Inhibition of Pseudomonas aeruginosa biofilms with a glycopeptide dendrimer containing D-amino acids†
Emma M. V.
Johansson
a,
Rameshwar U.
Kadam
a,
Gabriele
Rispoli
a,
Shanika A.
Crusz
b,
Kai-Malte
Bartels
c,
Stephen P.
Diggle
b,
Miguel
Cámara
b,
Paul
Williams
b,
Karl-Erich
Jaeger
c,
Tamis
Darbre
a and
Jean-Louis
Reymond
*a
aDepartment of Chemistry and Biochemistry, University of Berne, Freiestrasse 3, CH-3012, Berne, Switzerland. E-mail: jean-louis.reymond@ioc.unibe.ch; Fax: +41 31 631 80 57; Tel: +41 31 631 43 25
bSchool of Molecular Medical Sciences, University of Nottingham, Nottingham, NG7 2RD, UK
cInstitute of Molecular Enzyme Technology, Heinrich-Heine-University of Duesseldorf, Research Centre Juelich, D-52425, Juelich, Germany
First published on 10th March 2011
Abstract
The C-fucosyl peptide dendrimer D-FD2 (CFuc-lys-pro-leu)4(Lys-phe-lys-leu)2Lys-his-leu-NH2 was prepared as the D-amino acid containing stereoisomer of the Pseudomonas aeruginosa biofilm inhibitor FD2. D-FD2 showed lower affinity for the fucose specific lectin LecB, was also active as a biofilm inhibitor and in addition showed high stability towards proteolysis with pure proteases and in human serum.
The opportunistic human pathogen Pseudomonas aeruginosa is regarded as a major cause of death in cystic fibrosis, cancer, and for immunocompromised patients. The refractory nature of chronic infections with this bacterium can be attributed primarily to the formation of multi-antibiotic tolerant and host defence resistant biofilms.1 Inhibition of key biofilm mediators such as the galactose-specific lectin LecA (PA-IL)2 and the fucose-specific lectin LecB (PA-IIL),3,4 in particular using multivalent glycoclusters,5–8 represents a promising strategy for controlling P. aeruginosa biofilm-centered infections.9,10
Recently we reported that glycopeptide dendrimer11–14FD2 (CFucLysProLeu)4(LysPheLysIle)2LysHisIleNH2 selected from a combinatorial library15 is a strong inhibitor of the fucose specific lectin LecB and of P. aeruginosa biofilms.16 In our efforts to further develop this ligand,17 we herein report dendrimer D-FD2, a stereoisomer of FD2 containing D-amino acids in its branches. D-FD2 is potent against P. aeruginosa biofilms, and in addition shows complete resistance to proteolysis. This represents the first example of stabilizing a peptide dendrimer by using D-amino acids while retaining its biological activity, a strategy also used for linear peptides.18
The biofilm inhibitor FD2 contains several proteolysis sites in its sequence, rendering it potentially unstable towards proteolysis despite of its branched structure.19 Considering that peptide dendrimers are conformationally flexible and lack a precisely folded structure,20 we set out to investigate if a stereoisomer of FD2 incorporating D-amino acids might retain its biological activity while acquiring resistance to proteolysis. Dendrimer D-FD2 was thus designed by substituting D-amino acids for L-amino acids in the branches of FD2. A conservative exchange of isoleucines to leucines was also performed to simplify the sequence. Dendrimer D-FD2 was prepared by solid-phase peptide synthesis, followed by coupling of the protected C-fucosyl groups, deacetylation, acidic deprotection, cleavage from the resin, and purification by RP-HPLC (Fig. 1).
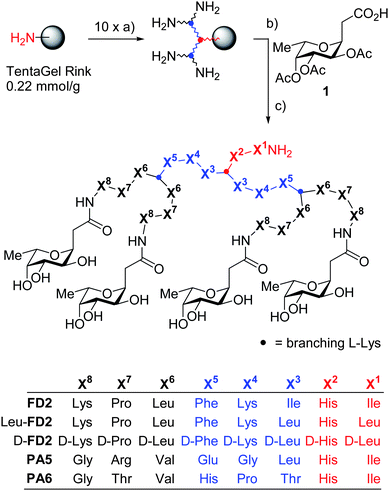 | ||
| Fig. 1 Synthesis and structure of glycopeptide dendrimer ligands of LecB. Conditions: a) FmocAAOH, PyBOP, DIEA, NMP,1–3 h (2x), then 20% piperidine in DMF, (2 × 10 min); b) 1, HCTU, DIEA, NMP, 12 h; c) MeOH, NH3, H2O, 24 h; d) CF3CO2H, TIS, H2O, 4 h, then precipitation from methyl tert-butyl ether and preparative RP-HPLC. The synthesis and characterization of PA5 and PA6 is described in ref. 16b. | ||
Lectin binding by D-FD2 was investigated using an enzyme-linked lectin assay (ELLA). The IC50 of D-FD2 for binding to LecB was 4.7-fold larger than that of FD2, showing that the binding affinity was dependent to some extent to amino acid chirality (Table 1). The conservative Ile → Leu exchange itself had no effect on binding as shown by the IC50 of Leu-FD2. The affinity of D-FD2 for LecB was comparable to that of dendrimers PA5 (reported relative potency r.p. = 14.7) but higher than that of PA6 (reported r.p. = 36.7)16b which have quite different amino acid sequences but identical topology and C-fucosyl endgroups.
| Cpd | IC50 (μM)a | nb | r.p.c | r.p./n |
|---|---|---|---|---|
| a Average of three independent IC50 determinations with standard deviation. See supporting information for experimental conditions. b n = number of C-fucosyl endgroups. c Relative potency (r.p.) = IC50(L-fucose)/IC50(cpd). | ||||
| L-fucose | 11 ± 1.5 | 1 | 1 | 1 |
| α-NPF | 5.27 ± 0.55 | 1 | 2.1 | 2.1 |
| FD2 | 0.14 ± 0.035 | 4 | 78.6 | 19.7 |
| Leu-FD2 | 0.15 ± 0.082 | 4 | 73.3 | 18.3 |
| D-FD2 | 0.66 ± 0.12 | 4 | 16.7 | 4.2 |
The activity of D-FD2 on P. aeruginosa biofilms was measured using the steel coupon assay in which the formation of biofilms in culture suspension is revealed by staining cells attached to the steel surface with acridine orange.10Dendrimer FD2 and the small molecule ligand α-nitrophenyl fucoside (α-NPF) served as positive controls. While α-NPF was only marginally active, D-FD2 reduced P. aeruginosabiofilm formation to background levels with the wild-type strain, as observed with FD2 (Fig. 2A). D-FD2 was also similarly active in disrupting already established biofilms, which are the major therapeutic target. On the other hand D-FD2 showed reduced activity compared to FD2 when tested against three additional P. aeruginosa clinical isolates (Fig. 2B). Nevertheless, the biofilm inhibitory effect of D-FD2 was remarkable considering that dendrimers PA5 and PA6, which had similar affinities to LecB as D-FD2, were much less active as biofilm inhibitors.
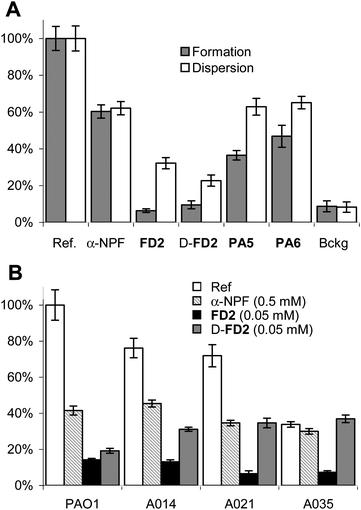 | ||
| Fig. 2 P. aeruginosa biofilm inhibition assay. A. Inhibition of P. aeruginosaPAO1 biofilms. B. Biofilm inhibition of PAO1 and three P. aeruginosa clinical isolates. Biofilm inhibition was performed on the wild-type strain PAO1 with added α-NPF (0.5mM) or dendrimers (0.05 mM). The values are normalized so that biofilm formation or dispersal without any inhibitor is set to 100%; Bckg is a background measurement with no added bacteria. The values are shown with the standard error from triplicate measurements. See supporting information for experimental conditions. Data for FD2, PA5 and PA6 from ref. 16b. | ||
Proteolytic studies showed that D-DF2 was highly resistant towards proteolytic degradation. D-FD2 was unaffected by treatment with trypsin or chymotrypsin for 48 h (100 mM Tris pH 7.5) Under the same conditions FD2 was completely degraded in 30 min. at the expected cleavage sites within its branches (see supporting information). The branching L-lysines themselves were unreactive to proteolysis confirming a previous model study.19 Similarly D-FD2 was completely stable in human serum, while FD2 underwent slow degradation (t1/2 ∼ 60 h).
The CD-spectra of D-FD2 and FD2 in aqueous buffer indicated random coil conformations of the peptide chains in both cases. Addition of 20% v/v trifluoroethanol (TFE) induced 20% α-helix content in FD2 but only 10% for D-FD2, suggesting differences in their conformations (Fig. 3AB). To further investigate this point, molecular dynamic simulations (MD) were performed with both dendrimers considering preformed α-helical conformation as starting point. During the course of the simulation for 10 ns at 300 K using NPT as ensemble, both dendrimers showed mainly random coil structures, with 20–40% retention of α-helix in FD2. FD2 had on average a smaller radius of gyration and a larger number of intramolecular hydrogen bonds compared to D-FD2, resulting in a significantly more compact structure than D-FD2 (Fig. 3C–F and supporting information). These data suggest that the conformational space occupied by these stereoisomeric dendrimers is significantly different, which might explain the weaker LecB binding and biofilm disrupting activity of D-FD2 compared to FD2.
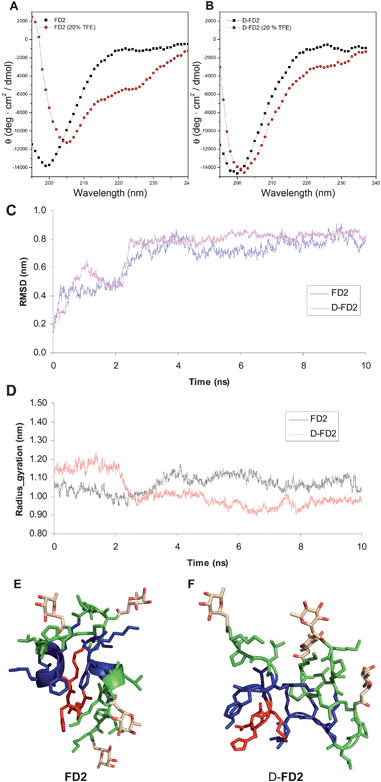 | ||
Fig. 3 Conformations of FD2 and D-FD2. CD spectra of A: FD2 and B: D-FD2 in 10 mM phosphate pH 7.0 (■) and with 20% TFE as cosolvent ( ) C: All-atom RMSD and D: radius of gyration (Rg) as function of time for FD2 and D-FD2 over 10 ns of MD. E: Snapshot of FD2 after 10 ns MD. F: Snapshot of D-FD2 after 10 ns MD. MD performed using the OPLS-AA (Optimized Potentials for Liquid Simulations-All Atom)21 force field in desmond molecular dynamics system and maestro-desmond interoperability tool, version 2.0.22 ) C: All-atom RMSD and D: radius of gyration (Rg) as function of time for FD2 and D-FD2 over 10 ns of MD. E: Snapshot of FD2 after 10 ns MD. F: Snapshot of D-FD2 after 10 ns MD. MD performed using the OPLS-AA (Optimized Potentials for Liquid Simulations-All Atom)21 force field in desmond molecular dynamics system and maestro-desmond interoperability tool, version 2.0.22 | ||
In summary substituting D-amino acids for L-amino acids in the branches of dendrimer FD2 produced peptide dendrimer D-FD2. The chirality exchange altered the binding affinity to LecB but retained the P. aeruginosa biofilm inhibitory properties, while providing complete resistance to proteolysis. Amino acid chirality exchanges might provide a general strategy to stabilize peptide dendrimers towards proteolysis while retaining their biological activity.
Acknowledgements
This work was supported financially by the University of Berne, the Swiss National Science Foundation, and Swiss Federal Office for Science and Education, and the COST action D34. SAC was in receipt of a Wellcome Trust Clinical Research Training Fellowship, award reference 071313/Z/03/Z.Notes and references
- V. E. Wagner and B. H. Iglewski, Clin. Rev. Allergy Immunol., 2008, 35, 124–134 Search PubMed.
- E. Mitchell, C. Houles, D. Sudakevitz, M. Wimmerova, C. Gautier, S. Perez, A. M. Wu, N. Gilboa-Garber and A. Imberty, Nat. Struct. Biol., 2002, 9, 918–921 CrossRef CAS.
- R. Loris, D. Tielker, K. E. Jaeger and L. Wyns, J. Mol. Biol., 2003, 331, 861–870 CrossRef CAS.
- A. Imberty, M. Wimmerova, E. P. Mitchell and N. Gilboa-Garber, Microbes Infect., 2004, 6, 221–228 CrossRef CAS.
- Y. C. Lee and R. T. Lee, Acc. Chem. Res., 1995, 28, 321–327 CrossRef CAS.
- M. Mammen, S. K. Choi and G. M. Whitesides, Angew. Chem., Int. Ed., 1998, 37, 2755–2794 CrossRef.
- J. J. Lundquist and E. J. Toone, Chem. Rev., 2002, 102, 555–578 CrossRef CAS.
- R. Roy, Trends in Glycosci. and Glycotechnol., 2003, 15, 291–310 Search PubMed.
- D. Tielker, S. Hacker, R. Loris, M. Strathmann, J. Wingender, S. Wilhelm, F. Rosenau and K. E. Jaeger, Microbiology, 2005, 151, 1313–1323 CrossRef CAS.
- S. P. Diggle, R. E. Stacey, C. Dodd, M. Camara, P. Williams and K. Winzer, Environ. Microbiol., 2006, 8, 1095–1104 CrossRef CAS.
- L. Crespo, G. Sanclimens, M. Pons, E. Giralt, M. Royo and F. Albericio, Chem. Rev., 2005, 105, 1663–1681 CrossRef CAS.
- K. Sadler and J. P. Tam, J. Biotechnol., 2002, 90, 195–229 CAS.
- (a) A. Esposito, E. Delort, D. Lagnoux, F. Djojo and J. L. Reymond, Angew. Chem., Int. Ed., 2003, 42, 1381–1383 CrossRef CAS; (b) T. Darbre and J. L. Reymond, Acc. Chem. Res., 2006, 39, 925–934 CrossRef CAS; (c) T. Darbre and J. L. Reymond, Curr. Top. Med. Chem., 2008, 8, 1286–1293 CrossRef CAS.
- (a) D. Lagnoux, T. Darbre, M. L. Schmitz and J. L. Reymond, Chem.–Eur. J., 2005, 11, 3941–3950 CrossRef CAS; (b) E. M. V. Johansson, J. Dubois, T. Darbre and J. L. Reymond, Bioorg. Med. Chem., 2010, 18, 6589–6597 CrossRef CAS.
- (a) A. Clouet, T. Darbre and J. L. Reymond, Angew. Chem., Int. Ed., 2004, 43, 4612–4615 CrossRef CAS; (b) N. Maillard, A. Clouet, T. Darbre and J. L. Reymond, Nat. Protoc., 2009, 4, 132–142 Search PubMed.
- (a) E. Kolomiets, E. M. Johansson, O. Renaudet, T. Darbre and J. L. Reymond, Org. Lett., 2007, 9, 1465–1468 CrossRef CAS; (b) E. M. V. Johansson, S. A. Crusz, E. Kolomiets, L. Buts, R. U. Kadam, M. Cacciarini, K. M. Bartels, S. P. Diggle, M. Camara, P. Williams, R. Loris, C. Nativi, F. Rosenau, K. E. Jaeger, T. Darbre and J. L. Reymond, Chem. Biol., 2008, 15, 1249–1257 CrossRef CAS.
- E. Kolomiets, M. A. Swiderska, R. U. Kadam, E. M. V. Johansson, K. E. Jaeger, T. Darbre and J. L. Reymond, ChemMedChem, 2009, 4, 562–569 CrossRef CAS.
- (a) I. L. Karle, S. K. Awasthi and P. Balaram, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 8189–8193 CrossRef CAS; (b) T. S. Haque and S. H. J. Gellman, J. Am. Chem. Soc., 1997, 119, 2303–2304 CrossRef CAS; (c) J.-P. Briand, N. Benkirane, G. Guichard, J. F. E. Newman, M. H. V. van Regen Mortel, F. Brown and S. Muller, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 12545–12550 CrossRef CAS; (d) J. F. Espinosa and S. H. Gellman, Angew. Chem., Int. Ed., 2000, 39, 2330–2333 CrossRef CAS; (e) R. Tugyi, K. Uray, D. Ivan, E. Fellinger, A. Perkins and F. Hudecz, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 413–418 CrossRef CAS.
- P. Sommer, V. S. Fluxa, T. Darbre and J. L. Reymond, ChemBioChem, 2009, 10, 1527–1536 CrossRef CAS.
- (a) S. Javor, A. Natalello, S. M. Doglia and J. L. Reymond, J. Am. Chem. Soc., 2008, 130, 17248–17249 CrossRef CAS; (b) S. Javor and J. L. Reymond, J. Org. Chem., 2009, 74, 3665–3674 CrossRef CAS.
- W. L. Jorgensen, D. Maxwell and J. Tirado-Rives, J. Am. Chem. Soc., 1996, 118, 11225–11236 CrossRef CAS.
- K. J. Bowers, et al., 2006, Proceedings of the ACM/IEEE Conference on Supercomputing (SC06) (ACM Press, New York) Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of the synthesis and bioassays. See DOI: 10.1039/c0md00270d |
| This journal is © The Royal Society of Chemistry 2011 |
