Diphenylbutylpiperidine-based cell autophagy inducers: Design, synthesis and SAR studies†
Gang
Chen
a,
Hongguang
Xia
a,
Yu
Cai
a,
Dawei
Ma
*a,
Junying
Yuan
*ab and
Chengye
Yuan
*a
aState Key Laboratory of Bio-organic and Natural Product Chemistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, Shanghai, 200032, China. E-mail: yuancy@mail.sioc.ac.cn; Fax: +86 21 5492 5379
bDepartment of Cell Biology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA
First published on 28th February 2011
Abstract
A novel series of diphenylbutylpiperidine derivatives in which a benzo-five or six membered heterocycle was linked at the 4-position of the piperidine moiety were designed and synthesized. Structure–activity relationship (SAR) studies of these compounds indicated that some molecules show promising autophagy inducing activity. Replacement of the fluorine atom by a CF3 group on the diphenyl part resulted in significant enhancement of the autophagy inducing effect. In the addition, a group of diphenylpentyl nitriles also showed similar bioactivities.
Introduction
Autophagy is a cellular catabolic mechanism mediating the turnover of intracellular organelles and proteins through a lysosome-dependent degradative pathway.1Autophagy functions as an essential survival mechanism for unicellular organisms such as yeast under starvation conditions by recycling intracellular material for macromolecular synthesis and energy production.2 Current studies have suggested that dysregulation of autophagy is involved in multiple major human diseases including cancer, cardiomyopathy and neurodegenerative disorders.3Autophagy is also involved in ageing, for example, spermidine can promote longevity by inducing autophagy.4We have previously carried out a high-throughput image-based screening and identified seven FDA-approved drugs which can induce autophagy without causing cell death.5 More recently, we have described the synthesis and SAR study of diphenylbutylpiperidines (DPBPs) based on fluspirilene and penfluridol as cell autophagy inducers (Fig. 1). To our knowledge, these molecules are the first DPBPs acting on autophagy. Extensive SAR studies resulted in improved derivatives with a 10-fold increase in autophagy inducing activity.6
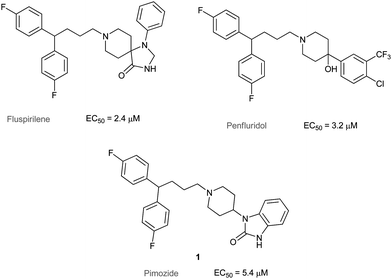 | ||
| Fig. 1 DPBP autophagy regulators. | ||
In this communication, we wish to report the result of our recent investigation of another class of DPBPs based on pimozide as autophagy inducers. We designed and synthesized some bioisosteres of lead compound as shown in Fig. 2, namely, a benzo-five or six membered heterocycle linked at the 4-position of the piperidine ring of DPBPs. These novel DPBPs acting on autophagy give us new insights into the structural and functional relationship of this class of compounds as cell autophagy inducers.
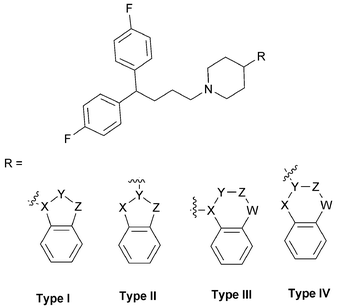 | ||
| Fig. 2 DPBPs with benzo-heterocycles at different bonding sites. | ||
Results and discussion
All of the diphenylbutylpiperidine derivatives in this paper were prepared by substitution reaction of diphenylbutyl halideviaSN2 reaction with piperidines6,7 and tested carefully by counting the number of spots of LC3-GFP in H4-LC3 cells, as described previously.6Synthetic variations on the benzimidazole scaffold (Type I and II compounds) are summarized in Scheme 1 and Scheme 2. Reductive amination of N-Boc-4-piperidone with o-aminophenol afforded the compound 2, subsequent cyclization with triphosgene, followed by deprotection gave compound 3.8 Substitution of methylsulfonyl group of piperidine derivative 4 with different 6,5-bicyclic compounds followed by removal of the Boc group using TFA afforded the desired compounds 5,86 and 7,9 respectively. Reductive amination of N-Boc-4-piperidone with indoline and deprotection using standard conditions led to compound 8. Treatment of piperidone with indole gave compound 9, which was converted to compound 10viahydrogenation with Pd/C.10
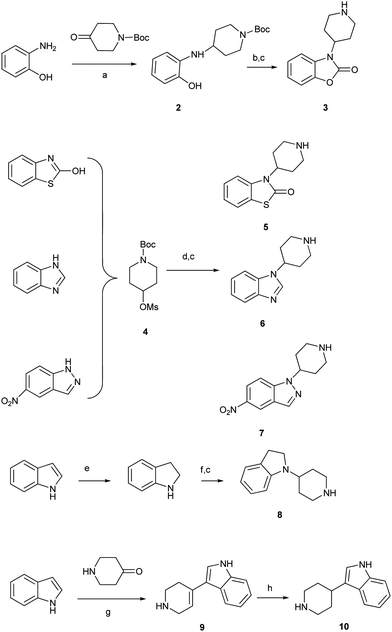 | ||
| Scheme 1 Reagents and conditions: (a) NaBH(OAc)3, DCE, RT; (b) (Cl3CO)3CO, py., THF; (c) TFA, CH2Cl2, 0 °C–RT; (d) NaH, DMF, 90 °C; (e) NaBH3CN, AcOH; (f) N-Boc-4-piperidone, NaBH(OAc)3, DCE, RT; (g) KOH, MeOH, reflux; (h) Pd-C, H2, EtOH, RT. | ||
 | ||
| Scheme 2 Reagents and conditions: (a) BH3-THF; (b) N-Boc-4-piperidone, NaBH(OAc)3, DCE, RT; (c) TFA, CH2Cl2, 0 °C–RT; (d) DMF, 90 °C; (e) DEAD, Ph3P, THF; (f) PPA, 180 °C | ||
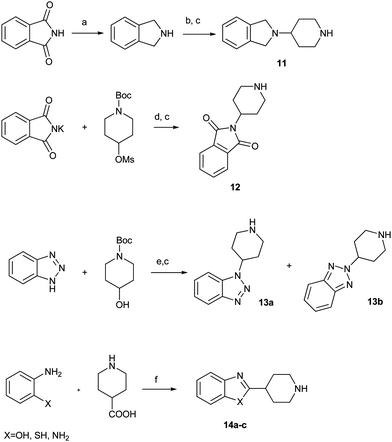
Compound 11 was synthesized from phthalimide by the same method as compound 8. Substitution of the methylsulfonyl group of piperidine with phthalimide potassium salt followed by the removal of the Boc group afforded the desired compound 12. Treatment of 4-hydroxyl piperidine with 1,2,3-benzotriazole under Mitsunobu reaction conditions followed by hydrolysis of the Boc group led to two compounds 13a and 13b.11Cyclization of different o-substituted anilines with 4-piperidine carboxylic acid gave the desired compounds 14a–c.12
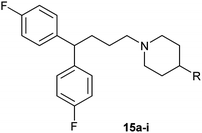
Table 1 showed the biological activity results of Type I compounds. First, it can be seen from SAR information of bioisosteres 15a–c that the activities decreased accordingly when the nitrogen atom (15a) was replaced by an oxygen (15b) or sulfur atom (15c), indicating that hydrogen bonding might play a significant role in the biological system. Next, interesting results were obtained when the benzimidazol moiety was replaced by other heterocyclic groups (15d–i); among them, compounds 15d, 15e and 15g exhibited a little increase in activity as compared to lead compound 1, however, a notable loss of inducing activity was seen with compounds 15f and 15i indicating that too many nitrogen atoms or an oxygen atom are unfavorable in this system.
|
|
||
|---|---|---|
| R | Compound | EC50 (μM) |
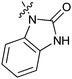
|
15a (1) | 5.4 |
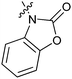
|
15b | 8.2 |
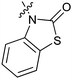
|
15c | >50 |
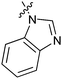
|
15d | 4.2 |
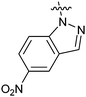
|
15e | 4.5 |
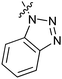
|
15f | >50 |
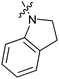
|
15g | 3.8 |
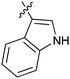
|
15h | 6.4 |
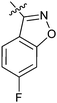
|
15i | >50 |
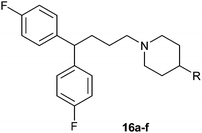
The biological activities of Type II compounds are shown in Table 2. Compounds 16a and 16d demonstrated increased potency compared to 1, however compounds 16b and 16c were found to be inactive. Among the bioisosteres 16d–f, 16d (with two nitrogen atoms) showed the best result in this series while 16e (with an oxygen atom) together with 16f (with a sulfur atom) proved to be less potent or even inactive. These results further supported our postulation that the hydrogen bonding might play an important role in the biological activity.
|
|
||
|---|---|---|
| R | Compound | EC50 (μM) |
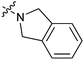
|
16a | 4.1 |
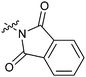
|
16b | >50 |
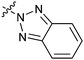
|
16c | >50 |
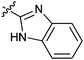
|
16d | 4.0 |
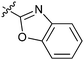
|
16e | 11.2 |
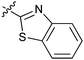
|
16f | 10.6 |
Encouraged by the positive results of Type I and Type II compounds (6,5-bicyclic system) as autophagy inducers, we also prepared Type III and Type IV compounds (6,6-bicyclic system) as shown in Scheme 3 and 4. Cyclization of anthranilamide with N-benzyl-4-piperidone afforded spiro compound 17, hydrogenation of 17 by Pd(OH)2/C gave the desired compound 18. On the other hand, reduction of 17 with LiAH4 afforded the intermediate compound 19, subsequent cyclization of which with different reagents followed by removal of the Bn group led to the desired compounds 20 and 21.13,14
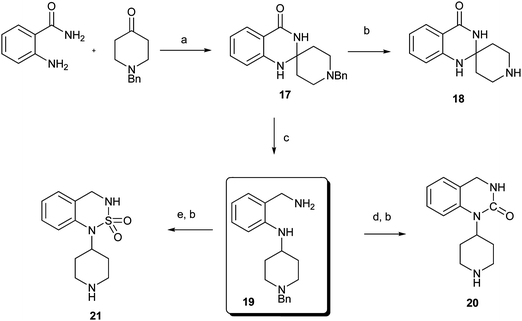 | ||
| Scheme 3 Reagents and conditions: (a) TsOH, toluene; (b) Pd(OH)2/C, H2, MeOH; (c) LiAlH4, dioxane; (d) CDl, CH3CN, reflux; (e) Sulfamide, pyridine, reflux. | ||
 | ||
| Scheme 4 Reagents and conditions: (a) NaBH4, NiCl2, MeOH; (b) N-Boc piperidone, NaBH(OAc)3, DCE, RT; (c) TFA, CH2Cl2, 0 °C–RT; (d) NaBH4, MeOH; (e) CDl, CH3CN, reflux; (f) Pd(OH)2/C, H2, MeOH; (g) Pd/C, H2, EtOH; (h) Sulfamide, pyridine, reflux; (i) Et3N, dioxane, reflux; (j) NaNO2, concd HCl; (k) ClCO2Et, reflux; (l) KOH, EtOH, reflux; (m) 47% aq. HBr, reflux. | ||
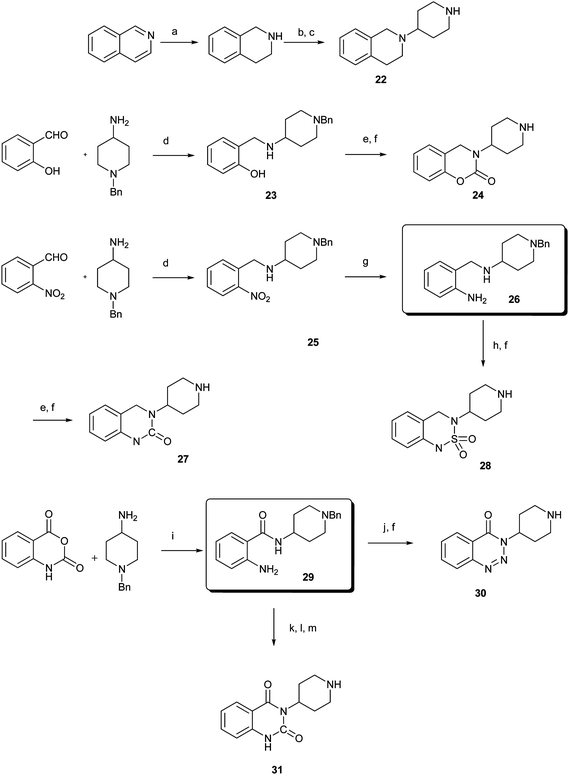
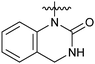
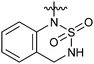
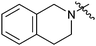
Reductive amination of N-Boc-4-piperidone with 1,2,3,4-tetrahydroquinoline and subsequent removal of the Boc group gave compound 22. Treatment of salicylaldehyde with 4-amino-1-benzylpiperidine afforded 23, subsequent cyclization with CDI then hydrogenation of the benzyl group led to compound 24.14 For the synthesis of 27 and 28, reductive amination of 2-nitrobenzaldehyde with 4-amino-1-benzylpiperidine and hydrogenation afforded the desired intermediate compound 26, while subsequent cyclization with different reagents followed by removal of the benzyl group resulted in the desired compounds 27 and 28.14 Compounds 30 and 31 were synthesized in the same manner as ring opening of isatoic anhydride with 4-amino-1-benzylpiperidine and followed by cyclization and removal of the benzyl group.14,15
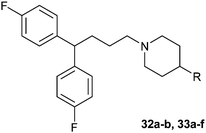
The activities of Type III and Type IV compounds are presented in Table 3. It could be found that compounds 32a, 33b, 33c and 33f showed more potency than that of lead compound 1 while compounds 32b and 33d (with sulfone group) showed decreased inducing activity, which also suggested that appropriate hydrogen bonding is of great significance. On the whole, the 4-position of the piperidine of Type III and IV compounds seem to have more tolerance of heterocycle groups than that of Type I and II compounds (33bvs.15b, 33evs.15f or 16c, 33cvs.16b).
|
|
||
|---|---|---|
| R | Compound | EC50 (μM) |
|
32a | 2.9 |
|
32b | 10.2 |
|
33a | 6.0 |
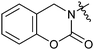
|
33b | 4.1 |
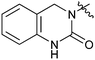
|
33c | 1.3 |
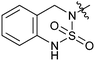
|
33d | 21.7 |
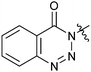
|
33e | 7.8 |
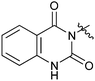
|
33f | 2.8 |
As previously reported by us, DPBPs with 4-CF3 in the diphenyl moiety have remarkably better autophagy inducing activity than the corresponding 4-F counterparts.6 As shown in Table 4, the data obtained for compounds 34a–e support such a tendency, compounds with R1 = CF3 showed 2.5–14 fold potent improved inducing activity compared to the corresponding compounds with R1 = F. Among them, compound 34c (R2 = indole moiety) showed the best result.
|
|
|||
|---|---|---|---|
| R2 | R3 = H | R1 = H, R3 = CN Compound/EC50 (μM) | |
| R1 = CF3 Compound/EC50 (μM) | R1 = F Compound/EC50 (μM) | ||
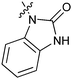
|
34a/1.2 | 15a (1)/5.4 | 35a/8.2 |
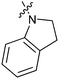
|
34b/1.2 | 15g/3.8 | 35b/4.5 |
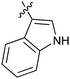
|
34c/0.45 | 15h/6.4 | 35c/2.6 |
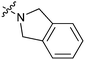
|
34d/1.5 | 16a/4.1 | 35d/7.1 |
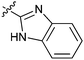
|
34e/0.90 | 16d/4.0 | 35e/8.5 |
As reported by Leurs et al.,7,16 a group of small molecules derived from diphenylpentyl nitriles has significant inhibitory effect on HIV-1. Due to the similarity of these compounds to the lead structure studied in this work, we were interested to examine the structural effect of this group of compounds as cell autophagy inducers. As shown in Table 4, these compounds also showed a high autophagy inducing effect, among them, compounds 35a, 35b, 35d and 35e exhibited diminished activities compared to the lead compound while compound 35c was around 2-fold more potent than the lead structure.
Meanwhile, it is worth pointing out the autophagy inducing effect of compounds 36 and 37, as shown in Fig. 3. As reported before,6 the replacement of a piperidine ring with a piperazine ring is unfavorable, however, compound 3617 with a piperazine ring showed some autophagy inducing activity. On the other hand, compound 37 with a spiro ring, which can be easily prepared and has better solubility in organic solvents than that of fluspirilene, showed improved autophagy inducing activity compared to that of fluspirilene.
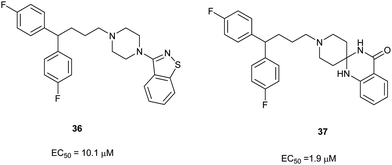 | ||
| Fig. 3 EC50 induction for compounds 36 and 37. | ||
Conclusion
In summary, we have designed and synthesized a new class of diphenylbutylpiperidines (DPBPs) bearing benzo-five and six membered heterocycles as autophagy inducers. SAR studies result in the identification of several promising compounds and suggested that 4-position of the piperidine of DPBPs can tolerate other bicyclic groups. Moreover, compounds 34c and 34e (R1 = CF3) displayed 10 and 5-fold more potency, respectively, than the lead structure. These compounds together with 37 (EC50 = 1.9 μM) have been selected for further investigation.Acknowledgements
This project was supported from the National Natural Science Foundation of China (no. 21072212 to C.Y.) and the National Institute of Health of US (to J. Y.). Zhongliang Sun and Xun Wang were involved in part of the synthetic work.Notes and references
- (a) D. J. Klionsky and S. D. Emr, Science, 2000, 290, 1717 CrossRef CAS; (b) B. Levine and J. Yuan, J. Clin. Invest., 2005, 115, 2679 CrossRef CAS.
- J. J. Lum, R. J. DeBerardinis and C. B. Thompson, Nat. Rev. Mol. Cell Biol., 2005, 6, 439 CrossRef CAS.
- T. Shintani and D. J. Klionsky, Science, 2004, 306, 990 CrossRef.
- (a) T. Eisenberg, et al, Nat. Cell Biol., 2009, 11, 1305 CrossRef CAS; (b) F. Madeo, T. Eisenberg, S. Büttner, C. Ruckenstuhl and G. Kroemer, Autophagy, 2010, 6, 160 Search PubMed.
- L. Zhang, J. Yu, H. Pan, P. Hu, Y. Hao, W. Cai, H. Zhu, A. Yu, D. Ma and J. Yuan, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 19023 CrossRef CAS.
- G. Chen, H. Xia, Y. Cai, D. Ma, J. Yuan and C. Yuan, Bioorg. Med. Chem. Lett., 2011, 21, 234 CrossRef CAS.
- J. W. Hulshof, P. Casarosa, M. P. B. Wiro, L. M. Kuusisto, H. van der Goot, M. J. Smit, I. J. P. de Esch and R. Leurs, Bioorg. Med. Chem., 2006, 14, 7213 CrossRef CAS.
- K. Flyren, L. O. Bergquist, V. M. Castro, C. Fotsch, L. Johansson, D. J. St. Jean, L. Sutina and M. Williamsa, Bioorg. Med. Chem. Lett., 2007, 17, 3421 CrossRef CAS.
- D. Zhang, D. Kohlman, J. Krushinski, S. Liang, B. Ying, J. E. Reilly, S. R. Dinn, D. B. Wainscott, S. Nutter, W. Gough, D. L. G. Nelson, J. M. Schaus and Y. Xu, Bioorg. Med. Chem. Lett., 2004, 14, 6011 CrossRef CAS.
- S. Fonquerna, M. Miralpeix, L. Pagés, C. Puig, A. Cardús, F. Antón, A. Cárdenas, D. Vilella, M. Aparici, E. Calaf, J. Prieto, J. Gras, J. M. Huerta, G. Warrellow, J. Beleta and H. Ryder, J. Med. Chem., 2004, 47, 6326 CrossRef CAS.
- T. Ruckle, M. Biamonte, T. Grippi-Valloton, S. Arkinstall, Y. Cambet, M. Camps, C. Chabert, D. J. Church, S. Halazy, X. Jiang, I. Martinou, A. Nichols, W. Sauer and J. P. Gotteland, J. Med. Chem., 2004, 47, 6921 CrossRef.
- Y. He, J. Yang, D. Robinson, K. Sprankle, P. Kung, K. Lowery, V. Mohan, S. Hofstadler, E. E. Swayze and R. Griffey, Bioorg. Med. Chem. Lett., 2004, 14, 695 CrossRef CAS.
- H. Takai, H. Obase, N. Nakamizo, M. Teranishi, K. Kubo, K. Shuto, Y. Kasuya, K. Shigenobl, M. Hashikami and N. Karashima, Chem. Pharm. Bull., 1985, 33, 1116 CAS.
- H. Takai, H. Obase, N. Nakamizo, M. Teranishi, K. Kubo, K. Shuto and T. Hashimoto, Chem. Pharm. Bull., 1985, 33, 1104 CAS.
- H. Takai, H. Obase, N. Nakamizo, M. Teranishi, A. Karasawa, K. Kubo, K. Shuto and T. Kasuya, Chem. Pharm. Bull., 1986, 34, 1907 CAS.
- J. W. Hulshof, P. Casarosa, W. M. P. B. Menge, L. M. S. Kuusisto, H. van der Goot, M. J. Smit, I. J. P. de Esch and R. Leurs, J. Med. Chem., 2005, 48, 6461 CrossRef CAS.
- The right side of compound 36 is commercially available.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c0md00236d |
| This journal is © The Royal Society of Chemistry 2011 |