Spatially controlled hydrogel mechanics to modulate stem cell interactions
Ross A.
Marklein
and
Jason A.
Burdick
*
Department of Bioengineering, University of Pennsylvania, 240 Skirkanich Hall, 210 South 33rd Street, Philadelphia, PA 19104, USA. E-mail: burdick2@seas.upenn.edu
First published on 27th October 2009
Abstract
Local control of the stem cell microenvironment with biomaterial design is of critical importance for tissue engineering. Matrix mechanics is one aspect of biomaterial design that has received considerable attention recently due to the effect of mechanics on stem cell proliferation, morphology, and differentiation. In order to investigate the effect of locally controlled mechanics on human mesenchymal stem cells (hMSCs), a sequentially crosslinked hyaluronic acid hydrogel system was developed that permits spatial patterning of mechanics (distinct patterns and gradients). Methacrylated hyaluronic acid was synthesized to allow for crosslinking via both Michael-type addition using a dithiol and radical polymerization using light. By varying the initial methacrylate consumption through addition crosslinking, restricting UV light to specified regions, and varying UV exposure time, a wide range of mechanics (from ∼3 kPa to ∼100 kPa) was possible in both uniform and patterned hydrogels. hMSCs exhibited increased spreading and proliferation on stiffer gels compared to cells cultured on softer gels. Furthermore, cells grown on gels with patterned mechanics exhibited spreading and proliferation behavior that correlated with the local mechanics. This method to spatially control matrix mechanics represents a novel hydrogel system to tune the stem cell microenvironment.
Introduction
The ability of stem cells to interact with and respond to their environment is being increasingly investigated both in native tissues and in synthetic systems.1 For example, it is now clear that cells respond to the mechanical properties of their surroundings, which was originally investigated in somatic cells such as fibroblasts and endothelial cells2,3 and more recently in stem cells, including the effects of mechanics in specifying lineage commitment.4 Native tissues can vary in stiffness (e.g., 0.1–1 kPa in brain tissue, ∼10 kPa in relaxed muscle, and >30 kPa for pre-mineralized bone5) and stem cells differentiate down tissue specific lineages based on these properties.6,7 Thus, a clear understanding of this behavior may be useful in the design of materials for applications in tissue engineering or for better understanding of cellular behavior in disease states. For instance, stem cells in fibrotic myocardium after injury, where mechanics are greater than in healthy tissue, may differentiate and mineralize their surrounding matrix.8Tissue engineering strategies have begun to incorporate matrix mechanics as a means to control stem cell behavior, including morphology, proliferation, and extracellular matrix (ECM) secretion.9 Coupled with other differentiation cues such as growth factors or adhesive ligands, an engineered biomimetic approach to tissue repair and regeneration may be possible by controlling the inherent mechanical properties of the engineered scaffold. However, one limitation of current biomaterial systems used in these investigations is the inability to spatially control the network properties of the scaffold. Due to the heterogeneous nature of tissues, it is necessary to design scaffolds that reflect these differences in spatial and temporal matrix properties in order to facilitate proper cell behavior and tissue integration. Spatial differences in local mechanics are also relevant in certain pathologies10,11 and wound healing processes,8,12 and therefore the characterization and understanding of cell responses to these complex microenvironments are critical for better understanding of fundamental stem cell behavior and developing an effective tissue engineering strategy.
Only a few examples exist where hydrogel properties are controlled spatially. Much of this is dependent on the use of light, due to the precise control that light affords. Photopolymerization with UV light is a commonly employed technique that involves radical polymerization using methacrylate or acrylate functionalized polymers.13 By restricting this light to certain regions, complex patterns of exposed and non-exposed regions can be imparted in hydrogels to spatially control cell behavior.14–16 Beyond patterning, gradients are useful in many applications and are found in many tissues and can direct cellular migration.17 Hydrogel gradients can be formed using specific mixing devices18 or microfluidic chambers,19,20 but these techniques rely on the use of complex systems or only permit gradients of a certain magnitude. Thus, a need exists for a hydrogel system that can be manipulated in space with respect to mechanical properties.
Hyaluronic acid (HA) is a polysaccharide that is present in native tissue and is also intimately involved in processes such as wound healing, cell motility, embryogenesis, and inflammation.21,22 HA possesses properties desirable for tunable scaffolds as a wide range of molecular weights can be obtained, as well as the presence of chemically modifiable groups (hydroxyl and carboxyl groups) on the backbone. Functionalized HA with reactive groups such as methacrylates and acrylates has been utilized to form HA-based hydrogels for controlling stem cell differentiation.21–26 These systems allow for uniform gel properties and effective cell encapsulation, but do not allow for local control of the spatial and temporal properties of the network. Recently, a hydrogel system based on acrylated HA demonstrated spatial control of human mesenchymal stem cells (hMSCs) by utilizing two crosslinking steps involving the acrylate functional group on the modified HA backbone.27 This allowed for differences in crosslinking in regions that were further exposed to UV light compared to regions that only utilized the initial crosslinking step. In these patterned 3D environments that only exhibited minor changes in mechanical properties, hMSCs adopted a rounded morphology in the dual crosslinked regions and a spread morphology in the single crosslinked regions.
In the current study, a similar dual crosslinking method that supports major changes in mechanics (from several kPa to ∼100 kPa) was utilized to explore the effects of mechanics on 2D hMSC behavior, namely spreading and proliferation. Additionally, the spatial patterning of mechanics was realized by regionally restricting light exposure during the second crosslinking. The ability of patterned and gradient mechanics to control hMSC behavior was also investigated. Although this is only a preliminary step towards the utility of these systems for actual tissue engineering constructs, this novel system allows for spatial control of matrix mechanics for the purposes of driving stem cell behavior.
Experimental
Methacrylated hyaluronic acid (MeHA) synthesis
MeHA was synthesized as described previously.23,28 Briefly, sodium hyaluronate (Lifecore, 59 kDa) was dissolved at 1 wt% in deionized water and methacrylic anhydride (MA) was added dropwise (∼2.4 mL MA per g HA) while stirring at 4 °C. The pH was maintained above 8 by adding 5 N NaOH for 8 h, followed by overnight reaction and further addition of MA (∼1.2 mL MA per g HA) and pH maintenance for 4 h the following day. The macromer solution was dialyzed against deionized water (SpectraPor, MW cutoff 6000–8000 Da) for 3 days, frozen at −80 °C, lyophilized, and stored frozen in powder form. 1H NMR revealed ∼100% modification of the hydroxyl groups on the HA backbone, resulting in the MeHA structure shown in Fig. 1A.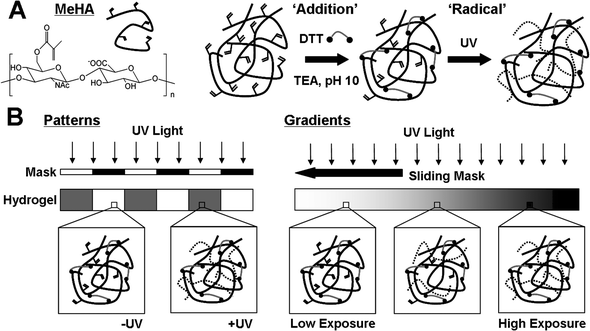 | ||
| Fig. 1 (A) Chemical structure of methacrylated hyaluronic acid (MeHA) and dual crosslinking mechanism. Michael addition of methacrylates with DTT (dithiol crosslinker) induces partial crosslinking of a solution of MeHA and TEA at pH 10. Remaining methacrylates undergo radical polymerization when exposed to UV light in the presence of a photoinitiator (dotted lines are kinetic chains) to increase crosslinking density (i.e., mechanics). (B) Spatial variations in hydrogel mechanics can be introduced by restricting UV light to certain regions of the addition crosslinked gel using a photomask to create patterns (left) or varying the time of UV exposure (via a sliding mask) to create gradients (right). | ||
Methacrylated slide preparation
In order to easily handle and process the thin gels, slides were methacrylated to allow for covalent attachment of the gels to the glass. 22 mm × 22 mm coverslips were first plasma coated for 3 min to activate the surface for methacrylation. Next, 100 µL of 3-(trimethoxysilyl)propyl methacrylate (Sigma) were placed on each activated slide and reacted at 100 °C for 1 h followed by 110 °C for 10 min. Finally, the slides were rinsed with deionized water and ethanol and allowed to dry.MeHA hydrogel crosslinking
MeHA hydrogels were formed using one- or two-step crosslinking processes (Fig. 1A). In the first step, Michael-type ‘addition’ crosslinking occurs via introduction of dithiothreitol (DTT, Sigma) to a 3 wt% solution of MeHA in PBS buffer containing 0.2 M triethanolamine (TEA, Sigma) and 0.05 wt% I2959 (Irgacure). Various amounts of DTT were added to vary the theoretical molar consumption of methacrylates (12, 15, 20, 30, 50, and 100%) to achieve a range of initial mechanics during this first step. The oligopeptide GCGYGRGDSPG was added prior to the DTT crosslinking step to allow for coupling of the well-established RGD adhesion moiety to the network. Gels were formed between slides with 150 µm spacing and methacrylated slides were used on one side to allow for covalent gel attachment. After mixing, solutions were reacted for 1 h at 37 °C to complete the ‘addition’ crosslinking step (“−UV” gels).Gels could be further exposed to UV light in order to initiate ‘radical’ crosslinking of the remaining unconsumed methacrylates. Collimated 10 mW cm−2 365 nm UV light (Omnicure S1000 UV Spot Cure System, Exfo Life Sciences Division, Mississauga, Ontario, Canada) was used to uniformly expose the entire gels for 4 min (“+UV” gels). This step could be performed to create uniform gels or to create gels with spatially controlled mechanics (Fig. 1) by either (a) restricting UV light with a photomask or (b) varying the UV exposure time using a sliding photomask. For patterns, photomasks consisted of printed transparencies containing 500 µm stripe patterns that were created using Adobe Photoshop and printed at a resolution of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 DPI. Gradient gels were formed by passing a photomask over the surface of the gel at a constant linear velocity (10 mm min−1 using a syringe pump) to create a range of varied exposure times and crosslinking (0–90 s) across a 15 mm distance.
000 DPI. Gradient gels were formed by passing a photomask over the surface of the gel at a constant linear velocity (10 mm min−1 using a syringe pump) to create a range of varied exposure times and crosslinking (0–90 s) across a 15 mm distance.
Characterization of hydrogel mechanics
Hydrogel surface mechanics were quantified using atomic force microscopy (AFM, Veeco Bioscope I). A silicon bead AFM tip with a spring constant of 0.06 N m−1 was used to obtain force curves for individual points on the gels (15 points chosen for each condition) from which a local elastic modulus was calculated. For patterned gels, points were chosen at regular intervals along the distance of the gel (500 µm for stripe patterns or every 1.5 mm for gradient patterns).Cell seeding
hMSCs were obtained from Lonza Corporation (Wakersville, MD) and used at low passage for all studies (passages 2–5). Cells were expanded and cultured in standard hMSC growth medium (α-MEM, 10% fetal bovine serum (FBS), 1% L-glutamine and penicillin–streptomycin) and seeded at a density of 5k cells per cm2 on gel surfaces. For most studies, an RGD concentration of 1 mM was used to promote hMSC adhesion and spreading. In order to determine whether the swelling of the less crosslinked gels (lowest mechanics) resulted in an effective diluting of the RGD ligand, we tested several ligand densities (1, 2, and 5 mM) to elucidate the influence of ligand density on cellular spreading. Gels were sterilized under a germicidal lamp for 2 h in sterile PBS prior to cell seeding.Cell imaging and quantification
Cell spread area was calculated after 24 h for both uniform and patterned gels using an inverted microscope (Axiovert 200, Carl Zeiss Inc.). ImageJ (NIH) was used to calculate average cell spread area (>50 cells per group) for each uniform gel condition (−/+UV), as well as on regions of both striped and gradient gels. Cells were stained with calcein AM (Invitrogen) for imaging on photopatterned gels. Further staining was performed by fixing cells with 4% formalin followed by permeabilization with 0.25% v/v Triton-X (Sigma) and cell nuclei staining with 2 µg mL−1 DAPI (Invitrogen). Cell proliferation was quantified by counting cell nuclei on 5 images at 10× magnification on days 1, 4, and 7 for uniform gels.Statistical analysis
Values are reported as means and standard deviations (mechanics, proliferation) or standard errors of the mean (cell spreading). Statistical differences (p < 0.05) were determined using a Student's t-test (JMP software) to compare either mechanics, cell spreading, or cell proliferation on −/+UV gels.Results and discussion
MeHA hydrogel characterization and cellular response
Hydrogels with uniform mechanics were formed by a multi-step crosslinking procedure, where addition crosslinking (via DTT) is performed initially (−UV group) to consume all or a fraction of reactive groups and then followed by radical crosslinking (+UV group) to further consume reactive groups. In this case, the reactivity was due to methacrylates on HA that can react with thiols (on DTT) via an addition reaction or with each other during a radical polymerization to form kinetic chains in the presence of light and a photoinitiator. A highly functionalized HA (∼100% modified) was used to allow for large changes in mechanics at a uniform concentration (3 wt%); however, these parameters can be varied to alter overall gel properties. Notably, the second step uses light that can be controlled spatially to obtain gels with regionally diverse properties.AFM mechanical testing allowed for local probing of the surface mechanical properties, which is representative of what a cell would sense when interacting with the material. Fig. 2A illustrates the wide range of mechanics (nearly three orders of magnitude) achieved using this dual crosslinking system. By varying the initial methacrylate consumption via molar ratio of DTT added, the hydrogel modulus ranged from 2.3 kPa (12% DTT −UV) to 84 kPa (100% DTT −UV). Furthermore, the exposure of these gels to UV light resulted in an increase in modulus to ∼100 kPa in all dual crosslinked hydrogels, which agreed well with the hydrogel polymerized using only the radical crosslinking step (Fig. 2A, 0% DTT +UV). Excluding the 100% −/+UV hydrogels, there were significant differences (**p < 0.001) in the mechanics between −/+UV hydrogels in all cases.
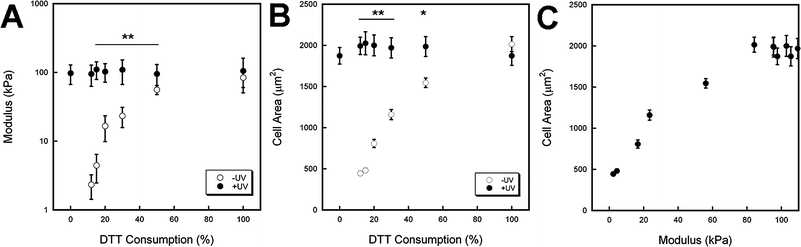 | ||
| Fig. 2 Characterization of mechanics (assessed with AFM) and hMSC response to hydrogels with uniform properties. (A) Hydrogel modulus for variable DTT consumption (theoretical values shown, based on molar ratio of thiols on DTT to methacrylates on MeHA) before (−UV, white) and after (+UV, black) light exposure. The mechanics can be tailored over two orders of magnitude with this system and result in a peak modulus of ∼100 kPa. (B) hMSC spread area 24 h after seeding for the same hydrogel systems. Spreading increased with increasing DTT consumption and UV exposed gels show similar spread area responses. (C) hMSC spread area 24 h after seeding versus mechanics shows increased cell area with increasing mechanics until a plateau is reached at ∼80 kPa. Significant differences (**p < 0.001, *p < 0.01) were found between −UV and +UV gels. | ||
The ability to sequentially crosslink hydrogels has been used in a similar addition–radical sequential crosslinking system27 and in other systems containing multi-component interpenetrating networks in which the two networks crosslink by different means.16,29–31 However, none of these systems exhibit the wide range of mechanics achievable with this dual crosslinkable MeHA system. While multi-component hydrogel systems allow for incorporation of multiple cell recognition sites and spatio-temporal control over mechanics and degradation, the complexity of these microenvironments makes determination of factors influencing stem cell behavior more difficult. In our system we use a constant polymer and ligand concentration while only altering the crosslinking of the same macromer, and thus mechanics. Therefore, any observed differences in hMSC behavior should be attributed to mechanics and not regional differences in ligand density and matrix components.
Fig. 2B shows the spread area of hMSCs on hydrogels with a range of DTT consumptions for hydrogels −/+UV after 24 h. Again, significant differences (**p < 0.001, *p < 0.01) were found between the cell responses on −/+UV hydrogels for all cases except the 100% −/+UV (where there was not a significant change in mechanics). The spread cell area is also plotted as a function of hydrogel mechanics in Fig. 2C. This demonstrates a clear dependence of hMSC spreading on the mechanics of the substrate as spreading increases until it plateaus. Increases in cell area with increasing mechanics have also been shown in studies using other substrates4,32 and other cell types.2,33,34 The ability of stem cells to mechanosense has been linked to integrin binding and coupling of the cytoskeleton to these adhesion sites, which is responsible for development of cellular tension and is stiffness dependent for adhesion dependent cells.2,35,36 The presence of the RGD motif allows for binding with α1β5 integrins, which have been implicated in stem cell morphology and fate decisions.37–39 The importance of RGD is further exemplified in negative controls consisting of HA gels without coupled RGD, which showed no cell spreading on both soft and stiff substrates (data not shown).
Histogram analysis of 12% DTT −/+UV hydrogels shows two distinct populations of hMSCs in terms of spread area (Fig. 3). Cells on the “soft” 12% −UV hydrogels exhibited a spherical morphology with very few extensions, while cells on the “stiff” +UV hydrogels showed a much more spread morphology and a wider distribution. Inset images are representative of cells in each condition after 24 h. These differences in morphology (due to mechanics) could specify further lineage commitment as stem cells have shown differentiation responses to controlled and restricted cell morphologies where less spread cells undergo adipogenesis while more spread cells undergo osteogenesis at a constant ligand density.40 However, these previous studies have been performed on substrates with uniform mechanics.
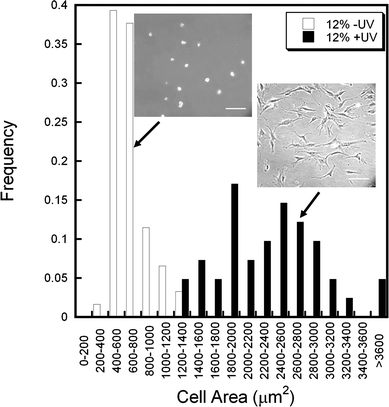 | ||
| Fig. 3 Distribution of hMSC spreading on “soft” (12% DTT −UV, white) and “stiff” (12% DTT +UV, black) hydrogels 24 h after seeding. Two distinct populations are evident as the stiffer hydrogel promotes greater cell spreading and a wider distribution of spread areas. Representative images are shown for each population, scale bar = 200 µm. | ||
RGD concentration dependence
While we were able to show orders of magnitude differences in mechanics for −/+UV hydrogels, it was necessary to demonstrate that the lack of spreading on 12% DTT −UV hydrogels was a result of mechanics and not an effective diluting of the RGD due to swelling. Large changes in surface area due to swelling were not observed, potentially due to the gel binding to the substrate, yet it is important to investigate how minor changes may influence outcomes. This potential decrease in surface ligand density could result in hMSCs not forming sufficient integrin binding sites to allow for spreading on soft substrates. To investigate this, cells were seeded on “soft” hydrogels (12% DTT −UV) containing 1, 2, and 5 mM RGD to see if the increase in ligand density would result in spreading. Due to the high modification of HA used in this system, the percentages of methacrylates consumed by the RGD coupling were ∼1.5, 3, and 7.5% for 1 mM, 2 mM, and 5 mM, respectively. This low percentage of methacrylates consumed by the RGD coupling would therefore not result in competition with the DTT crosslinking step. As shown in Fig. 4, cells at all ligand concentrations exhibit the same rounded morphology, indicating that the lack of spreading is not due to potential ligand density issues arising from hydrogel swelling. The strength of α1β5 integrin binding to fibronectin (specifically RGD and its synergy sequences) is tension dependent and while the number of total integrin binding complexes is constant on hydrogels of different mechanics, the adhesive strength of these complexes is stiffness dependent.41 Although the cell may be forming more or less adhesive bonds to the MeHA hydrogels at different RGD concentrations, these “relaxed bonds” do not allow the cell to develop sufficient tension to spread on these soft substrates. Based on these findings, 1 mM RGD was used for the remaining studies. | ||
| Fig. 4 Representative images (top, scale bar = 200 µm) and histogram (bottom) of the relationship between hMSC spreading and adhesive ligand density for “soft” (12% DTT −UV) hydrogels 24 h after seeding. No significant differences in cell area or morphology were observed with increasing ligand density of 1 (white), 2 (black) and 5 (gray) mM RGD. This provides evidence that the spreading is due to mechanical differences, rather than potential changes in ligand density. | ||
Stiffness effects on long term cell behavior
After determining the effects of matrix mechanics on short-term cell morphology, we investigated the long-term cell response to 12% DTT −/+UV hydrogels by monitoring cell morphology and proliferation at several time points. Fig. 5A shows the dramatic differences in cell proliferation over 7 days for the −/+UV hydrogels. Cells on the +UV stiffer hydrogels proliferated much more than their −UV counterparts over the course of 7 days. Representative images indicate that the cells on the softer hydrogels maintained their rounded morphology while cells on the stiffer hydrogels remained highly spread and fully confluent after 7 days. | ||
| Fig. 5 (A) hMSC proliferation for up to 7 days on “soft” (12% DTT −UV, white) and “stiff” (12% DTT +UV, black) hydrogels. Cells on stiffer gels proliferate at a higher rate than cells on softer gels. (B) Representative images with culture time for both hydrogels reveal the qualitative differences in hMSC number and morphology over 7 days, scale bar = 200 µm. Significant differences (**p < 0.001, *p < 0.01) were found between −UV and +UV gels. | ||
Cell proliferation has been shown to be dependent on matrix mechanics in several notable studies.38,42 Highly spread cells possess a greater proportion of phosphorylated focal adhesion kinase (FAK), which has been shown to increase intracellular tension through Rho signaling. This maintenance of intracellular tension has significant consequences on whether a cell proliferates, differentiates, remains quiescent, or undergoes apoptosis.43–45 The effects of spreading and mechanics have also been shown to result in changes in nuclear volume and chromatin condensation. In one study, increases in endothelial cell spreading led to an increase in nuclear volume and a greater proportion of cells in the S Phase of cell division.46 Our findings show a similar behavior in hMSCs as proliferation was significantly higher on the stiffer substrates.
Photopatterned mechanics and stem cell response
The ability to control stem cell spreading and proliferation has been demonstrated on substrates with uniform mechanics; however, spatial control of these behaviors is necessary due to the heterogeneous nature of many tissues. This is useful for initial steps towards advanced tissue engineering approaches, as well as to understand multi-phenotype differentiation from a single cell population. Fig. 6A shows the differences in mechanics on photopatterned regions of non-exposed (∼6 kPa) and exposed (∼31 kPa) stripes of a width of 500 µm. After 24 h, cells acquire morphologies reminiscent of the uniform gels on the corresponding mechanical environments (i.e., rounded on “soft” regions and highly spread on “stiff” regions, Fig. 6B). This is observed in representative images of photopatterned stripes with cells showing the spatial control of cell morphology based on the local elasticity. It is clear that cells maintain a rounded morphology on the softer −UV regions and are highly spread on the stiffer +UV regions (Fig. 6C). Interestingly, many cells aligned along the soft/stiff interface just as NIH3T3 fibroblasts do on similar mechanical interfaces.47,48 Cell migration due to durotaxis can also take place in these interfacial regions as cell adhesion sites on the stiffer regions result in greater tension, and subsequent greater adhesion strength, which allows the cells to migrate from soft to stiff regions. After 7 days, cells became confluent in the “stiff” regions, but not in the “soft” regions (Fig. 6D). These large differences in confluence could be due to cells proliferating, as well as cells migrating from the softer to stiffer regions. | ||
| Fig. 6 Spatially controlled mechanics (A) and hMSC spreading (B) on photopatterned stripes (500 µm width) on 12% DTT hydrogels. The mechanics vary with space across the hydrogel depending on whether it was exposed to light and is correlated to hMSC response. The cellular morphology on patterns after 1 (C) and 7 (D) days illustrates the importance of mechanics on cellular behavior, including spreading and proliferation. Scale bar = 400 µm. Statistically significant differences in gel modulus and cell area *p < 0.001. | ||
In another photopattern strategy, the extent of exposure was linearly varied by passing a photomask across the surface of the preliminarily crosslinked gel (see schematic in Fig. 1C). Locally probing the modulus at regular intervals along the length of the gradient allowed for correlation of matrix mechanics with distance (i.e., time of light exposure). As shown in Fig. 7A, the modulus gradually increases from a minimum of ∼6 kPa to ∼25 kPa over regions with up to a minute of exposure followed by a sharp increase in mechanics for regions with an additional 30 s of exposure up to a maximum elasticity comparable to the 12% DTT +UV uniform gel (∼90 kPa). Other dual crosslinked systems saw similar rapid secondary crosslinking upon exposure of UV to radically polymerize remaining photoreactive groups.27,30 As expected, the hMSC spreading increased locally along the length of the gradient, reaching a spreading plateau in regions of the gel that had been exposed for greater than 60 s.
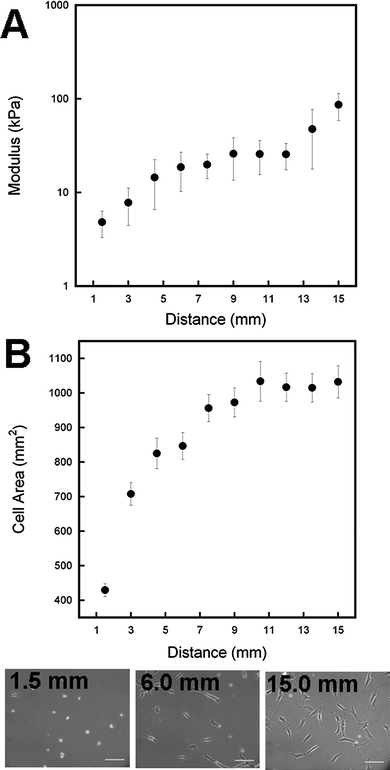 | ||
| Fig. 7 Mechanical gradients were achieved using a sliding photomask to locally vary the light exposure time and thus mechanics nearly two orders of magnitude across a single gel (A). hMSC response to the mechanical gradient shows increased cell spreading with increasing exposure until a maximal spread area is achieved (B). Representative images at different distances (exposures) along the gradient gel. Scale bar = 200 µm. | ||
These photopatterning studies not only indicate that we can control matrix mechanics in a binary fashion (−/+UV), but also in a gradient fashion with the capability to create a wide range of mechanics using the same base HA network composition. However, further investigation is needed to understand how this behavior translates to features such as differentiation and to understand how cells in different local environments affect each other through factors such as paracrine signaling. This same system could also be translated in a 3-D system by polymerizing the gel around a desired template (such as sintered polymethyl methacrylate (PMMA) microspheres14) to create a macroporous hydrogel with locally controlled mechanics.
Conclusions
We have successfully developed a mechanically tunable sequentially crosslinked hydrogel system, which represents a novel method for spatially controlling hMSC morphology and proliferation by varying matrix mechanics. By changing the initial crosslinker concentration, elastic moduli over several orders of magnitude were obtained that could be significantly increased once the secondary radical crosslinking mechanism was incorporated. Furthermore, the ability to spatially control mechanics was possible by restricting light to regions of the gel or by varying the light exposure time in space. This allowed for patterned cell responses, as hMSCs cultured on these patterned gels exhibited morphologies corresponding well with the local substrate mechanics. Stem cell morphology and proliferation were highly dependent on mechanics, as cells became more spread and more proliferative on substrates of higher mechanics. While these outputs are not necessarily indicative of stem cell differentiation, morphology and proliferation can be determinants and effectors of differentiation in both 2D and 3D microenvironments.32,38,42,49 Since mechanical differences are relevant in certain pathologies the characterization and understanding of cell responses to mechanics in a controlled manner are critical for developing effective tissue engineering strategies.Acknowledgements
The authors gratefully acknowledge funding from the David and Lucile Packard Foundation (JAB) and the National Science Foundation CAREER Award (JAB) and Graduate Research Fellowship Program (RAM).References
- R. A. Marklein and J. A. Burdick, Adv. Mater., 2009 DOI:10.1002/adma.200901055 , in press.
- T. Yeung, P. Georges, L. Flanagan, B. Marg, M. Ortiz, M. Funaki, N. Zahir, W. Ming, V. Weaver and P. Janmey, Cell Motil. Cytoskeleton, 2004, 60, 24–34.
- R. J. Pelham and Y. Wang, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 13661–13665 CrossRef CAS.
- A. Engler, S. Sen, H. Sweeney and D. Discher, Cell, 2006, 126, 677–689 CrossRef CAS.
- F. Rehfeldt, A. Engler, A. Eckhardt, F. Ahmed and D. Discher, Adv. Drug Delivery Rev., 2007, 59, 1329–1339 CrossRef CAS.
- K. Saha, A. J. Keung, E. F. Irwin, Y. Li, L. Little, D. V. Schaffer and K. E. Healy, Biophys. J., 2008, 95, 4426–4438 CrossRef CAS.
- A. J. Engler, M. A. Griffin, S. Sen, C. G. Bönnemann, H. L. Sweeney and D. E. Discher, J. Cell Biol., 2004, 166, 877–887 CrossRef CAS.
- D. Discher, D. Mooney and P. Zandstra, Science, 2009, 324, 1673–1677 CrossRef CAS.
- J. A. Burdick and G. Vunjak-Novakovic, Tissue Eng. Pt. A, 2008, 5, 205–219 Search PubMed.
- K. R. Johnson, J. L. Leight and V. M. Weaver, Methods Cell Biol., 2007, 83, 547–583 Search PubMed.
- R. Wells, Hepatology, 2008, 47, 1394–1400 CrossRef.
- G. Forte, F. Carotenuto, F. Pagliari, S. Pagliari, P. Cossa, R. Fiaccavento, A. Ahluwalia, G. Vozzi, B. Vinci, A. Serafino, A. Rinaldi, E. Traversa, L. Carosella, M. Minieri and P. Di Nardo, Stem Cells, 2008, 26, 2093–2103 Search PubMed.
- J. L. Ifkovits and J. A. Burdick, Tissue Eng., 2007, 13, 2369–2385 CrossRef CAS.
- S. J. Bryant, J. L. Cuy, K. D. Hauch and B. D. Ratner, Biomaterials, 2007, 28, 2978–2986 CrossRef CAS.
- S. Lee, J. Moon and J. West, Biomaterials, 2008, 29, 2962–2968 CrossRef CAS.
- S. Suri and C. E. Schmidt, Acta Biomater., 2009, 5, 2385–2397 CrossRef CAS.
- M. Singh, C. Berkland and M. S. Detamore, Tissue Eng., Part B: Rev., 2008, 14, 341–366 Search PubMed.
- S. A. DeLong, A. S. Gobin and J. West, J. Controlled Release, 2005, 109, 139–148 CrossRef CAS.
- J. A. Burdick, A. Khademhosseini and R. Langer, Langmuir, 2004, 20, 5153–5156 CrossRef.
- N. Zaari, P. Rajagopalan, S. Kim, A. Engler and J. Wong, Adv. Mater., 2004, 16, 2133–2137 CrossRef CAS.
- C. Chung and J. A. Burdick, Tissue Eng., Part A, 2009, 15, 243–254 Search PubMed.
- M. A. Serban and G. D. Prestwich, Methods, 2008, 45, 93–98 CrossRef CAS.
- I. E. Erickson, A. H. Huang, C. Chung, R. T. Li, J. A. Burdick and R. L. Mauck, Tissue Eng., Part A, 2009, 15, 1041–1052 Search PubMed.
- N. E. Fedorovich, M. H. Oudshoorn, D. van Geemen, W. E. Hennink, J. Alblas and W. J. Dhert, Biomaterials, 2009, 30, 344–353 CrossRef CAS.
- N. S. Hwang, S. Varghese, P. Theprungsirikul, A. Canver and J. Elisseeff, Biomaterials, 2006, 27, 6015–6023 CrossRef CAS.
- J. Kutty, E. Cho, J. Soolee, N. Vyavahare and K. Webb, Biomaterials, 2007, 28, 4928–4938 CrossRef CAS.
- S. Khetan, J. S. Katz and J. A. Burdick, Soft Matter, 2009, 5, 1601–1606 RSC.
- C. Chung, J. Mesa, G. J. Miller, M. A. Randolph, T. J. Gill and J. A. Burdick, Tissue Eng., 2006, 12, 2665–2673 CrossRef CAS.
- Y. Liu and M. B. Chan-Park, Biomaterials, 2009, 30, 196–207 CrossRef.
- S. A. Zawko, S. Suri, Q. Truong and C. E. Schmidt, Acta Biomater., 2009, 5, 14–22 CrossRef CAS.
- A. M. Rokstad, I. Donati, M. Borgogna, J. Oberholzer, B. L. Strand, T. Espevik and G. Skjåk-Braek, Biomaterials, 2006, 27, 4726–4737 CrossRef CAS.
- A. Rowlands, P. George and J. Cooper-White, Am. J. Physiol.: Cell Physiol., 2008, 295, C1037–C1044 CrossRef CAS.
- A. Engler, L. Bacakova, C. Newman, A. Hategan, M. Griffin and D. Discher, Biophys. J., 2004, 86, 617–628 CrossRef CAS.
- X. Q. Brown, K. Ookawa and J. Y. Wong, Biomaterials, 2005, 26, 3123–3129 CrossRef CAS.
- F. J. Byfield, R. K. Reen, T. P. Shentu, I. Levitan and K. J. Gooch, J. Biomech., 2009, 42, 1114–1119 CrossRef.
- J. Solon, I. Levental, K. Sengupta, P. Georges and P. Janmey, Biophys. J., 2007, 93, 4453–4461 CrossRef CAS.
- D. S. Benoit, A. R. Durney and K. S. Anseth, Biomaterials, 2007, 28, 66–77 CrossRef CAS.
- W. A. Comisar, N. H. Kazmers, D. Mooney and J. J. Linderman, Biomaterials, 2007, 28, 4409–4417 CrossRef CAS.
- M. M. Martino, M. Mochizuki, D. A. Rothenfluh, S. A. Rempel, J. A. Hubbell and T. H. Barker, Biomaterials, 2009, 30, 1089–1097 CrossRef CAS.
- R. McBeath, D. M. Pirone, C. M. Nelson, K. Bhadriraju and C. S. Chen, Dev. Cell, 2004, 6, 483–495 CrossRef CAS.
- J. C. Friedland, M. H. Lee and D. Boettiger, Science, 2009, 323, 642–644 CrossRef CAS.
- S. X. Hsiong, P. Carampin, H. J. Kong, K. Y. Lee and D. J. Mooney, J. Biomed. Mater. Res., 2008, 85, 145–156 Search PubMed.
- C. S. Chen, M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, Science, 1997, 276, 1425–1428 CrossRef CAS.
- R. Assoian and E. Klein, Trends Cell Biol., 2008, 18, 347–352 CrossRef CAS.
- A. L. Zajac and D. E. Discher, Curr. Opin. Cell Biol., 2008, 20, 609–615 CrossRef CAS.
- P. Roca-Cusachs, J. Alcaraz, R. Sunyer, J. Samitier, R. Farré and D. Navajas, Biophys. J., 2008, 94, 4984–4995 CrossRef CAS.
- S. Chou, C. Cheng and P. R. Leduc, Biomaterials, 2009, 30, 3136–3142 CrossRef CAS.
- C. M. Lo, H. B. Wang, M. Dembo and Y. Wang, Biophys. J., 2000, 79, 144–152 CrossRef CAS.
- D. Dikovsky, H. Bianco-Peled and D. Seliktar, Biophys. J., 2007, 94, 2914–2925.
| This journal is © The Royal Society of Chemistry 2010 |
