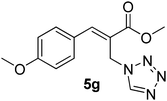
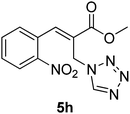
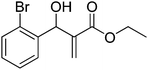
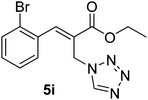
Download mol file of compoundH2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), rt; (iv) HC(OEt)3, NaN3 AcOH, 90 °C R = H, R1 = Me (5a); R = p-Cl, R1 = Et (5b); R = p-Me, R1 = Me (5c); R = p-F, R1 = Me (5d); R = p-Br, R1 = Et (5e); R = p-NO2, R1 = Me (5f); R = p-OMe, R1 = Me (5g); R = o-NO2, R1 = Me (5h); R = o-Br, R1 = Et (5i). 1), rt; (iv) HC(OEt)3, NaN3 AcOH, 90 °C R = H, R1 = Me (5a); R = p-Cl, R1 = Et (5b); R = p-Me, R1 = Me (5c); R = p-F, R1 = Me (5d); R = p-Br, R1 = Et (5e); R = p-NO2, R1 = Me (5f); R = p-OMe, R1 = Me (5g); R = o-NO2, R1 = Me (5h); R = o-Br, R1 = Et (5i). | |
 |
| | Scheme 2 Reagents and conditions. (i) CH3COCl, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundPyridine, DCM, 0 °C–rt, (ii) NaN3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMSO, rt, (iii) Zn, NH4Cl, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundEtOH/COMPOUND LINKS
Read more about this on ChemSpider
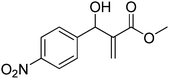
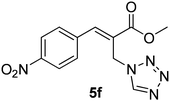
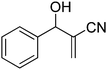
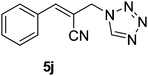
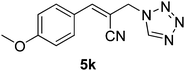
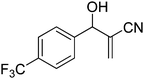
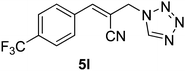
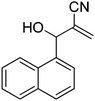
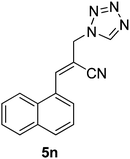
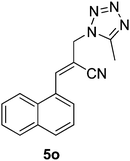
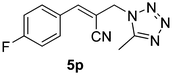
Download mol file of compoundH2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), rt; (iv) HC(OEt)3/CH3C(OEt)3, NaN3 AcOH, 90 °C R′ = H, X = H (5j); R′ = p-OMe, X = H (5k); R′ = p-CF3, X = H (5l); R′ = p-Me, X = H (5m); R′ = Nap, X = H (5n); R′ = Nap, X = Me (5o); R′ = p-F, X = Me (5p). 1), rt; (iv) HC(OEt)3/CH3C(OEt)3, NaN3 AcOH, 90 °C R′ = H, X = H (5j); R′ = p-OMe, X = H (5k); R′ = p-CF3, X = H (5l); R′ = p-Me, X = H (5m); R′ = Nap, X = H (5n); R′ = Nap, X = Me (5o); R′ = p-F, X = Me (5p). | |
The amines are treated with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtriethyl orthoformate and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundsodium azide in the presence of acetic acid23 to get the dipolar cycloaddition product, 1-substituted 1H-1,2,3,4-tetrazole. 1,5-Disubstituted tetrazoles are obtained while using COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtriethyl orthoacetate instead of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtriethyl orthoformate. The various tetrazole compounds prepared are arranged in Table 1. Thus synthesized tetrazoles were screened for in vitro anticancer activity against five cancer cell lines and promising results are obtained.
Stereochemistry of obtained tetrazoles having ester functionality is predicted as E and nitrile functionality as Z-configuration. The discussion of stereochemistry is started with the formation of azides from acetates of Baylis–Hillman adducts. The stereochemistry is continued/retained from the azides to amines and then retained to the last heterocyclization step. There are a few reports in the literature regarding preparation of azides from Baylis–Hillman adducts and their stereochemistry.20 Following their reports and according to the 1H NMR data of the prepared azides, we finally envisaged that the stereochemistry of ester containing functional groups is E and nitrile containing functional groups is Z. The configuration of double bond and structure is also confirmed by single X-ray crystallographic data24 for the compounds 5h and 5m (Fig. 2).
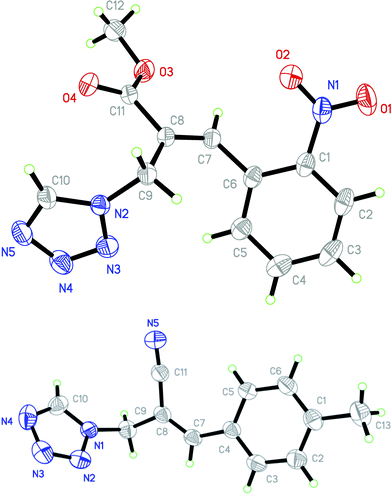 |
| | Fig. 2 ORTEP diagrams of 5h and 5m. | |
Biological evaluation
The origin of the cell lines used in the current study and the culture media are fully detailed in reference 25. The in vitro cytotoxicity of tetrazole derivatives were determined in selected human cancer cell lines of liver carcinoma (Hep-G2), lung adenocarcinoma (A-549), breast (MDA-MB-231), prostate carcinoma (DU-145), neuroblastoma (SK-N-SH) origin. We have evaluated the IC50in vitrogrowth inhibitory values of the compounds under study by means of the sulforhodamine B (SRB) colorimetric assay.26 The cells were grown in MEM (Minimum Essential Medium) containing 10% fetal bovine serum. Cells were seeded at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/well in 96-well microtiter plates. These 96-well microtiter plates were incubated at 37 °C, 5% CO2 and 100% relative humidity for 24 h prior to addition of experimental drugs. Test compounds were added to the appropriate microtiter wells to get the required final drug concentrations. Plates were incubated further for 48 h, and the assay was terminated by the addition of 30 μL of cold COMPOUND LINKS
000 cells/well in 96-well microtiter plates. These 96-well microtiter plates were incubated at 37 °C, 5% CO2 and 100% relative humidity for 24 h prior to addition of experimental drugs. Test compounds were added to the appropriate microtiter wells to get the required final drug concentrations. Plates were incubated further for 48 h, and the assay was terminated by the addition of 30 μL of cold COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtrichloroacetic acid (TCA) (final concentration, 10% TCA) and plates were again incubated for 60 min at 40 °C. The plates were washed five times with distilled COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater and air-dried. SRB solution (50 μL) at 0.4% (w/v) in 1% COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetic acid was added to each of the wells, and plates were incubated for 20 min at room temperature. The residual dye was removed by washing five times with 1% COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetic acid. The plates were air-dried. Bound stain was subsequently eluted with 10 mM COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundTris base, and the absorbance was read on an ELISA plate reader at a wavelength of 540 nm. Percent growth was calculated on a plate-by-plate basis for test wells relative to control wells.
The IC50 values of all the 16 compounds are listed in the Table 2. The data in Table 2 also reveal that out of 16 compounds under study, the compounds 5b, 5f, 5l and 5o displayed the highest in vitro anticancer activity (1.0–4.0 μM) on both liver carcinoma (Hep G2) and lung adenocarcinoma (A 549) cell lines. Compounds 5e, 5f, 5g and 5o displayed significant activity (4.0–7.0μM) against prostate (DU 145) cancer cell line. The substituents having p-trifluoromethyl, p-chloro and p-nitro on the phenyl ring and 1,5-disubstituted tetrazole moiety displayed more activity compared to other compounds. All the compounds have shown moderate (15.0–70.0 μM) to good (7.0–15.0 μM) anticancer activity on both breast and neuronal cell lines.
| |
Cell lines (IC50 values; μM ± SDa) |
| Compound |
HepG2 |
A549
|
MDA-MB-231
|
DU145
|
SK-N-SH
|
|
SD: Standard deviation. All experiments were independently performed three times.
|
|
5a
|
10.73 ± 1.2 |
7.91 ± 0.94 |
16.35 ± 1.8 |
9.02 ± 1.14 |
31.6 + 5.43 |
|
5b
|
3.0 ± 0.34 |
3.18 ± 0.43 |
23.9 ± 2.21 |
8.44 + 0.76 |
28.2 ± 3.5 |
|
5c
|
10.73 ± 1.2 |
16.03 ± 2.1 |
62.64 ± 1.4 |
8.42 + 0.93 |
71.67 ± 1.3 |
|
5d
|
12.07 ± 1.25 |
6.9 ± 0.59 |
15.9 ± 1.3 |
8.57 ± 0.82 |
18.33 ± 2.4 |
|
5e
|
4.89 ± 0.6 |
4.78 ± 0.64 |
18.94 ± 2.04 |
5.5 ± 0.64 |
27.34 ± 3.37 |
|
5f
|
2.07 ± 0.41 |
2.84 ± 0.9 |
9.14 ± 1.26 |
5.5 ± 0.84 |
22.26 ± 1 9 |
|
5g
|
6.65 ± 0.87 |
3.83 ± 1.2 |
21.03 ± 2.41 |
5.6 ± 0.48 |
7.9 ± 1.3 |
|
5h
|
4.33 ± 0.65 |
6.88 ± 0.84 |
43.67 ± 5.2 |
10.18 ± 1.23 |
19.82 + 1.7 |
|
5i
|
3.33 ± 0.42 |
5.94 ± 0.49 |
14.78 ± 1.84 |
7.79 ± 0.87 |
16.26 ± 2.1 |
|
5j
|
5.83 ± 0.43 |
9.99 ± 1.32 |
18.35 ± 2.1 |
9.45 ± 1.3 |
23.26 ± 3.02 |
|
5k
|
27.39 ± 3.7 |
23.79 ± 2.86 |
20.3 ± 1.9 |
19.41 ± 2.37 |
64.2 ± 2.1 |
|
51
|
1.65 ± 0.42 |
3.51 ± 0.58 |
8.7 ± 1.05 |
5.92 ± 0.54 |
11.79 ± 1.54 |
|
5m
|
6.56 ± 0.82 |
7.95 ± 0.93 |
25.65 ± 2.38 |
7.3 ± 0.85 |
20.12 ± 2.7 |
|
5n
|
7.11 ± 0.85 |
5.7 ± 0.63 |
31.4 ± 3.42 |
7.36 ± 0.91 |
16.86 ± 1.95 |
|
5o
|
2.60 + 0.61 |
2.95 ± 0.52 |
14.1 ± 1.62 |
5.76 ± 0.68 |
13.7 ± 1.5 |
|
5p
|
7.29 ± 0.94 |
6.4 ± 0.82 |
28.76 ± 3.2 |
7.19 ± 0.62 |
20.16 ± 2.8 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundNocodazole
|
<0.1 |
<0.1 |
<0.1 |
<0.1 |
<0.1 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundPodophyllotoxin
|
<0.1 |
<0.1 |
<0.1 |
<0.1 |
<0.1 |
DNA binding studies
DNA binding studies are important for the rational design and construction of new and more efficient drugs targeted to DNA.27 A variety of small molecules interact reversibly with double-stranded DNA, primarily through three modes: (i) electrostatic interactions with the negatively charged nucleic sugar–phosphate structures, which are along the external DNA double helix and do not possess selectivity; (ii) binding interactions with two grooves of DNA double helix; and (iii) intercalation between the stacked base pairs of native DNA. Heterocyclic dyes, such as ethidium, anthracyclines, phenothiazines and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacridine derivatives interact through intercalation with the planar, aromatic group stacked between base pairs.28 To explore the binding affinity of the tetrazole to DNA, we have planned the following experiments.
Absorption spectroscopy is one of the most convenient tools to investigate the interaction between drugs and DNA. In order to address the DNA binding ability of this tetrazole derivative, the UV-vis absorption spectral titration of the compound 5o with ct DNA in Tris-HCl buffer was performed. Representative spectra of the compound in the absence and presence of ct DNA is given in Fig. 3 (a). The tetrazole compound showed a clear decrease in the absorption intensity in the presence of ct DNA, while DNA did not absorb light in this region. This hypochromic effect (at λmax 314 nm) indicated that this compound can bind with DNA and form stable complexes through the interactions of the tetrazole chromophore and DNA. Additionally, during the UV-vis absorption titrations with ct DNA, one well-resolved isosbestic point at 301 nm was also observed (Fig. 3 (a)). The intrinsic binding constant (Kb) for the tetrazole 5o with DNA was determined by the half-reciprocal plot method29 (Fig. 3(b)) and found to be 0.75 × 105 M−1.
![(a) UV-vis absorption spectra of tetrazole 5o in the presence of increasing amounts of ct DNA: [DNA] = 0, 10, 20, 40, 60, 80, 100 μM. (b) The plot of D/Δεap as a function of DNA concentration as determined from the absorption spectral data.](/image/article/2011/MD/c0md00263a/c0md00263a-f3.gif) |
| | Fig. 3 (a) UV-vis absorption spectra of tetrazole 5o in the presence of increasing amounts of ct DNA: [DNA] = 0, 10, 20, 40, 60, 80, 100 μM. (b) The plot of D/Δεap as a function of DNA concentration as determined from the absorption spectral data. | |
Fluorescence studies
In order to confirm the interaction of compound 5o with ct DNA, the binding properties were also investigated by the fluorescence spectroscopy. The compound naphthyl tetrazole 5o in buffer solution has fluorescence at 432 nm. Therefore one can directly perform a DNA binding affinity experiment by observing the intrinsic fluorescence of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtetrazole (4 × 10−5 mol L−1) at 432 nm in the absence or presence of DNA. Fig. 4 shows fluorescence emission spectra (λexc 315 nm) of 5o at various ct DNA concentrations. We found that the fluorescence intensities at 432 nm increase gradually as the ct DNA concentration increases (Fig. 4). This phenomenon indicated the interaction between the compound 5o and ct DNA. The stronger enhancement of intensity may be due to the increase of the molecular planarity of the compound and the decrease of the collisional frequency of the solvent molecules.30 The results of the above absorption and fluorescence experiments indicate that the compound is interacting with the DNA.
Conclusions
In conclusion, we have disclosed efficient reaction conditions for the synthesis of new tetrazole derivatives from Baylis–Hillman allyl amines. The compounds were found to exert cytotoxic activity on the five human cancer cell lines. Compounds 5b, 5f, 5l, and 5o are particularly more active than remaining compounds against liver carcinoma (Hep G2) and lung adenocarcinoma (A 549) cancer cell lines. Compounds 5e, 5f, 5g and 5o displayed significant activity against prostate (DU 145) cancer cell line. The hit compound 5o could bind to DNA and form a stable complex and may act as a potential genotoxic agent for cancer therapy. Overall, these compounds are promising agents since only a very few tetrazole derivatives are described in the literature as having anticancer activity. Efforts are continuing to establish the mechanism of action, modify the structures and determine the exact mode of binding with ct DNA.
Acknowledgements
The authors thank the Director, IICT and Head, Organic-II Division for encouragement. This work is the Main Lab Project of IICT. CHNSSP and AS thank CSIR, New Delhi for research fellowship. Dr Shasivardhan Kalivendi, IICT is gratefully acknowledged for the cytotoxic data.
Notes and references
-
(a) L. H. Hurley, J. Med. Chem., 1989, 32, 2027 CrossRef CAS;
(b) G. Damia and M. Broggini, Eur. J. Cancer, 2004, 40, 2550 CrossRef CAS;
(c) R. Langer, Science, 2001, 293, 58 CrossRef CAS.
- A. Di Marco, Antibiot. Chemother., 1978, 23, 216 Search PubMed.
- P. B. Dervan, Science, 1986, 232, 464 CrossRef CAS.
- L. Seymore, Cancer Treat. Rev., 1999, 25, 301 CrossRef.
-
(a)
R.-Y. Yang, S. M. Ali, M. A. Ashwell, E. Kelleher, R. Palma and N. Westlund, US Patent, 2009, 0130117A1;
(b) V. H. Bhaskar and P. B. Mohite, J. Optoelectron. Biomed. Mater., 2010, 2, 249 Search PubMed.
-
(a) B. Schmidt and B. Schieffcr, J. Med. Chem., 2003, 46, 2261 CrossRef CAS;
(b) A. A. Bekhit, O. A. El-Sayed, E. Aboulmagd and J. Y. Park, Eur. J. Med. Chem., 2004, 39, 249 CrossRef CAS.
-
(a) V. V. Zarubaev, E. L. Golod, P. M. Anfimov, A. A. Shtro, V. V. Saraev, A. S. Gavrilov, A. V. Logvinov and O. I. Kiselev, Bioorg. Med. Chem., 2010, 18, 839 CrossRef CAS;
(b) Sankyo Co.Ltd., Jpn. Kokai Tokkyo Koho 80 108, 878 (C1 C07D 501/57), 1980, Chem. Abstr., 1981, 94, 30774;
(c)
T. D. Connor, A. P. Young and M. V. Strandtmann, US Patent, 1980, 4, 225, 722; T. D. Connor, A. P. Young and M. V. Strandtmann, Chem. Abstr., 1981, 94, 84137b Search PubMed;
(d) C. L. Mitchell, Toxicol. Appl. Pharmacol., 1964, 6, 23 CrossRef CAS;
(e) K. Raman, S. S. Parmar and S. P. Singh, J. Heterocycl. Chem., 1980, 17, 1137 CrossRef CAS.
- For reviews on the chemistry of tetrazoles, see:
(a) L. V. Myznikov, A. Hrabalek and G. I. Koldobskii, Chem. Heterocycl. Compd., 2007, 43, 1 CrossRef CAS;
(b)
H. R. Meier, H. Heimgartner, In Methoden der Organischen Chemie (Houben-Weyl); Schumann, E., Ed.; Georg Thieme: Stuttgart, 1994, Vol. E8d, 664 Search PubMed;
(c)
R. N. Butler, in Comprehensive Heterocyclic Chemistry; A. R. Katritzky, C. W. Rees, ed.; Pergamon: Oxford, 1984; Vol. 5, 791 Search PubMed.
-
(a) H. Singh, A. S. Chawla, V. K. Kapoor, D. Paul and R. K. Malhotra, Prog. Med. Chem., 1980, 17, 151 CAS;
(b) M. J. Genin, D. A. Allwine, D. J. Anderson, M. R. Barbachyn, D. E. Emmert, S. A. Garmon, D. R. Graber, K. C. Grega, J. S. Hester, D. K. Hutchinson, J. Morris, R. J. Reischer, C. W. Ford, G. E. Zurenko, J. C. Hamel, R. D. Schaadt, D. Stapert and B. H. Yagi, J. Med. Chem., 2000, 43, 953 CrossRef CAS;
(c) P. Ward, D. R. Armour, D. E. Bays, B. Evans, G. M. P. Giblin, N. Hernon, T. Hubbard, K. Liang, D. Middlemiss, J. Mordaunt, A. Naylor, N. A. Pegg, M. V. Vinader, S. P. Watson, C. Bountra and D. C. Evans, J. Med. Chem., 1995, 38, 4985 CrossRef CAS.
-
(a) A. Rajasekaran and P. P. Thampi, Eur. J. Med. Chem., 2004, 39, 273 CrossRef CAS;
(b) A. P. Kozikowski, J. Zhang, F. Nan, P. A. Petukhov, E. Grajkowska, J. T. Wroblewski, T. Yamamoto, T. Bzdega, B. Wroblewska and J. Neale, J. Med. Chem., 2004, 47, 1729 CrossRef CAS.
- R. S. Upadhayaya, S. Jain, N. Sinha, N. Kishore, R. Chandra and S. K. Arora, Eur. J. Med. Chem., 2004, 39, 579 CrossRef CAS.
- A.-M. Faucher, P. W. White, C. Brochu, C. Grand-Maitre, J. Rancourt and G. Fazal, J. Med. Chem., 2004, 47, 18 CrossRef CAS.
- A. A. Bekhit, O. A. El-Sayed, T. A. K. Al-Allaf, H. Y. Aboul-Enein, M. Kunhi, S. M. Pulicat, K. Al-Hussain, F. Al-Khodairy and J. Arif, Eur. J. Med. Chem., 2004, 39, 499 CrossRef CAS.
-
(a) R. J. Herr, Bioorg. Med. Chem., 2002, 10, 3379 CrossRef CAS;
(b) S. J. Wittenberger, Org. Prep. Proced. Int., 1994, 26, 499 CrossRef CAS.
-
(a) A. J. A. Cobb, D. M. Shaw and S. V. Ley, Synlett, 2004, 558 CAS;
(b) V. Wascholowski, K. R. Knudsen, C. E. T. Mitchell and S. V. Ley, Chem.–Eur. J., 2008, 14, 6155 CrossRef CAS.
- P. N. Gaponik, S. V. Voitekhovich and O. A. Ivashkevich, Russ. Chem. Rev., 2006, 75, 507 Search PubMed.
-
(a) P. Narender, M. Ravinder, P. S. Sadhu, B. China Raju, Ch. Ramesh and V. Jayathirtha Rao, Helv. Chim. Acta, 2009, 92, 959 CrossRef CAS;
(b) M. Ravinder, P. S. Sadhu and V. Jayathirtha Rao, Tetrahedron Lett., 2009, 50, 4229 CrossRef CAS;
(c) M. Ravinder, P. S. Sadhu, A. Santhoshi, P. Narender, G. Y. S. K. Swamy, K. Ravi Kumar and V. Jayathirtha Rao, Synthesis, 2010, 573 CAS;
(d) P. Narender, B. Gangadasu, M. Ravinder, U. Srinivas, G. Y. S. K. Swamy, K. Ravikumar and V. Jayathirtha Rao, Tetrahedron, 2006, 62, 954 CrossRef CAS;
(e) P. Narender, U. Srinivas, M. Ravinder, B. Anand Rao, Ch. Ramesh, K. Harakishore, B. Gangadasu, U. S. N. Murthy and V. Jayathirtha Rao, Bioorg. Med. Chem., 2006, 14, 4600 CrossRef CAS;
(f) C.h. Srinivas, C.h. N. S. S. P. Kumar, B. China Raju, V. Jayathirtha Rao, V. G. M. Naidu, S. Ramakrishna and P. V. Diwan, Bioorg. Med. Chem. Lett., 2009, 19, 5915 CrossRef CAS.
-
(a) D. Basavaiah, A. J. Rao and T. Satyanarayana, Chem. Rev., 2003, 103, 811 CrossRef CAS;
(b) V. Sing and S. Batra, Tetrahedron, 2008, 64, 4511 CrossRef CAS;
(c) K. Y. Lee, S. Gowrisankar and J. N. Kim, Bull. Korean Chem. Soc., 2005, 26, 1481 CrossRef CAS;
(d) D. Basavaiah, P. S. Rao and R. S. Hyma, Tetrahedron, 1996, 52, 8001 CrossRef CAS.
-
(a) S. Nag, S. Madapa and S. Batra, Synthesis, 2008, 101 CAS;
(b) S. Nag, R. Pathak, M. Kumar, P. K. Shukla and S. Batra, Bioorg. Med. Chem. Lett., 2006, 16, 3824 CrossRef CAS;
(c) R. Pathak and S. Batra, Tetrahedron, 2007, 63, 9448 CrossRef CAS.
- M. Sa Marcus, M. D. Ramos and L. Fernadez, Tetrahedron, 2006, 62, 11652 CrossRef CAS and references cited therein.
- D. Basavaiah, M. Krishnamacharyulu, R. S. Hyma, P. K. S. Sarma and N. Kumaragurubaran, J. Org. Chem., 1999, 64, 1197 CrossRef CAS.
- W. Lin, X. Zhang, Z. He, Y. Jin, L. Gong and A. Mi, Synth. Commun., 2002, 32, 3279 CrossRef CAS.
- P. N. Gaponik, V. P. Karavai and Y. V. Grigor'ev, Chem. Heterocycl. Compd., 1985, 21, 1255 CrossRef.
-
(a)
Bruker, 2001. SAINT (Version 6.28a) & SMART (Version 5.625). Bruker AXS Inc., Madison, Wisconsin, USA Search PubMed;
(b) G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112 ; Crystal data for 5h. C12H11N5O4. M = 289.26, triclinic, space groupP
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , a = 6.6504(6) Å, b = 6.7965(6) (7) Å, c = 14.9061(13)Å, α = 94.940(1)°, β = 90.556(1)°, γ = 103.864(1)°, V = 651.37(10) Å3, Z = 2, Dc = 1.475 Mg m−3, λ = 0.71073Å; μ(Mo Kα) = 0.114 mm−1, F000 = 300, T = 294(2) K. Total number of measured reflections is 6267. Final refinement to convergence on F2 gave R = 0.0347 (2079 obs. data only) and Rw = 0.0916, GOF = 1.044. CCDC reference number 803412.
Crystal data for 5m. C12H11N5. M = 225.26, monoclinic, space groupP21/c, a = 13.2759(14) Å, b = 6.9131(7) Å, c = 12.9123(13) Å, β = 93.332(2)°, V = 1183.1(2) Å3, Z = 4, Dc = 1.265 Mg m−3, λ = 0.71073Å; μ(Mo Kα) = 0.082 mm−1, F000 = 292, T = 294(2) K. Total number of measured reflections is 4672. Final refinement to convergence on F2 gave R = 0.0377 (1785 obs. data only) and Rw = 0.1136, GOF = 1.064. CCDC reference number 803411.
, a = 6.6504(6) Å, b = 6.7965(6) (7) Å, c = 14.9061(13)Å, α = 94.940(1)°, β = 90.556(1)°, γ = 103.864(1)°, V = 651.37(10) Å3, Z = 2, Dc = 1.475 Mg m−3, λ = 0.71073Å; μ(Mo Kα) = 0.114 mm−1, F000 = 300, T = 294(2) K. Total number of measured reflections is 6267. Final refinement to convergence on F2 gave R = 0.0347 (2079 obs. data only) and Rw = 0.0916, GOF = 1.044. CCDC reference number 803412.
Crystal data for 5m. C12H11N5. M = 225.26, monoclinic, space groupP21/c, a = 13.2759(14) Å, b = 6.9131(7) Å, c = 12.9123(13) Å, β = 93.332(2)°, V = 1183.1(2) Å3, Z = 4, Dc = 1.265 Mg m−3, λ = 0.71073Å; μ(Mo Kα) = 0.082 mm−1, F000 = 292, T = 294(2) K. Total number of measured reflections is 4672. Final refinement to convergence on F2 gave R = 0.0377 (1785 obs. data only) and Rw = 0.1136, GOF = 1.064. CCDC reference number 803411.
-
(a) E. Van Quaquebeke, G. Simon, A. Andre, J. Dewelle, M. E. Yazidi, F. Bruyneel, J. Tuti, O. Nacoulma, P. Guissou, C. Decaestecker, J. C. Braekman, R. Kiss and F. Darro, J. Med. Chem., 2005, 48, 849 CrossRef;
(b) L. Ingrassia, F. Lefranc, J. Dewelle, L. Pottier, V. Mathieu, S. Spiegl-Kreinecker, S. Sauvage, M. El Yazidi, M. Dehoux, W. Berger, E. Van Quaquebeke and R. Kiss, J. Med. Chem., 2009, 52, 1100 CrossRef CAS;
(c) D. Lamoral-Theys, A. Andolfi, G. Van Goietsenoven, A. Cimmino, B. Le Calvé, N. Wauthoz, V. Mégalizzi, T. Gras, C. Bruyère, J. Dubois, V. Mathieu, A. Kornienko, R. Kiss and A. Evidente, J. Med. Chem., 2009, 52, 6244 CrossRef CAS.
-
(a) V. Vichai and K. Kirtikara, Sulforhodamine B colorimetric assay for cytotoxicity screening, Nat. Protoc., 2006, 1, 1112 CrossRef CAS;
(b) P. Houghton, R. Fang, I. Techatanawat, G. Steventon, P. J. Hylands and C. C. Lee, Methods, 2007, 42, 377 CrossRef CAS.
-
M. J. Waring, in: G. C. K. Roberts (Ed.), Drug Action at the Molecular Level, Macmillan, London, 1977, 167 Search PubMed.
-
(a) C. Gao, F. Liu, X. Luan, C. Tan, H. Liu, Y. Xie, Y. Jin and Y. Jiang, Bioorg. Med. Chem., 2010, 18, 7507 CrossRef CAS;
(b) M. J. Waring, J. Mol. Biol., 1965, 13, 269 CrossRef CAS;
(c) L. P. G. Wakelin, G. J. Atwell, G. W. Rewcastle and W. A. Denny, J. Med. Chem., 1987, 30, 855 CrossRef CAS;
(d) J. B. Chaires, N. Duttagupta and D. M. Crothers, Biochemistry, 1982, 21, 3933 CrossRef CAS.
-
(a) S. Bhattacharya and S. S. Mandal, Chem. Commun., 1996, 1515 RSC;
(b) P. Chaudhuri, H. K. Majumder and S. Bhattacharya, J. Med. Chem., 2007, 50, 2536 CrossRef CAS.
- C.-M. Che, M. Yang, K.-H. Wong, H.-L. Chan and W. Lam, Chem.–Eur. J., 1999, 5, 3350 CrossRef CAS.
|
| This journal is © The Royal Society of Chemistry 2011 |
Click here to see how this site uses Cookies. View our privacy policy here. 
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), rt; (iv) HC(OEt)3, NaN3 AcOH, 90 °C R = H, R1 = Me (5a); R = p-Cl, R1 = Et (5b); R = p-Me, R1 = Me (5c); R = p-F, R1 = Me (5d); R = p-Br, R1 = Et (5e); R = p-NO2, R1 = Me (5f); R = p-OMe, R1 = Me (5g); R = o-NO2, R1 = Me (5h); R = o-Br, R1 = Et (5i).
1), rt; (iv) HC(OEt)3, NaN3 AcOH, 90 °C R = H, R1 = Me (5a); R = p-Cl, R1 = Et (5b); R = p-Me, R1 = Me (5c); R = p-F, R1 = Me (5d); R = p-Br, R1 = Et (5e); R = p-NO2, R1 = Me (5f); R = p-OMe, R1 = Me (5g); R = o-NO2, R1 = Me (5h); R = o-Br, R1 = Et (5i).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), rt; (iv) HC(OEt)3/CH3C(OEt)3, NaN3 AcOH, 90 °C R′ = H, X = H (5j); R′ = p-OMe, X = H (5k); R′ = p-CF3, X = H (5l); R′ = p-Me, X = H (5m); R′ = Nap, X = H (5n); R′ = Nap, X = Me (5o); R′ = p-F, X = Me (5p).
1), rt; (iv) HC(OEt)3/CH3C(OEt)3, NaN3 AcOH, 90 °C R′ = H, X = H (5j); R′ = p-OMe, X = H (5k); R′ = p-CF3, X = H (5l); R′ = p-Me, X = H (5m); R′ = Nap, X = H (5n); R′ = Nap, X = Me (5o); R′ = p-F, X = Me (5p).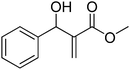

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/well in 96-well microtiter plates. These 96-well microtiter plates were incubated at 37 °C, 5% CO2 and 100% relative humidity for 24 h prior to addition of experimental drugs. Test compounds were added to the appropriate microtiter wells to get the required final drug concentrations. Plates were incubated further for 48 h, and the assay was terminated by the addition of 30 μL of cold trichloroacetic acid (TCA) (final concentration, 10% TCA) and plates were again incubated for 60 min at 40 °C. The plates were washed five times with distilled water and air-dried. SRB solution (50 μL) at 0.4% (w/v) in 1% acetic acid was added to each of the wells, and plates were incubated for 20 min at room temperature. The residual dye was removed by washing five times with 1% acetic acid. The plates were air-dried. Bound stain was subsequently eluted with 10 mM Tris base, and the absorbance was read on an ELISA plate reader at a wavelength of 540 nm. Percent growth was calculated on a plate-by-plate basis for test wells relative to control wells.
000 cells/well in 96-well microtiter plates. These 96-well microtiter plates were incubated at 37 °C, 5% CO2 and 100% relative humidity for 24 h prior to addition of experimental drugs. Test compounds were added to the appropriate microtiter wells to get the required final drug concentrations. Plates were incubated further for 48 h, and the assay was terminated by the addition of 30 μL of cold trichloroacetic acid (TCA) (final concentration, 10% TCA) and plates were again incubated for 60 min at 40 °C. The plates were washed five times with distilled water and air-dried. SRB solution (50 μL) at 0.4% (w/v) in 1% acetic acid was added to each of the wells, and plates were incubated for 20 min at room temperature. The residual dye was removed by washing five times with 1% acetic acid. The plates were air-dried. Bound stain was subsequently eluted with 10 mM Tris base, and the absorbance was read on an ELISA plate reader at a wavelength of 540 nm. Percent growth was calculated on a plate-by-plate basis for test wells relative to control wells.
![(a) UV-vis absorption spectra of tetrazole 5o in the presence of increasing amounts of ct DNA: [DNA] = 0, 10, 20, 40, 60, 80, 100 μM. (b) The plot of D/Δεap as a function of DNA concentration as determined from the absorption spectral data.](/image/article/2011/MD/c0md00263a/c0md00263a-f3.gif)
![Fluorescence emission spectra of tetrazole 5o in the presence of increasing amounts of ct DNA: [DNA] = 0, 50, 100, 150, 200 μM after excitation at λexc 315 nm.](/image/article/2011/MD/c0md00263a/c0md00263a-f4.gif)
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , a = 6.6504(6) Å, b = 6.7965(6) (7) Å, c = 14.9061(13)Å, α = 94.940(1)°, β = 90.556(1)°, γ = 103.864(1)°, V = 651.37(10) Å3, Z = 2, Dc = 1.475 Mg m−3, λ = 0.71073Å; μ(Mo Kα) = 0.114 mm−1, F000 = 300, T = 294(2) K. Total number of measured reflections is 6267. Final refinement to convergence on F2 gave R = 0.0347 (2079 obs. data only) and Rw = 0.0916, GOF = 1.044. CCDC reference number 803412.
Crystal data for 5m. C12H11N5. M = 225.26, monoclinic, space groupP21/c, a = 13.2759(14) Å, b = 6.9131(7) Å, c = 12.9123(13) Å, β = 93.332(2)°, V = 1183.1(2) Å3, Z = 4, Dc = 1.265 Mg m−3, λ = 0.71073Å; μ(Mo Kα) = 0.082 mm−1, F000 = 292, T = 294(2) K. Total number of measured reflections is 4672. Final refinement to convergence on F2 gave R = 0.0377 (1785 obs. data only) and Rw = 0.1136, GOF = 1.064. CCDC reference number 803411.
, a = 6.6504(6) Å, b = 6.7965(6) (7) Å, c = 14.9061(13)Å, α = 94.940(1)°, β = 90.556(1)°, γ = 103.864(1)°, V = 651.37(10) Å3, Z = 2, Dc = 1.475 Mg m−3, λ = 0.71073Å; μ(Mo Kα) = 0.114 mm−1, F000 = 300, T = 294(2) K. Total number of measured reflections is 6267. Final refinement to convergence on F2 gave R = 0.0347 (2079 obs. data only) and Rw = 0.0916, GOF = 1.044. CCDC reference number 803412.
Crystal data for 5m. C12H11N5. M = 225.26, monoclinic, space groupP21/c, a = 13.2759(14) Å, b = 6.9131(7) Å, c = 12.9123(13) Å, β = 93.332(2)°, V = 1183.1(2) Å3, Z = 4, Dc = 1.265 Mg m−3, λ = 0.71073Å; μ(Mo Kα) = 0.082 mm−1, F000 = 292, T = 294(2) K. Total number of measured reflections is 4672. Final refinement to convergence on F2 gave R = 0.0377 (1785 obs. data only) and Rw = 0.1136, GOF = 1.064. CCDC reference number 803411.