New insight into the mode of action of vancomycin dimers in bacterial cell wall synthesis†
Osamu
Yoshida
b,
Jun
Nakamura
a,
Hidenori
Yamashiro
b,
Kenji
Miura
b,
Sayaka
Hayashi
a,
Kosei
Umetsu
a,
Shu
Xu
a,
Hideki
Maki
*b and
Hirokazu
Arimoto
*a
aGraduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Sendai, 980-8577, Japan. E-mail: arimoto@biochem.tohoku.ac.jp; Fax: +81-(0)22-217-6204; Tel: +81-(0)22-217-6201
bDiscovery Research Laboratories, Shionogi & Co., Ltd., Osaka, 561-0825, Japan. E-mail: hideki.maki@shionogi.co.jp
First published on 11th February 2011
Abstract
The emergence of vancomycin-resistant bacteria has created an urgent need for new active analogues of vancomycin. We previously reported vancomycin dimers with in vivo antibacterial activity. Here, we provide the first experimental insights into their inhibitory actions in bacterial cell wall synthesis.
The emergence of vancomycin-resistant enterococci (VRE) and vancomycin-resistant Staphylococcus aureus (VRSA)1 is a serious concern in clinical practice, since vancomycin is commonly used for the treatment of Gram-positive multidrug-resistant microorganisms such as methicillin-resistant S. aureus (MRSA).2 Some new drugs, such as linezolid, are available for the treatment of diseases caused by vancomycin-resistant bacteria, but pathogens resistant to linezolid have already appeared.3 It is worthy of comment that these new anti-VRE drugs target bacterial protein synthesis and thus they are different from vancomycin-class glycopeptide antibiotics with respect to their mode of action (vide infra). Vancomycin exhibits its antibacterial activity by inhibiting the reactions of bacterial cell wall (peptidoglycan, PG) biosynthesis.4 Specific affinity of vancomycin to the D-alanyl-D-alanine terminus of the PG precursor results in the inhibition of enzymatic reactions in which the precursor is used as a substrate (Fig. 1). When vancomycin-resistance is induced by exposure to vancomycin, the D-alanyl-D-alanine residue is replaced with D-alanyl-D-lactate by a set of enzymes expressed by the van cluster, the resistance genes. Consequently, the affinity of vancomycin to the PG precursor within resistant organisms is reduced to one thousandth of that in non-resistant organisms,5 which results in the loss of its antibacterial activity (Fig. 1).6
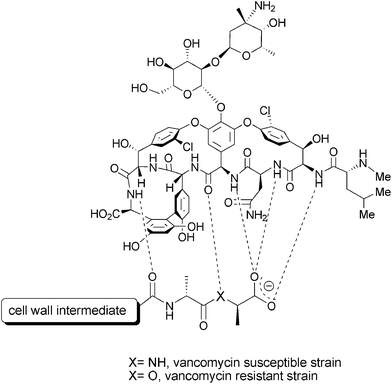 | ||
| Fig. 1 Interaction of vancomycin with the bacterial cell wall intermediate. | ||
Two major approaches to enhancing anti-VRE activity have emerged in recent years: one is based on hydrophobic modification of vancomycin (monomeric derivative),7 the other on covalent linking of two or more vancomycin molecules (vancomycin-dimers and -polymers).8 The first approach has resulted in the recent approval of telavancin by the FDA in the USA.9 The latter approach, dimers of vancomycin, has also been extensively examined for use in obtaining compounds active against vancomycin-resistant strains. Vancomycin dimers have been designed on the assumption that they would bind with enhanced avidity through multi-valent interactions with D-alanyl-D-lactate existing in cell-wall precursors of resistant bacteria.8a,8e If this was the case, the dimers might be able to show activity against resistant strains by an antibacterial mechanism similar to that of vancomycin against susceptible strains. However, that these dimers affect the cell wall synthesis of VRE has never been validated.
On the other hand, some research groups have questioned the above hypothetical mechanism for the antibacterial activity of covalently linked vancomycin dimers based on their analysis using a structure–activity relationship approach. Ellman reported that the antibacterial activity of the dimers is not likely to be due primarily to L-Lys-D-Ala-D-Lac binding.10 Griffin proposed that the anti-VRE activity of their vancosamine-linked dimers was derived from their having added hydrophobic substituents to the disaccharide moiety of vancomycin, because some monomeric vancomycin derivatives with a hydrophobic appendage at the disaccharide moiety were also known to possess anti-VRE activity.8c
These groups also did not examine the inhibitory effect of their dimers on bacterial cell wall synthesis. Thus, to address these issues, an effort needs to be made to evaluate the inhibitory activity of dimers on the cell wall synthesis of resistant bacteria using whole cells or a well-designed in vitro assay. Here, we describe the results of our biochemical investigations into the action of the vancomycin dimers, which provide the first experimental insights into their inhibitory actions in bacterial cell wall synthesis.
We reported previously that vancomycin dimers 3 and 4 (Fig. 2) exhibited excellent activity against vancomycin-resistant bacteria.11 Two vancomycin units are connected in these dimers via a rigid phenoxazone linker between vancosamine sugar moieties. In order to examine the effect of linkage orientation and length on the antibacterial mechanism of the dimers, new derivatives 1 and 2 (Fig. 2), which were linked through the carboxylate terminals of their vancomycin units,12 were also prepared with the phenoxazone linker. Table 1 summarizes the minimum inhibitory concentration (MIC) of the compounds employed in this study. The data of a monomeric derivative, Van-M-02 (Fig. 2),13 is also included in the table for comparison. S. aureus RN4220 is a vancomycin-susceptible strain. E. faecium SR7940 and SR23598 are VREs with different phenotypes (VanA and VanB).13 Dimers 1–4 exhibited a good-to-excellent level of activity against VREs. However, the newly synthesized 1 and 2 had significantly reduced activity against vancomycin-susceptible S. aureus (16 and 8 μg mL−1, respectively). Edman degradation of vancomycin, 4, and Van-M-02 removed the N-terminal leucine, and gave ΔN-vancomycin, 5, and ΔN-Van-M-02.13 Among these, only ΔN-vancomycin showed a complete loss of activity against all three of the bacterial strains tested. The degraded dimer 5 lost the activity against VREs, but retained that against S. aureus. A degraded monomer derivative ΔN-Van-M-02 showed weaker yet still good antibacterial potency against both S. aureus and VREs. The observation of different effects of N-terminal degradation on antibacterial activity may imply differences in the molecular modes of action of vancomycin, vancomycin dimer, and Van-M-02. The effects of the N-terminal degradation of vancomycin derivatives on their binding to D-alanyl-D-alanine are well known.5,7a At this stage, however, caution is needed in interpreting such results due to the lack of proof that the dimers exhibit their activities by inhibiting the cell wall synthesis of VREs.
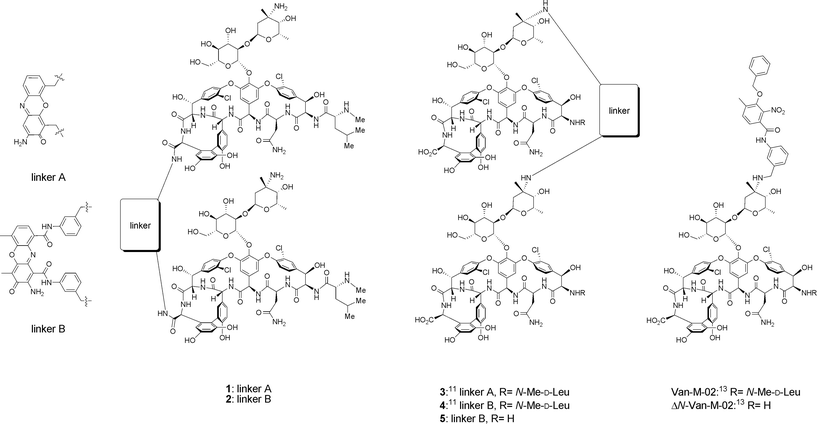 | ||
| Fig. 2 Vancomycin derivatives with different linking patterns. | ||
| Compounds | MIC (μg mL−1)a | ||
|---|---|---|---|
| S. aureus | E. faecium | E. faecium | |
| X= NHe | X= Oe | X= Oe | |
| Susceptible | Resistant (VanA) | Resistant (VanB) | |
| a Minimum Inhibitory Concentration [μg mL−1]. b RN4220. c SR7940. d SR23598. e See Fig. 1. f ref. 11. g ref. 13. | |||
| Vancomycin | 1 | >64 | >64 |
| ΔN-Vancomycin | >64 | >64 | >64 |
| 1 | 16 | 4 | 1 |
| 2 | 8 | 16 | 16 |
| 3 | 2 | 8 | 1 |
| 4 | 1 | 8 | 2 |
| 5 | 4 | >64 | 32 |
| Van-M-02g | 0.125 | 1 | 0.25 |
| ΔN-Van-M-02g | 2 | 8 | 2 |
We next examined the primary site of action of vancomycin dimers against VREs. The inhibition of bacterial biomacromolecules (protein, RNA, DNA, peptidoglycan, and lipid biosynthesis) by the dimers 2 and 4 was evaluated by monitoring the incorporation of radio-isotope-labeled precursors (Fig. 3). Despite their different linking positions, both dimers 2 and 4 showed selective inhibition of bacterial PG synthesis. To our knowledge, this is the first experimental evidence that vancomycin dimers exhibit antibacterial activity against VRE through the suppression of cell wall synthesis. Under similar conditions, the parent vancomycin did not show inhibition of any of these biomacromolecule syntheses of VREs (data not shown).
![Dimers inhibiting the synthesis of peptidoglycan. (Strain) E. faecium SRM1101 (const. VanB); (RI labeling) [14C]: leucine - protein synthesis, [14C]: thymidine - DNA synthesis, [3H]: uracil - RNA synthesis, [14C]: acetic acid - fatty acid synthesis, [3H]: N-acetyl-d-glucosamine (GlcNAc) - peptidoglycan synthesis; (procedure) Antibacterial and precursors labeled with radio-isotope were added to the culture fluid at the logarithmic growth phase. Amount of each precursor taken into bacteria was quantitated after cultivation for fifteen minutes at 37 °C.](/image/article/2011/MD/c0md00230e/c0md00230e-f3.gif) | ||
| Fig. 3 Dimers inhibiting the synthesis of peptidoglycan. (Strain) E. faecium SRM1101 (const. VanB); (RI labeling) [14C]: leucine - protein synthesis, [14C]: thymidine - DNA synthesis, [3H]: uracil - RNA synthesis, [14C]: acetic acid - fatty acid synthesis, [3H]: N-acetyl-D-glucosamine (GlcNAc) - peptidoglycan synthesis; (procedure) Antibacterial and precursors labeled with radio-isotope were added to the culture fluid at the logarithmic growth phase. Amount of each precursor taken into bacteria was quantitated after cultivation for fifteen minutes at 37 °C. | ||
In order to obtain more detailed information about the cell wall synthesis inhibition, we conducted semi-quantitative analysis using an in vitro cell wall synthesis assay. In the assay (Fig. 4),13 a membrane fraction capable of in vitro peptidoglycan synthesis was prepared from S. aureus, and UDP-MurNAc-pentapeptide (L-Ala-γ-D-Glu-L-Lys-D-Ala-D-Ala) isolated from the same strain was used as a starting material. Through a series of enzymatic reactions in this system, a pentaglycinated lipid intermediate (LI, Fig. 4) is synthesized, and could be quantified by the radioactivity derived from 14C-labeled glycine. The transglycosylase domain of penicillin-binding protein (PBP) catalyzes the last steps of the PG synthetic reactions, and forms 1,4-glycosyl bonds among labeled LIs to generate immature PG. Then the transpeptidase domain of PBP connects the polyglycine terminus to the amino terminus of a neighboring residue producing mature PG, the final product of this system (Fig. 4). The combined amount of immature and mature PG is also quantified by radioactivity. The effect of compounds on in vitro LI synthesis and PG synthesis (Fig. 4) of vancomycin-susceptible S. aureus was thus evaluated. Because VRSA and VRE employ the same precursor, UDP-MurNAc-pentadepsipeptide (L-Ala-γ-D-Glu-L-Lys-D-Ala-D-Lactate), the use of the precursor isolated from VRE, along with the cell membrane of S. aureus allowed us to estimate the effects of vancomycin derivatives on the in vitro cell wall synthesis of VRSA.
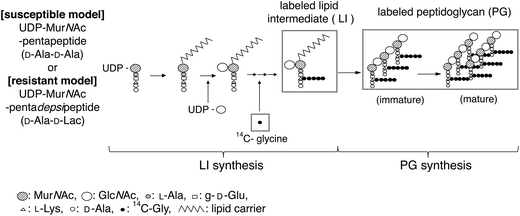 | ||
| Fig. 4 In vitro inhibition assay of PG synthesis in the bacterial membrane of S. aureus. 14C-glycine, Sup Mix, UDP-GlcNAc (200 mM), Tris-HCl (pH 8.5), UDP- MurNAc-pentapeptide (100 mM), MgCl2, β- ME, ATP, DMSO. | ||
Dimers 1 (carboxyl terminus linked dimer), 3, 4 (vancosamine terminus linked dimers), and 5 (damaged dimer) were tested in this assay (Table 2). In both the resistant and susceptible models, the dimers inhibited both LI and PG synthesis (Fig. 4) notwithstanding their different linking styles. This suggested that the dimers have different type of mechanism comparing to vancomycin because it can hardly inhibit LI synthesis. In the resistant model, regardless of their linking style, the dimers inhibited both LI and PG synthesis, and showed more intensive inhibitory activity in PG synthesis than in LI synthesis. This result was in contrast to that of Van-M-02, which has been shown to inhibit both LI and PG synthesis in a resistant model in a similar range.2 Although a report by Griffin proposed that the dimers forming a link between vancosamine moieties may have a similar antibacterial mechanism with the monomeric derivatives modified at the same sugar moieties,8c dimers 1, 3, and 4 showed a quite different pattern of inhibition from Van-M-02. Thus, the mechanism of these dimers against resistant bacteria should be different from that of Van-M-02.
| Compounds | IC50 (μg mL−1)a | |||
|---|---|---|---|---|
| Susceptible model | Resistant model | |||
| LI step | PG step | LI step | PG step | |
| a 50% Inhibitory Concentration. b ref. 13. | ||||
| Vancomycinb | 180 ± 46 | 4.5 ± 0.4 | 2,300 ± 1,400 | 85 ± 28 |
| 1 | 64 ± 0.5 | 1.2 ± 0.1 | 200 ± 28 | 21 ± 10 |
| 3 | 98 ± 36 | 2.2 ± 0.6 | 700 ± 220 | 31 ± 11 |
| 4 | 74 ± 7.7 | 11 ± 0.7 | 300 ± 23 | 61 ± 14 |
| Van-M-02b | 32 ± 7.0 | 4.2 ± 0.1 | 76 ± 0.6 | 36 ± 4.2 |
The inhibitory activity of compound 5 was evaluated, because removal of the N-terminal leucine has been shown to reduce the avidity of glycopeptides to D-alanyl-D-alanine or D-alanyl-D-lactate (Table 3).5 Although Table 1 showed large differences in MIC of compounds 4 and 5 for resistant strains, the data in Tables 2 and 3 show minimal differences in inhibitory activity of compounds 4 and 5 for the resistant model. Further studies will thus be needed to decipher the detail mode of actions of these compounds.
Conclusions
We have demonstrated for the first time that vancomycin dimers inhibit cell wall synthesis of vancomycin-resistant bacteria. The dimers strongly inhibited the polymerization step of peptidoglycan in an in vitro assay. This observation is in contrast to the findings reported for monomeric Van-M-02 in a previous study, and may reflect a unique molecular mode of action of vancomycin dimers. Because the cell wall polymerization step appears to be the target of vancomycin dimers, further analyses using purified bacterial enzymes are warranted.Acknowledgements
This work was supported in part by Grant-in-Aid for Scientific Research from MEXT, Japan (Nos. 17035039, 18032010, and 21310136) and by funds from the Uehara Memorial Foundation and Mochida Foundation.Notes and references
- (a) D. Miller, V. Urdaneta, A. Weltman and S. Park, JAMA, J. Am. Med. Assoc., 2002, 288, 2116; (b) H. K. Tiwari and M. R. Sen, BMC Infect. Dis., 2006, 6 Search PubMed Article 156; (c) M. Aligholi, M. Emaneini, F. Jabalameli, S. Shahsavan, H. Dabiri and H. Sedaght, Med. Princ. Pract., 2008, 17, 432–434.
- D. Kahne, C. Leimkuhler, W. Lu and C. Walsh, Chem. Rev., 2005, 105, 425–448 CrossRef CAS.
- I. A. Herrero, N. C. Issa and R. Patel, N. Engl. J. Med., 2002, 346, 867–869.
- (a) D. H. Williams, M. P. Williamson, D. W. Butcher and S. J. Hammond, J. Am. Chem. Soc., 1983, 105, 1332–1339 CrossRef CAS; (b) P. Reynolds, Eur. J. Clin. Microbiol. Infect. Dis., 1989, 8, 943–950 CAS.
- P. H. Popieniek and R. F. Pratt, Anal. Biochem., 1987, 165, 108–113 CAS.
- (a) C. T. Walsh, S. L. Fisher, I. S. Park, M. Prahalad and Z. Wu, Chem. Biol., 1996, 3, 21–28 CrossRef CAS; (b) T. D. H. Bugg, G. D. Wright, S. Dutka-Malen, M. Arthur, P. Courvalin and C. T. Walsh, Biochemistry, 1991, 30, 10408–10415 CrossRef CAS; (c) R. Xu, G. Greiveldinger, L. E. Marenus, A. Cooper and J. A. Ellman, J. Am. Chem. Soc., 1999, 121, 4898–4899 CrossRef CAS.
- (a) R. Nagarajan, J. Antibiot., 1993, 46, 1181–1195 CAS; (b) M. Ge, Z. Chen, H. R. Onishi, J. Kohler, L. L. Silver, R. Kerns, S. Fukuzawa, C. Thompson and D. Kahne, Science, 1999, 284, 507–511 CrossRef CAS; (c) R. Kerns, S. D. Dong, S. Fukuzawa, J. Carbeck, J. Kohler, L. Silver and D. Kahne, J. Am. Chem. Soc., 2000, 122, 12608–12609 CrossRef CAS; (d) Y. Nakama, O. Yoshida, M. Yoda, K. Araki, Y. Sawada., J. Nakamura, S. Xu, K. Miura, H. Maki and H. Arimoto, J. Med. Chem., 2010, 53, 2528–2533 CAS.
- (a) J. Rao and G. M. Whitesides, J. Am. Chem. Soc., 1997, 119, 10286–10290 CrossRef CAS; (b) K. C. Nicolaou, R. Hughes, S. Y. Cho, N. Winssinger, H. Labischinski and R. Endermann, Chem.–Eur. J., 2001, 7, 3824–3843 CrossRef CAS; (c) J. H. Griffin, M. S. Linsell, M. B. Nodwell, Q. Chen, J. L. Pace, K. L. Quast, K. M. Krause, L. Farrington, T. X. Wu, D. L. Higgins, T. E. Jenkins, B. G. Christensen and J. K. Judice, J. Am. Chem. Soc., 2003, 125, 6517–6531 CrossRef CAS; (d) H. Arimoto, K. Nishimura, T. Kinumi, I. Hayakawa and D. Uemura, Chem. Commun., 1999, 1361–1362 RSC; (e) U. N. Sundram, T. I. Nicas and J. H. Griffin, J. Am. Chem. Soc., 1996, 118, 13107–13108 CrossRef CAS.
- (a) A. King, I. Phillips and K. Kaniga, J. Antimicrob. Chemother., 2004, 53, 797–803 CrossRef CAS; (b) D. L. Higgins, R. Chang, D. V. Debabov, J. Leung, T. Wu, K. M. Krause, E. Sandvik, J. M. Hubbard, K. Kaniga, D. E. Schmidt Jr, Q. Gao, R. T. Cass, D. E. Karr, B. M. Benton and P. P. Humphrey, Antimicrob. Agents Chemother., 2005, 49, 1127–1134 CrossRef CAS.
- R. K. Jain, J. Trias and J. A. Ellman, J. Am. Chem. Soc., 2003, 125, 8740–8741 CrossRef CAS.
- J. Lu, O. Yoshida, S. Hayashi and H. Arimoto, Chem. Commun., 2007, 251–253 RSC.
- U. N. Sundram, T. I. Nicas and J. H. Griffin, J. Am. Chem. Soc., 1996, 118, 13107–13108 CrossRef CAS.
- K. Miura, H. Yamashiro, K. Uotani, S. Kojima, T. Yutsudo, J. Lu, O. Yoshida, Y. Yamano, H. Maki and H. Arimoto, Antimicrob. Agents Chemother., 2010, 54, 960–962 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures; 1H NMR data and spectra; HRMS; compound purity. See DOI: 10.1039/c0md00230e |
| This journal is © The Royal Society of Chemistry 2011 |
