Online parallel fragment screening and rapid hit exploration for nicotinic acetylcholine receptors†
Gerdien E.
de Kloe‡
a,
Jeroen
Kool‡
*b,
Rene
van Elk
c,
Jacqueline E.
van Muijlwijk-Koezen
a,
August B.
Smit
c,
Henk
Lingeman
a,
Hubertus
Irth
b,
Wilfried M. A.
Niessen
b and
Iwan J. P.
de Esch
a
aLeiden/Amsterdam Center for Drug Research (LACDR), Division of Medicinal Chemistry, Faculty of Sciences, VU University Amsterdam, De Boelelaan 1083, 1081 HV, Amsterdam, The Netherlands
bLeiden/Amsterdam Center for Drug Research (LACDR), Division of BioMolecular Analysis, Faculty of Sciences, VU University Amsterdam, De Boelelaan 1083, 1081 HV, Amsterdam, The Netherlands. E-mail: j.kool@vu.nl; Tel: +31-205987542
cDepartment of Molecular and Cellular Neurobiology, Center for Neurogenomics and Cognitive Research, Neuroscience Campus Amsterdam, VU University Amsterdam, De Boelelaan 1085, 1081 HV, Amsterdam, The Netherlands
First published on 21st April 2011
Abstract
An online bioaffinity analysis system was used to screen our in-house fragment library on two related proteins, Ls- and Ac-AChBP, model proteins for nAChRs, in particular the α7 subtype. An efficient protocol for medium throughput fragment screening, hit validation and affinity ranking after single concentration injections was developed. The screening of the fragment library and the good correlation between online estimated pKi values (derived from a single injection) and pKi values measured with a radioligand binding assay (RBA, full range concentration curve) have proven the value of the online fluorescence enhancement assay in FBDD. The online bioaffinity system was also used for rapid hit exploration using single point injections of combinatorial libraries at 96-well format. This led to the discovery of an optimized hit with micromolar affinity towards the α7 nAChR.
Introduction
The nicotinic acetylcholine receptors (nAChR) belong to the family of ligand-gated ion channels and comprise potentially new pharmaceutical targets in multiple brain disorders. They are nowadays one of the most intensively studied type of ion channels.1–3 One of the major challenges is formed by the large diversity of receptor subtypes, originating from multiple homo- and heteromeric combinations of subunits forming pentameric ion channels.4 For drug discovery, compounds that selectively inhibit one nAChR subtype are of great interest.1 In particular, selective ligands targeting the α7 nAChR subtype are expected to be of use in Schizophrenia and Alzheimer's disease.5–7The use of Fragment-Based Drug Discovery (FBDD) for efficient screening and optimization of ligands is nowadays widely accepted.8 In order to apply FBDD on nAChRs, we recently developed and validated an online fluorescence enhancement assay for the water-soluble acetylcholine binding protein (AChBP),9 a model protein for the extracellular ligand-binding domain of nAChRs.10,11AChBP is a stable protein which was first crystallized in 2001.12AChBP assembles as a pentamer and has ligand binding pharmacology most similar to the α7 nAChR.13–17 Several homologous proteins were found in different snail species, amongst which Lymnaea stagnalis (Ls)12 and Aplysia californica (Ac).15 Ls- and Ac-AChBP share only 33% of their amino acids, and have different ligand selectivity profiles. The majority of small ligands for nAChRs, like endogenous acetylcholine, nicotine and epibatidine, are Ls-AChBP selective.17 Lobeline, however, is highly selective towards Ac-AChBP.14,17 Several cone snail derived bioactive toxin peptides (conotoxins) have comparable affinities for both AChBPs, but the α7 selective conotoxin α-ImI has a >6000-fold higher affinity for Ac-AChBP over Ls-AChBP.15 Since the literature is not decisive in which one of the two AChBPs is best suitable to find hit fragments for the α7 nAChR, we decided to screen our in-house fragment library simultaneously on Ls- and Ac-AChBP.
Our recently published online fluorescence enhancement assay9 is ideally suited for parallel screening in medium throughput mode. To this end, the Ls-AChBP fluorescence enhancement assay9 was optimized for Ac-AChBP. The fragment library was then screened on both proteins. The validated hits allowed for the determination of detailed selectivity profiles for both proteins. A subset of these hits was evaluated for binding to the α4β2 and α7 subtype nAChRs. Next, one of the α7 selective hits was optimized rapidly by combinatorial chemistry in 96-well format. In our previous paper,9 we showed that the online bioanalysis system is very accurate and reproducible in predicting pKi values at single concentration injections. We now use this system for the screening of two generations of focused combinatorial libraries, allowing a rapid exploration of hit fragments.
Results and discussion
Parallel screening assay and conditions
A schematic setup of the online bioaffinity analysis system is shown in Fig. 1. The bioaffinity analysis assays make use of the tracer ligand DAHBA (shown in Fig. 2A), which displays fluorescence enhancement upon binding to AChBP.9 For screening of the fragment library, the setup was operated in flow injection analysis (FIA) mode enabling the injected compounds to proceed to the bioaffinity analysis assays and UV detection directly after a 3-split in the eluent flow from the autoinjector. In the bioaffinity analysis assays, ligands interact with continuously infused AChBPvia the superloops operated by pumps P1 (Ls-AChBP) and P3 (Ac-AChBP) in the first set of reaction coils. In the second set of reaction coils, the tracer ligand DAHBA is continuously infused (via the superloops operated by pumps P2 and P4) and interacts with the remaining free binding sites of AChBP. The resulting fluorescence enhancement is monitored by a fluorescence detector.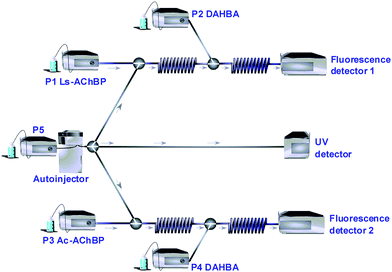 | ||
| Fig. 1 General schematic setup of the online bioaffinity analysis system. P1–P5 = HPLC pumps. Injection with split to the two online assays and the UV detector. Online addition of AChBP (P1 and P3) and the tracer ligand (P2 and P4), via reaction coils to fluorescence detectors. | ||
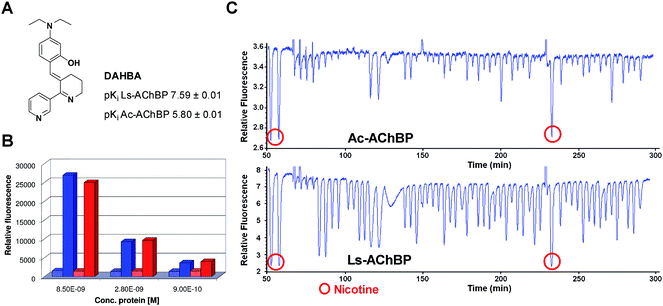 | ||
| Fig. 2 (A) Structure and affinities of DAHBA for Ls- and Ac-AChBP. (B) Assay window for Ls- and Ac-AChBP. Blue: Ls-AChBP and red: Ac-AChBP. Dotted bars: epibatidine displacement of DAHBA, filled bars: relative fluorescence of DAHBA in the active site. (C) Typical assay results (hit validation), showing fluorescence enhancement traces for Ls- and Ac-AChBP. Negative peaks indicate DAHBA displacement of an injected ligand thus representing affinity. | ||
The assay conditions of Ls-AChBP, which were previously optimized and validated,9 were used as a starting point for Ac-AChBP assay optimization, since Ls- and Ac-AChBP are closely related. The Ac-AChBP conditions were adjusted to the fluorescence enhancement properties and affinity of DAHBA for Ac-AChBP. The affinity of DAHBA for Ac-AChBP is 60 times lower than for Ls-AChBP. However, in order to keep the background fluorescence signal as low as possible, the concentration of DAHBA in the Ac-AChBP assay was chosen only 20 times higher than in the Ls-AChBP assay. To determine the assay windows for both assays (fluorescence signal DAHBA minus background signal), the fluorescence signal of DAHBA was measured at three different concentrations of Ls- and Ac-AChBP (Fig. 2B). Full displacement of DAHBA with the high-affinity ligand epibatidine was used to measure the background signal. The assay window was approximately equal for both proteins under the chosen conditions.
Fragment library screening
The commercially available IOTA MedChem Diverse fragment library consists of 1010 structurally diverse fragments and obeys general fragment library criteria, i.e., the number of heavy atoms ≤ 22, log P < 3, the number of H-bond donors ≤ 3 and the number of H-bond acceptors ≤ 3, the number of rotatable bonds ≤ 5 (adjusted rule of three).18 The fragments had to contain at least one ring structure, and compounds containing reactive functional groups were removed.19 The scaffold diversity analysis by Xu20 was used, amongst others, to assess the structural diversity of the library. In this analysis, each compound is divided into scaffolds and side chains following defined rules.20 The complexity value of the scaffolds is calculated using six descriptors,20 the cyclicity value is the ratio between scaffolds and side chains. To visualize the scaffold diversity, complexity is plotted versus cyclicity, as presented in Fig. 3.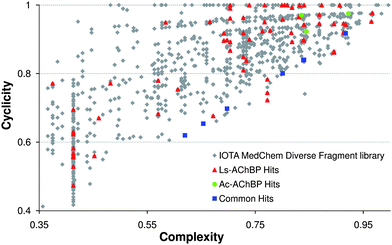 | ||
| Fig. 3 Screening results depicted in scaffold analysis plots. Overview of hits for Ls- and Ac-AChBP. The fragment at complexity = 0.18, cyclicity = 0.28 is not visualized to allow a clearer illustration. | ||
The 1010 fragments, along with regular injections of nicotine and lobeline as control ligands, were injected as 5 × 10−4 M concentrations (40 µL) at 1.5 minute intervals. Signal-to-noise ratios were determined by dividing the signal of a 100% displacing concentration of nicotine by the noise. The noise was determined by measuring the baseline for two minutes. Resulting S/N ratios were around 20 for Ls-AChBP and around 10 for Ac-AChBP. The hit criterion was a signal of at least three times the noise. The hit rates were 7.7% for Ls-AChBP (78 hits) and 1.1% for Ac-AChBP (11 hits). Of these 11 Ac-AChBP hits, 8 were also identified as a hit in the Ls-AChBP assay. A total of 81 hits were thus obtained.
In Fig. 3, the hits are highlighted in a scaffold diversity plot, to show the diversity of the hits resulting from the screening, and to analyze the relation between affinity/selectivity and complexity. For Ls-AChBP, a balanced distribution is seen with respect to complexity and cyclicity, indicating structurally diverse hits. For Ac-AChBP, the hits have higher complexities (represented at the right side of the plot), or have lower cyclicity values (closer to the bottom of the plot), indicating that these scaffolds are more complex than the mean of the fragment library.
Hit validation on AChBP
Of the hit set, 51 diverse fragments were considered for further evaluation. Representatives were chosen for duplicate scaffolds. Fragments with high similarity scores (Tc ≥ 0.8 based on ECFP4 fingerprints using Pipeline Pilot21) to known compounds for the α7 receptor (ChEMBL database22) were disregarded. Fresh 10−2 M solutions were made in DMSO. These solutions were injected, with injection volumes of 2 µL and injection intervals of 3.5 minutes. The very small injection volumes of 2 µL are immediately diluted in the eluent stream after injection. For both assays, a higher protein concentration was used to provide a larger assay window for validation of this small set of selected hits (typical S/N ratios: for Ls-AChBP ∼75, for Ac-AChBP ∼45). A typical screening result is depicted in Fig. 2C. As actual active protein concentrations differed per batch and due to the freeze–thaw process prior to use,9 assay results were always taken relative to the full displacement, achieved by injection of a high concentration of nicotine. The hit validation was performed in duplicate to improve hit ranking and to provide more accurate Ls- vs.Ac-AChBP selectivity information. For compounds with (almost) full displacement, appropriate dilutions were made and re-injected.Estimated pKi values from single concentration injections
The displacement data at a single concentration, relative to the 100% displacement of nicotine, were used to estimate pKi values, as described in our previous paper.9 The list of Ac-/Ls-AChBP affinities and selectivities for the 51 validated hits is presented in Table S1† of the ESI. From these 51 fragments, only two fragments showed minor selectivity for Ac-AChBP; all other compounds were found to be selective for Ls-AChBP. Interestingly, 35% of the compounds have a 10 to 60-fold more affinity for Ls- over Ac-AChBP. Together with the observation that hits for Ac-AChBP tend to be larger and more complex fragments (Fig. 3), this leads to the suggestion that Ac-AChBP apparently is not so easy to target.23 For this reason, we decided to focus on Ls-AChBP in the remaining part of this study.To validate our screening procedure, we compared the ‘estimated pKi values’ with pKi values obtained with a [3H]-epibatidine radioligand binding assay ((RBA), n = 3). As a proof-of-principle, RBA pKi values were determined for 23 randomly picked fragments and the correlation found demonstrated the validity of our single concentration pKi estimations. The correlation (R2 = 0.80) between these full curve values and the estimated pKi values for Ls-AChBP is shown in Fig. 4. It should be stressed that these estimated pKi values were obtained from duplicate injections of a single concentration per fragment; the good correlation indicates that a reliable ranking of the (low affinity) fragment hits can be obtained this way.
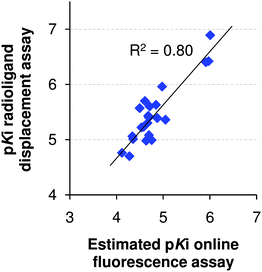 | ||
| Fig. 4 Correlation for Ls-AChBP of estimated pKis from a single concentration measurement in the online fluorescence assay versus full curve pKis from a radioligand displacement assay. | ||
Evaluation of a subset of hits on human nAChRs
A subset of ten hit fragments was taken for evaluation of affinities for two important nAChR subtypes, namely α7 and α4β2 nACh receptors. The selection of these ten fragments was based on scaffold diversity and distinct Ls- and Ac-AChBP affinity profiles. The diversity of this subset of hits is demonstrated with a similarity matrix, Table S2† in the ESI. The Tanimoto scores were calculated based on ECFP4 fingerprints using Pipeline Pilot.21 According to the published similarity cutoff of Tc ≥ 0.26, 93% of the fragment subset pairs are dissimilar.24 The highest Tc in the set was 0.37 between fragments 9 and 10.The pharmacological results for the subset are presented in Table 1. Only one of the fragments had affinity for the α4β2 receptor, with a 20-fold selectivity for this receptor over the α7 receptor. Six out of ten fragments showed affinity for the α7 receptor; five of these hits were α7 selective over the α4β2 receptor. This result is in line with the hypothesis that the AChBP ligand binding site is more similar to the α7 than the α4β2 binding site.
| No. | MW | clog P | Ls-AChBP affinitya | LEb Ls | Ac-AChBP affinityc | LE b Ac | Ls/Ac ratiod | α7 affinitye | LE b α7 | α4β2 affinityf | LE b α4β2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a Competition binding assay of [3H]-epibatidine using isolated Ls-AChBP. b Ligand efficiencies (LE) were calculated using the formula of Hopkins et al.:22LE = ΔG/the number of heavy atoms and are given in kcal mol−1HA−1. c Competition binding assay of [3H]-epibatidine using isolated Ac-AChBP. d As determined by dividing Ki values of Ls- and Ac-AChBP. e Competition binding assay of [3H]-MLA using membranes of SH-SY5Y cells expressing the nAChR α7 receptor. f Competition binding assay of [3H]-epibatidine using membranes of HEK293t cells expressing the nAChR α4β2 receptor. | |||||||||||
| 1 | 213.3 | 2.68 | 5.63 ± 0.14 | 0.48 | 4.58 ± 0.07 | 0.39 | 11.2 | <4 | — | <4 | — |
| 2 | 182.7 | 2.31 | 6.89 ± 0.02 | 0.79 | 4.04 ± 0.20 | 0.46 | 707.9 | 5.03 ± 0.36 | 0.58 | 6.30 ± 0.03 | 0.72 |
| 3 | 236.3 | 1.99 | 5.06 ± 0.17 | 0.39 | 5.32 ± 0.19 | 0.41 | 0.5 | 5.95 ± 0.10 | 0.45 | <4 | — |
| 4 | 278.4 | 2.79 | 5.96 ± 0.05 | 0.39 | <4 | — | >91.2 | <4 | — | <4 | — |
| 5 | 202.2 | 2.67 | 5.30 ± 0.06 | 0.49 | <4 | — | >20.0 | <4 | — | <4 | — |
| 6 | 157.2 | 0.60 | 4.98 ± 0.04 | 0.62 | <4 | — | >9.5 | 4.99 ± 0.17 | 0.62 | <4 | — |
| 7 | 221.3 | 2.35 | 6.08 ± 0.02 | 0.49 | 4.77 ± 0.01 | 0.39 | 20.4 | 4.67 ± 0.23 | 0.38 | <4 | — |
| 8 | 252.7 | 1.61 | 5.01 ± 0.05 | 0.41 | 5.21 ± 0.01 | 0.42 | 0.6 | 5.01 ± 0.04 | 0.41 | <4 | — |
| 9 | 228.3 | 1.74 | 4.76 ± 0.04 | 0.38 | 5.04 ± 0.02 | 0.40 | 0.5 | <4 | — | <4 | — |
| 10 | 270.4 | 1.03 | 6.40 ± 0.06 | 0.44 | 4.37 ± 0.14 | 0.30 | 107.2 | 4.67 ± 0.03 | 0.32 | <4 | — |
Focused combinatorial libraries
The online bioanalysis system is not only ideally suitable for primary fragment screening, but could be used for rapid hit exploration as well. Combinatorial chemistry in 96-well format, followed by single point injections, could potentially iteratively improve the affinity and evaluate growing directions within a few days. To prove the ability of rapid combinatorial hit expansion, one of the hits was chosen for optimization. The hit with the highest LE for the α7 receptor (compound 6, Fig. 5) was regarded as the best starting point for fragment growing, also because its hydroxylamine moiety offered good opportunities for combinatorial chemistry. Hydroxylamine bonds are easily formed under acidic conditions, and are quite stable under basic conditions. Next to hydroxylamines, hydrazines could be readily investigated as well.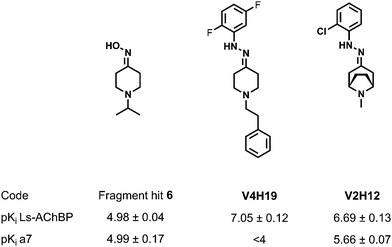 | ||
| Fig. 5 Chemical structures and pharmacological results for fragment hit and the two best compounds of the combinatorial libraries. | ||
Two generations of focused chemical libraries were constructed in 96-well format. The first generation contained three ketones (V1–V3, Fig. 6) and 11 hydroxylamines and hydrazines (H1–H11). For this first generation library, very diverse in house building blocks were chosen. The hydroxylamine/hydrazine with the highest estimated affinity was the 5-bromophenylhydrazine (H5), and was taken as the template for the selection of new building blocks for the second generation library (H5, H12–H21). Diverse substitution patterns were selected, as well as different electron densities in the aromatic ring. Four more ketones were selected as well; three of them were synthesized in house (V4–V6). Ketones V2 and V7 are known substructures for α7 ligands. Ketones V1–V4 and V6 were chosen based on in house data on fragment hits and a running structure-based program. Ketone V5 originates from fragment hit 6. All mixtures were screened on Ls- and Ac-AChBP, although minor displacement was obtained for Ac-AChBP (which was expected, for the hit compound had 16-fold selectivity for Ls-AChBP). The data presented in Fig. 6 are Ls-AChBP data.
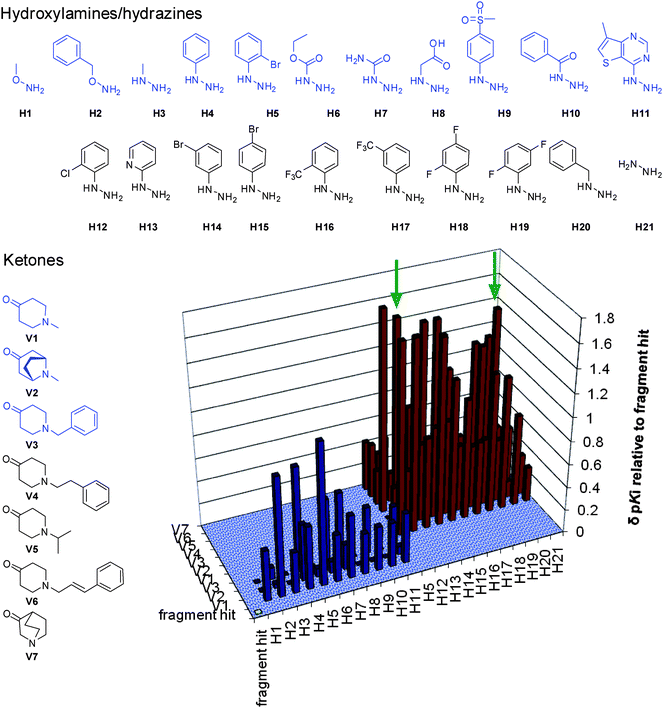 | ||
| Fig. 6 Chemical structures and results on Ls-AChBP of the focused combinatorial libraries. The structures presented in blue formed the first generation library, the results for this library are shown as blue bars. All ketones and the H5, H12–H21 formed the second generation library, the results are shown as red bars. The ligands presented in Fig. 6 are indicated with green arrows. | ||
Control experiments were performed to validate the experimental setup. The top-ranked compounds, with respect to the displacement of DAHBA, have been synthesized at larger scale in order to confirm both product formation by LC-MS and NMR and binding by full curve affinity determinations in plate reader format. These compounds were all formed with >95% purities. The best ligand (V4H19, Fig. 5) had a pKi of 7.05 ± 0.12 for Ls-AChBP, which stands for a 100-fold improvement with respect to the original fragment hit. The binding to the α7 receptor was also evaluated for these top-ranked ligands. The second-best ligand for Ls-AChBP (V2H12, Fig. 5, pKi of 6.69 ± 0.13) showed a 5-fold increase in affinity for the α7 receptor (pKi = 5.66 ± 0.07), with an affinity comparable to nicotine. The ligand has good lead-like properties (MW = 263.77 g mol−1, log P = 1.95, TPSA = 28.83 Å2, # rot. bonds = 2) and forms an excellent starting point for further optimization.
Conclusion
An online bioaffinity analysis system was used to screen our fragment library on two related proteins, Ls- and Ac-AChBP, model proteins for nAChRs, in particular the α7 subtype. The results showed that of these two proteins, Ls-AChBP is a better template for fragment screening, for many more hits were picked up for evaluation on the α7 nACh receptor. An efficient protocol for medium throughput fragment screening, hit validation and affinity ranking after single concentration injections was developed. The screening of the fragment library and the good correlation between online estimated pKi values (derived from a single injection) and pKi values measured with a RBA (full concentration curve) have proven the value of the online fluorescence enhancement assay in FBDD. The online bioaffinity system was also found to be perfectly accurate for rapid hit exploration using single point injections of combinatorial libraries at 96-well format. The technology is broadly applicable; it only needs a water-soluble protein and a good fluorescence enhancement tracer ligand.Abbreviations
| FBDD | Fragment-Based Drug Discovery |
| nAChR | nicotinic acetylcholine receptor |
| AChBP | acetylcholine binding protein |
| Ls | Lymnaea stagnalis |
| Ac | Aplysia californica |
| FIA | flow injection mode |
| UV | ultraviolet |
| RBA | radioligand binding assay |
| T c | Tanimoto score |
| NMR | Nuclear Magnetic Resonance |
Acknowledgements
ABS, RvE, GdE, and IJPdE received funding from Top Institute Pharma (grant D2-103). ABS and IJPdE received funding from the European Union Seventh Framework Programme under grant agreement no. HEALTH-F2-2007-202088 (“NeuroCypres” project).References
- A. Taly, P. J. Corringer, D. Guedin, P. Lestage and J. P. Changeux, Nicotinic receptors: allosteric transitions and therapeutic targets in the nervous system, Nat. Rev. Drug Discovery, 2009, 8, 733–750 Search PubMed.
- J. P. Changeux and A. Taly, Nicotinic receptors, allosteric proteins and medicine, Trends Mol. Med., 2008, 14, 93–102 Search PubMed.
- A. A. Jensen, B. Frolund, T. Lijefors and P. Krogsgaard-Larsen, Neuronal nicotinic acetylcholine receptors: structural revelations, target identifications, and therapeutic inspirations, J. Med. Chem., 2005, 48, 4705–4745 CrossRef CAS.
- C. Gotti, M. Moretti, A. Gaimarri, A. Zanardi, F. Clementi and M. Zoli, Heterogeneity and complexity of native brain nicotinic receptors, Biochem. Pharmacol., 2007, 74, 1102–1111 Search PubMed.
- S. Barak, M. Arad, A. De Levie, M. D. Black, G. Griebel and I. Weiner, Pro-cognitive and antipsychotic efficacy of the [alpha]7 nicotinic partial agonist SSR180711 in pharmacological and neurodevelopmental latent inhibition models of Schizophrenia, Neuropsychopharmacology, 2009, 34, 1753–1763 Search PubMed.
- S. N. Haydar and J. Dunlop, Neuronal nicotinic acetylcholine receptors—targets for the development of drugs to treat cognitive impairment associated with Schizophrenia and Alzheimers disease, Curr. Top. Med. Chem., 2010, 10, 144–152 Search PubMed.
- C. M. Hernandez, R. Kayed, H. Zheng, J. D. Sweatt and K. T. Dineley, Loss of {alpha}7 nicotinic receptors enhances {beta}-amyloid oligomer accumulation, exacerbating early-stage cognitive decline and septohippocampal pathology in a mouse model of Alzheimer's disease, J. Neurosci., 2010, 30, 2442–2453 Search PubMed.
- G. E. de Kloe, D. Bailey, R. Leurs and I. J. P. de Esch, Transforming fragments into candidates: small becomes big in medicinal chemistry, Drug Discovery Today, 2009, 14, 630–646 Search PubMed.
- J. Kool, G. E. de Kloe, B. Bruyneel, J. S. de Vlieger, K. Retra, M. Wijtmans, R. van Elk, A. B. Smit, R. Leurs, H. Lingeman, I. J. P. de Esch and H. Irth, Online fluorescence enhancement assay for the acetylcholine binding protein with parallel mass spectrometric identification, J. Med. Chem., 2010, 53, 4720–4730 Search PubMed.
- P. H. Celie, I. E. Kasheverov, D. Y. Mordvintsev, R. C. Hogg, P. van Nierop, R. van Elk, S. E. van Rossum-Fikkert, M. N. Zhmak, D. Bertrand, V. Tsetlin, T. K. Sixma and A. B. Smit, Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an alpha-conotoxin PnIA variant, Nat. Struct. Mol. Biol., 2005, 12, 582–588 CrossRef CAS.
- A. B. Smit, K. Brejc, N. Syed and T. K. Sixma, Structure and function of AChBP, homologue of the ligand-binding domain of the nicotinic acetylcholine receptor, Ann. N. Y. Acad. Sci., 2003, 998, 81–92 Search PubMed.
- K. Brejc, W. J. van Dijk, R. V. Klaassen, M. Schuurmans, J. van Der Oost, A. B. Smit and T. K. Sixma, Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors, Nature, 2001, 411, 269–276 CrossRef CAS.
- P. H. N. Celie, S. E. van Rossum-Fikkert, W. J. van Dijk, K. Brejc, A. B. Smit and T. K. Sixma, Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures, Neuron, 2004, 41, 907–914 CrossRef CAS.
- S. B. Hansen, G. Sulzenbacher, T. Huxford, P. Marchot, P. Taylor and Y. Bourne, Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations, EMBO J., 2005, 24, 3635–3646 CrossRef CAS.
- S. B. Hansen, T. T. Talley, Z. Radic and P. Taylor, Structural and ligand recognition characteristics of an acetylcholine-binding protein from Aplysia californica, J. Biol. Chem., 2004, 279, 24197–24202 CrossRef CAS.
- P. Rucktooa, A. B. Smit and T. K. Sixma, Insight in nAChR subtype selectivity from AChBP crystal structures, Biochem. Pharmacol., 2009, 78, 777–787 Search PubMed.
- P. Taylor, T. T. Talley, Z. Radic, S. B. Hansen, R. E. Hibbs and J. Shi, Structure-guided drug design: conferring selectivity among neuronal nicotinic receptor and acetylcholine-binding protein subtypes, Biochem. Pharmacol., 2007, 74, 1164–1171 Search PubMed.
- M. Congreve, R. Carr, C. Murray and H. Jhoti, A rule of three for fragment-based lead discovery?, Drug Discovery Today, 2003, 8, 876–877 CrossRef.
- T. I. Oprea, Property distribution of drug-related chemical databases, J. Comput.-Aided Mol. Des., 2000, 14, 251–264 CrossRef CAS.
- J. Xu, A new approach to finding natural chemical structure classes, J. Med. Chem., 2002, 45, 5311–5320 CrossRef CAS.
- Pipeline Pilot, Version 6.1, SciTegic Inc., San Diego, CA 92123-1365, http://accelrys.com/products/scitegic Search PubMed.
- ChEMBL, http://www.ebi.ac.uk/chembl/.
- P. J. Hajduk, J. R. Huth and S. W. Fesik, Druggability indices for protein targets derived from NMR-based screening data, J. Med. Chem., 2005, 48, 2518–2525 CrossRef CAS.
- A. Steffen, T. Kogej, C. Tyrchan and O. Engkvist, Comparison of molecular fingerprint methods on the basis of biological profile data, J. Chem. Inf. Model., 2009, 49, 338–347 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Table with data for the validated hits, similarity matrix of ten hits, and experimental procedures. See DOI: 10.1039/c1md00031d |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2011 |
