Microwave-assisted one-step rapid synthesis of folic acid modified gold nanoparticles for cancer cell targeting and detection†
Zhaowu
Zhang
,
Jing
Jia
,
Yanyan
Ma
,
Jian
Weng
,
Yanan
Sun
and
Liping
Sun
*
Department of Biomaterials, College of Materials, Research Center of Biomedical Engineering, Xiamen University, Xiamen, 361005, People's Republic of China. E-mail: sunliping@xmu.edu.cn; Fax: +86 (0)592-2185299; Tel: +86 (0)592-2183181
First published on 19th September 2011
Abstract
In this study, folic acid-protected gold nanoparticles (FA-GNPs) were fabricated by one-step reduction of HAuCl4 by folic acid under microwave irradiation. FA-GNPs were successfully used to target and detect human cervical carcinoma cells.
Introduction
Folic acid (FA) conjugated with gold nanoparticles (GNPs) has been successfully used in targeting and imaging cancer cells, because folate receptors are up-regulated on a variety of human cancers.1,2 Numerous attempts using polymers to develop biodegradable hydrophilic folate–nanoparticle conjugate have been reported.3,4 In our previous study, FA was coupled on the GNP surface through glutathione, and the targeting of FA-GSH-GNPs in human cervical carcinoma cells (HeLa cells) was confirmed.5 The limitations of these methods are that a linkage mediator is required to connect GNPs with FA, which makes the procedure complicated and time-consuming. To overcome these problems, we developed a facile one-step method to fabricate FA-GNPs under microwave irradiation.There are a large number of ways to synthesize GNPs. The reducing reagents could be either inorganic, such as sodium/potassium borohydrate, hydrazine and salts of tartrate, or organic, such as sodium citrate and ascorbic acid.6 Recently, more and more biomolecules have been used as reducing agents to prepare GNPs, including amino acids, chitosan, glucose, peptides, nucleotides and vitamin B2. These molecules can create stable GNPs of consistent sizes in a water-based environment.6–11 Moreover, biomolecule-functionalized GNPs are non-toxic and can be easily adapted for biological applications. FA is comprised of para-aminobenzoic acid linked at one end to a pteridine ring and at the other end to glutamic acid (Scheme 1). The 2-amino group (–NH2) and 4-hydroxyl group (–OH) on the pteridine ring possibly reduce the gold ion to the gold particle. And FA adsorbed on the surface of the GNPs provides colloidal stability owing to its negative charge. So in our reaction, FA acts as both the reducing and stabilizing agent.
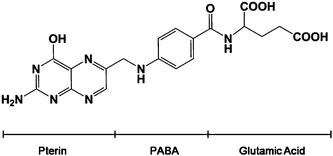 | ||
| Scheme 1 The chemical structure of folic acid. | ||
Microwave synthesis is an invaluable tool for medicinal chemistry and drug discovery applications since it is quite fast, simple and very energy efficient compared to conventional thermal convection.12,13 Many successful applications of microwave heating have been reported to yield platinum, silver, gold and other nanoparticles in solution.14–18 In this paper, we report a microwave irradiation-based strategy for the preparation of gold nanoparticles by direct microwave irradiation of a FA/HAuCl4 aqueous solution without the additional step of introducing other reducing agents and protective agents. Significantly, these FA-capped GNPs have precisely controlled sizes and are highly stable. And based on a simple spectroscopic method, FA-GNPs were successfully used to detect HeLa cells with a detection limit of 750 cells.
Results and discussion
For the synthesis of gold nanoparticles, FA solution (0.2 mL, 1.0 mM) was added to HAuCl4 (5 mL, 0.8 mM) solution under magnetic stirring. Due to the poor solubility of FA in acidic HAuCl4 solution, some precipitate appeared. For this reason, 1 mol L−1NaOH was subsequently added to obtain a clear orange solution (pH 10.60). The solution was stirred for 30 min and immediately placed in a sealed vessel and reacted in a microwave reactor. The reactions were run for 2 h at 100 °C. The resulting solution appeared wine red, indicating the formation of gold nanoparticles. This was further confirmed by UV-vis spectroscopy (Fig. 1A), which showed a surface plasmon band around 529 nm. The as-prepared FA functionalized GNPs (FA-GNPs) were characterized by high-contrast transmission electron microscopy (TEM), Fourier transform infrared spectroscopy (FTIR), X-ray photoelectron spectroscopy (XPS), and X-ray diffraction (XRD). | ||
| Fig. 1 UV-vis absorption spectrum (A) and TEM micrograph (B) of FA-GNPs. | ||
TEM images of the FA-GNPs displayed well-dispersed spherical nanoparticles with a size range of 15 ± 5 nm (Fig. 1B). The shadow around the spherical gold core could be free folic acid wrapped around the individual nanoparticles. UV-vis spectra showed that the FA-GNPs were stable in PBS buffer up to a concentration of 0.6 mM (Fig. S1†). The comparison of the FTIR spectra of the FA-GNPs and FA is shown in Fig. 2. The characteristic bands of folic acid at 1699 (amide I) and 1604 (amide II) are also shown in the FA-GNPs conjugate, indicating successful tailoring of the nanoparticle surface with FA.19 The –NH2 double bands of the pteridine ring (3530, 3415 cm−1) disappeared in the FA-GNPs conjugate. This may provide evidence that the –NH2group has been conjugated to the GNPs by forming a N–Au bond. The binding of FA to the GNPs is also demonstrated by the presence of C, N, O on the surface of gold nanoparticles (XPS data, Fig. S1†). The binding energy of Au 4f 5/2 and Au 4f 7/2 appears at 87.5 eV and 84.0 eV, which is consistent with the emission of 4f photoelectrons from Au0, thereby suggesting the successful formation of GNPs.10XRD results revealed the crystalline nature (face-centered cubic structure) of the FA-GNPs (Fig. S3†). The cytotoxicity of the FA-GNPs was evaluated by a WST-1 assay after incubation with FA-GNPs for 12, 24 and 36 h, respectively (Fig. S4†). HeLa cell viability remained over 80% after treatment with FA-GNPs for 36 h, implying that FA-GNPs are biocompatible at a concentration of up to 100 μg mL−1.
 | ||
| Fig. 2 FTIR spectra of (a) FA-GNPs, (b) pure folic acid. | ||
In order to obtain a quantitative measurement of GNPs in the cells, inductively coupled plasma mass spectroscopy (ICP-MS) was used to quantify the content of Au in each cell sample. HeLa cells, a human cervical carcinoma cell line that is known to overexpress folate receptors (FAR), were chosen for these experiments. To study the relationship between intracellular uptake and incubation time, cells were cultured for intervals of 1, 2, 3, 4, and 5 h. As shown in Fig. 3A, the amount of FA-GNPs or 13 nm citrate-capped GNPs in HeLa cells increases gradually with incubation time. The intracellular uptake of FA-GNPs is faster than that of GNPs. The uptake rate of the FA-GNPs reaches a maximum level at an incubation time of 3 h, while that of the GNPs reaches a plateau at 5 h. To confirm the receptor-mediated uptake, competition experiments were conducted. Fig. 3B shows intracellular uptake of nanoparticles after 3 h co-incubation of FA-GNPs or GNPs with HeLa cells. Cells were first saturated with free FA. The cellular uptake of FA-GNPs gradually decreases with increasing FA concentration, because the binding of FA to folate receptors in HeLa cells competitively prevents FA-GNPs binding to FAR. In contrast, no obvious changes of the cellular uptake of GNPs are found. These results clearly demonstrate the preferential accumulation of FA-GNPs in HeLa cells, and show that folate-receptor-targeting conjugation promoted this tumor preferential accumulation of particles. Scheme 2 illustrates that the receptor-mediated endocytosis enhances the internalization of FA-GNPs by target cells compared with citrate-capped GNPs (Scheme 2A, 2B), and folic acid competitively inhibits their binding to the folate receptor and reduces the uptake by cancer cells (Scheme 2C).
 | ||
| Fig. 3 Cellular uptake of FA-GNPs by HeLa cells in dependence on incubation time (A) and concentration of folic acid (B). | ||
 | ||
| Scheme 2 Schematic illustration of the intracellular uptake of nanoparticles by HeLa cells. | ||
Based on a simple spectroscopic method, FA-GNPs were successfully used to detect HeLa cells. FA-GNPs bound to cancer cells can be easily deposited together with the cells at low-speed centrifugation (1000 rpm), leading to a decrease in absorbance of the supernatant at 529 nm. ΔA was calculated according to the following formula: ΔA = A0 − A1 (A0 = Absorbance of blank, A1 = Absorbance of supernatant). Two cell lines were selected in this study, FAR-abundant HeLa cells (target cell line) and FAR-deficient fibroblasts (FB cells, control cell line). In Fig. 4A, four samples are as follows: FA-GNPs, FA-GNPs in PBS, FA-GNPs with 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 FB cells in PBS, and FA-GNPs with 30
000 FB cells in PBS, and FA-GNPs with 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 HeLa cells in PBS. After centrifugation, the absorbance of FA-GNPs with HeLa cells is significantly lower than that of FA-GNPs with FB cells. These results indicate that the FA-GNPs are binding selectively to the target cells and that the assembly of the FA-GNPs around the target cells causes a decrease in the absorption of the supernatant. UV-vis spectra for different amounts of target and control cells are shown in Fig. 4B and 4C, respectively. There is little change in the FA-GNP absorption spectra of the FB cell samples regardless of the amount of cells present. It is obvious that the higher the HeLa cell concentration, the higher the ΔA is (Fig. 4D), suggesting that more nanoparticles are being deposited at 1000 rpm by targeting HeLa cells. ΔA of the control cells (FB) has little change in comparison to the blank because FA-GNPs cannot bind to fibroblast cells owing to the lack of folate receptors. The nanoparticles still remain in the supernatant after low-speed centrifugation. In the range of 100 to 105cells mL−1, HeLa cells and FB can be distinguished clearly by a significant difference in ΔA of all groups (P < 0.05, Student's t-test for paired data). A detection limit of 100 cells mL−1 was obtained. This assay showed a linear correlation between ΔA and the amount of HeLa cells ranging from 102 to 105cells mL−1 (ΔA = 0.9168 + 0.0450 C, correlation coefficient of 0.9940). Based on these results, the assay has demonstrated excellent sensitivity and selectivity. Compared with our previous synthetic process of FA-GSH-GNPs,5 the preparation of FA-GNPs is more convenient, requiring only a one-step reaction. FA acts as both the reductant of HAuCl4 and the target molecule for folate receptors on the cancer cells. The sensitivity of FA-GNPs for cancer cell detection remains the same as that of FA-GSH-GNPs (100 cells mL−1). In addition, the size of FA-GNPs can be modulated by using a different mole ratio of [FA]/[HAuCl4].
000 HeLa cells in PBS. After centrifugation, the absorbance of FA-GNPs with HeLa cells is significantly lower than that of FA-GNPs with FB cells. These results indicate that the FA-GNPs are binding selectively to the target cells and that the assembly of the FA-GNPs around the target cells causes a decrease in the absorption of the supernatant. UV-vis spectra for different amounts of target and control cells are shown in Fig. 4B and 4C, respectively. There is little change in the FA-GNP absorption spectra of the FB cell samples regardless of the amount of cells present. It is obvious that the higher the HeLa cell concentration, the higher the ΔA is (Fig. 4D), suggesting that more nanoparticles are being deposited at 1000 rpm by targeting HeLa cells. ΔA of the control cells (FB) has little change in comparison to the blank because FA-GNPs cannot bind to fibroblast cells owing to the lack of folate receptors. The nanoparticles still remain in the supernatant after low-speed centrifugation. In the range of 100 to 105cells mL−1, HeLa cells and FB can be distinguished clearly by a significant difference in ΔA of all groups (P < 0.05, Student's t-test for paired data). A detection limit of 100 cells mL−1 was obtained. This assay showed a linear correlation between ΔA and the amount of HeLa cells ranging from 102 to 105cells mL−1 (ΔA = 0.9168 + 0.0450 C, correlation coefficient of 0.9940). Based on these results, the assay has demonstrated excellent sensitivity and selectivity. Compared with our previous synthetic process of FA-GSH-GNPs,5 the preparation of FA-GNPs is more convenient, requiring only a one-step reaction. FA acts as both the reductant of HAuCl4 and the target molecule for folate receptors on the cancer cells. The sensitivity of FA-GNPs for cancer cell detection remains the same as that of FA-GSH-GNPs (100 cells mL−1). In addition, the size of FA-GNPs can be modulated by using a different mole ratio of [FA]/[HAuCl4].
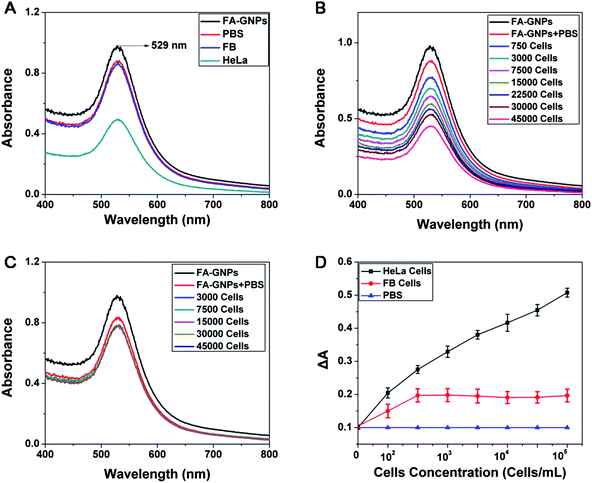 | ||
Fig. 4 A: UV-vis spectra of FA-GNPs, FA-GNPs in PBS, supernatant of FA-GNPs with 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 HeLa cells in PBS, FA-GNPs with 30 000 HeLa cells in PBS, FA-GNPs with 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 FB cells in PBS. B: Spectra for different amounts of HeLa cells. C: Spectra for different amounts of FB cells. D: ΔA of the target HeLa cells (black line) and fibroblasts (red line). The blank sample contains no cell (PBS with nanoparticles, blue line). 000 FB cells in PBS. B: Spectra for different amounts of HeLa cells. C: Spectra for different amounts of FB cells. D: ΔA of the target HeLa cells (black line) and fibroblasts (red line). The blank sample contains no cell (PBS with nanoparticles, blue line). | ||
Conclusions
In summary, a straightforward and rapid process has been developed to synthesise folic acid-conjugated gold nanoparticles. The ICP-MS analyses showed that these nanoparticles could successfully target tumor cells overexpressing the folate receptors. FA-GNPs have been demonstrated for the sensitive and selective detection of human HeLa cells through utilizing the unique spectral properties of gold nanoparticles and the excellent selectivity of folic acid. The results collected in this study indicate that FA-GNPs may be used as efficient materials for biomedical applications such as cancer diagnosis and therapy.Acknowledgements
This research has been supported by the National Natural Science Foundation of China (30900350 and 81171453).Notes and references
- G. A. Mansoori, K. S. Brandenburg and A. Shakeri-Zadeh, Cancers, 2010, 2, 1911–1928 CrossRef CAS.
- S. W. Tsai, J. W. Liaw, F. Y. Hsu, Y. Y. Chen, M. J. Lyu and M. H. Yeh, Sensors, 2008, 8, 6660–6673 CrossRef CAS.
- X. G. Shi, S. H. Wang, S. Meshinchi, M. E. Van Antwerp, X. D. Bi, I. H. Lee and J. R. Baker, Small, 2007, 3, 1245–1252 CrossRef CAS.
- V. Dixit, J. V. Bossche, D. M. Sherman, D. H. Thompson and R. P. Andres, Bioconjugate Chem., 2006, 17, 603–609 CrossRef CAS.
- Z. W. Zhang, J. Jia, Y. Q. Lai, Y. Y. Ma, J. Weng and L. P. Sun, Bioorg. Med. Chem., 2010, 18, 5528–5534 CrossRef CAS.
- A. Akbarzadeh, D. Zare, M. R. Mehrabi, D. Norouzian, S. Tangestaninejad, M. Moghadam and N. Bararpour, Am. J. Appl. Sci., 2009, 6, 691–695 CrossRef CAS.
- H. Z. Huang and X. R. Yang, Biomacromolecules, 2004, 5, 2340–2346 CrossRef CAS.
- P. Raveendran, J. Fu and S. L. Wallen, Green Chem., 2006, 8, 34–38 RSC.
- R. R. Bhattacharjee, A. K. Das, D. Haldar, S. Si, A. Banerjee and T. K. Mandal, J. Nanosci. Nanotechnol., 2005, 5, 1141–1147 CrossRef CAS.
- W. A. Zhao, F. Gonzaga, Y. F. Li and M. A. Brook, Adv. Mater., 2007, 19, 1766–1771 CrossRef CAS.
- N. N. Mallikarjuna, A. M. Wu, H. Kolla and S. K. Manohar, Abstr. Pap. Am. Chem. S., 2005, 22, U959–U959 Search PubMed.
- F. K. Liu, Y. C. Chang, F. H. Ko and T. C. Chu, Mater. Lett., 2004, 58, 373–377 CrossRef CAS.
- V. Polshettiwar and R. S. Varma, Chem. Soc. Rev., 2008, 37, 1546–1557 RSC.
- D. L. Boxall and C. M. Lukehart, Chem. Mater., 2001, 13, 806–810 CrossRef CAS.
- J. F. Zhu and Y. J. Zhu, J. Phys. Chem. B, 2006, 110, 8593–8597 CrossRef CAS.
- S. Komarneni, D. S. Li and B. Newalkar, Langmuir, 2002, 18, 5959–5962 CrossRef CAS.
- Z. Q. Tian, S. P. Jiang and Y. M. Liang, J. Phys. Chem. B, 2006, 110, 5343–5350 CrossRef CAS.
- M. N. Nadagouda and R. S. Varma, Green Chem., 2006, 8, 516–518 RSC.
- S. Mohapatra, S. K. Mallick, T. K. Maiti, S. K. Ghosh and P. Pramanik, Nanotechnology, 2007, 18, 385102–385110 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section and characterization details. See DOI: 10.1039/c0md00274g |
| This journal is © The Royal Society of Chemistry 2011 |
