Three-dimensional elemental bio-imaging of Fe, Zn, Cu, Mn and P in a 6-hydroxydopamine lesioned mouse brain†
Dominic J.
Hare
a,
Jessica L.
George
b,
Rudolph
Grimm
c,
Simon
Wilkins‡
b,
Paul A.
Adlard
bd,
Robert A.
Cherny
be,
Ashley I.
Bush
be,
David I.
Finkelstein§
bd and
Philip
Doble§
*a
aElemental Bio-imaging Facility, University of Technology Sydney, Australia
bThe Mental Health Research Institute, Parkville, Australia
cAgilent Technologies, Santa Clara, USA
dCentre for Neuroscience, University of Melbourne, Parkville, Australia
eDepartment of Pathology, University of Melbourne, Parkville, Australia
First published on 29th October 2010
Abstract
Three dimensional maps of iron (Fe), zinc (Zn), copper (Cu), manganese (Mn) and phosphorous (P) in a 6-hydroxydopamine (6-OHDA) lesioned mouse brain were constructed employing a novel quantitative laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS) imaging method known as elemental bio-imaging. The 3D maps were produced by ablating serial consecutive sections taken from the same animal. Each section was quantified against tissue standards resulting in a three dimensional map that represents the variation of trace element concentrations of the mouse brain in the area surrounding the substantia nigra (SN). Damage caused by the needle or the toxin did not alter the distribution of Zn, and Cu but significantly altered Fe in and around the SN and both Mn and Fe around the needle track. A 20% increase in nigral Fe concentration was observed within the lesioned hemisphere. This technique clearly shows the natural heterogeneous distributions of these elements throughout the brain and the perturbations that occur following trauma or intoxication. The method may applied to three-dimensional modelling of trace elements in a wide range of tissue samples.
Introduction
Elemental bio-imaging employs laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS) for quantitative in situ analysis of trace elements in biological tissues. Images are produced by directing a focused laser beam onto a thin tissue section (4–100 μm), where material is ablated from the tissue surface. The ablated material is then swept into the ICP-MS by an argon carrier gas, where its elemental composition is determined. The sample is moved across the laser beam in straight lines. In this way, an image can be constructed from multiple ablation lines, much the same as a dot matrix printer prints an image. Quantitative data is produced by representative ablation of prepared tissue standards. Since the ICP-MS is a sequential multi-element analyser, it is possible to build maps for multiple elements in a single experiment. We have recently applied elemental bio-imaging for the quantitative determination of the cortical distribution of copper (Cu), manganese (Mn), iron (Fe) and zinc (Zn) in an animal model of Parkinson's disease,1 as well as for the detection of melanoma in lymph nodes by imaging phosphorus,2 and the determination of calcium phosphate crystals in knee synovial fluid and cartilage obtained from arthritis patients.3 Matusch et al. recently described the application of elemental bio-imaging to Fe, Cu, Mn and Zn in an alternate animal model for Parkinson's disease.4Use of LA-ICP-MS in elemental bio-imaging has increased considerably in recent times. LA-ICP-MS offers a fast, accurate, quantitative isotope-specific imaging technique with μm resolution. The method has few matrix effects and has one of the lowest detection limits (<0.01 μg g−1) of commonly used elemental techniques.5 While LA-ICP-MS lacks the nm-scale resolution seen in synchrotron X-ray microspectroscopic methods,6,7 the technique offers a low-cost, more widely available option with the ability to analyse larger sections.
Parkinson's disease is the second most common neurodegenerative disorder and is characterised by a progressive degeneration of dopaminergic neurons by mechanisms which are poorly understood. There are many strategies employed to elucidate the possible risk factors and mechanisms underlying this currently incurable disease.8 One of the most widely used animal models for Parkinson's disease involves the direct injection of the neurotxoin 6-hydroxydopamine (6-OHDA) into the substantia nigra (SN). This results in the loss of dopaminergic cells, mimicking the loss of SN neurons observed in PD.9 The mechanism of 6-OHDA toxicity is thought to involve Fe, which plays a role in cell death and superoxide radical generation.10–12 Clioquinol, a drug with affinity for copper, zinc and iron, is effective at attenuating 6-OHDA lesion size in mice when administered on the same day as 6-OHDA.13 Therefore, methods for quantification of the regional distribution of Fe and other trace elements would be of benefit to not only elucidate mechanisms underlying the pathology present in the 6-OHDA lesion model, but to also probe the mechanism, treatment and progression of PD.
Parkinson's disease primarily affects the substantia nigra, which extends just over a cubic millimetre anteroposteriorly and medo-laterally in the mouse brain. As the ablation of a single 30 μm coronal section of a mouse SN does not necessarily provide a representative sample of metal concentrations present in the SN, due to the irregular shape and likely heterogeneous distribution of metals within the region, the construction of three-dimensional images of the SN would provide a more informative view of the trace metal concentration within and surrounding this area. Andersson et al. have described a protocol for using matrix-assisted laser desorption/ionisation imaging mass spectrometry (MALDI-IMS) to create three-dimensional reconstructions of proteins and peptides in the rat brain.14 By applying a similar processing procedure to elemental bio-imaging, it is possible to generate 3D images of trace elements in equivalent tissue sections.
This paper presents a novel method to construct quantified three-dimensional maps of Mn, Fe, Cu and Zn within the brain of a mouse model of Parkinson's disease encompassing the area of the substantia nigra. This is the first reported use of LA-ICP-MS for the construction of 3D element models from consecutive tissue sections.
Materials and methods
Laser ablation—inductively coupled plasma—mass spectrometry
LA-ICP-MS analysis was performed using a New Wave UP213 laser ablation unit (Kenelec Technlogies, Mitcham, Victoria, Australia) connected to an Agilent Technologies 7500 ce ICP-MS (Agilent Technologies, Forrest Hill, Victoria, Australia). The UP213 unit used a Nd![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) YAG emission source emitting a nanosecond laser pulse in the fifth harmonic at 213 nm. The standard New Wave ablation cell was used. Argon was used as the carrier gas, flowing through the chamber at 1.15 L min−1. The 7500 ce ICP-MS was fitted with a ‘cs’ lens system for enhanced sensitivity. Forward RF power was reduced to 1250 W for dry plasma conditions. Operational parameters for the LA-ICP-MS system is summarised in Table 1.
YAG emission source emitting a nanosecond laser pulse in the fifth harmonic at 213 nm. The standard New Wave ablation cell was used. Argon was used as the carrier gas, flowing through the chamber at 1.15 L min−1. The 7500 ce ICP-MS was fitted with a ‘cs’ lens system for enhanced sensitivity. Forward RF power was reduced to 1250 W for dry plasma conditions. Operational parameters for the LA-ICP-MS system is summarised in Table 1.
| Agilent 7500ce ICP-MS | New Wave UP213 Laser Ablation | ||
|---|---|---|---|
| RF Power | 1250 W | Wavelength | 213 nm |
| Cooling gas flow rate | 15 L min−1 | Repetition frequency | 20 Hz |
| Carrier gas flow rate | 1.15 L min−1 | Laser energy density | 0.3 J cm−2 |
| Sample depth | 4.0 mm | Spot size | 100 μm |
| QP Bias | −5 V | Scan rate | 100 μm s−1 |
| OctP Bias | −8 V | Line spacing | 100 μm |
| Scan mode | Peak hopping | Carrier gas | Ar |
| Dwell time | 0.1 s per m/z | ||
| Measured m/z | 13, 31, 55, 56, 63, 66 | ||
| Extracts 1, 2 | 6.8, −126 V | ||
The LA-ICP-MS system was tuned for sensitivity prior to each experiment using NIST 612 Trace Elements in Glass. Additionally, ICP-MS was also tuned to minimise the formation of oxides by monitoring the m/z 248/232 (232Th16O+/232Th+) ratio. A value of less than 0.2% for this ratio indicated the signal from potential matrix-based polyatomic interferences was negligible.
Preparation of tissue standards
Matrix-matched tissue standards were prepared for the quantification of selected trace elements in brain tissue. Chicken breast was approximately spiked with standard solutions of each element of interest and homogenised. Digests of the homogenised tissue were analysed by solution nebulisation (SN-ICP-MS to assess the homogeneity of the prepared standards (measured by the standard deviation of n = 6 digests per standard) and to determine the concentration accurately.These tissue standards were prepared as previously described.1 Chicken breast tissue was obtained from a local market and was stripped of all fatty and connective tissue using a PTFE-coated scalpel. Approximately 5 g portions of the tissue were spiked with appropriate volumes of 1000 mg L−1 Mn, Fe, Cu and Zn standards in 5% HNO3 (Choice Analytical, Thornleigh, NSW, Australia). A further volume of 5% Seastar Baseline grade HNO3 (Choice Analytical) was added to a final mass of 6 g to aid homogenisation. Concentration ranges were selected based on values in both developing rodent brains and 6-OHDA lesioned animals reported by Tarhoda et al.15,16 Homogenisation was performed using an OmniTech TH tissue homogeniser fitted with polycarbonate probes (Kelly Scientific, North Sydney, NSW, Australia). Six aliquots (ca. 100 mg) of accurately weighed homogenised tissue were then digested in 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 70% Baseline HNO3 (Choice Analytical) and Ultrapure 31% H2O2 (Merck, Kilsyth, Victoria, Australia) using a Milestone MLS 1200 closed vessel microwave digester. The digests were diluted to ca. 50 g with 18.2 mΩ deionised H2O and its weight accurately recorded.
1 70% Baseline HNO3 (Choice Analytical) and Ultrapure 31% H2O2 (Merck, Kilsyth, Victoria, Australia) using a Milestone MLS 1200 closed vessel microwave digester. The digests were diluted to ca. 50 g with 18.2 mΩ deionised H2O and its weight accurately recorded.
These solutions were analysed by solution nebulisation ICP-MS using the Agilent 7500 ce ICP-MS. In brief, mixed standards containing each element were prepared from fresh stocks (Choice Analytical) for calibration. A 250 ppb solution of 103Rh was used as the reference element that was added to each sample via a peristaltic pump and T-piece connector. A quartz concentric nebuliser and Scott-type spray chamber (Glass Expansion, Australia) were used. Helium was used as a collision gas to remove polyatomic interferences in solution ICP-MS experiments only. Concentration determined from SN-ICP-MS for each tissue standard is given in Table 2. Note that the spiked amounts were approximates; the accurate measured amounts by SN-ICP-MS were used for all quantitative imaging procedures.
| Standard | Approximate spiked amount (mg kg−1) | Measured amount by SN-ICP-MS (mg kg−1)% ± SD (n = 6) | |
|---|---|---|---|
| 1 | Mn | 1 | 0.96 ± 0.04 |
| Fe | 10 | 13.5 ± 2.3 | |
| Cu | 1 | 1.3 ± 0.2 | |
| Zn | 10 | 7.3 ± 1.5 | |
| 2 | Mn | 5 | 4.0 ± 0.2 |
| Fe | 20 | 17.1 ± 0.9 | |
| Cu | 10 | 7.9 ± 1.0 | |
| Zn | 20 | 26.1 ± 4.2 | |
| 3 | Mn | 10 | 8.5 ± 0.4 |
| Fe | 50 | 34.2 ± 2.7 | |
| Cu | 20 | 19.7 ± 1.8 | |
| Zn | 40 | 42.5 ± 1.5 | |
| 4 | Mn | 20 | 14.5 ± 0.7 |
| Fe | 100 | 107.6 ± 8.2 | |
| Cu | 50 | 47.9 ± 3.7 | |
| Zn | 60 | 51.9 ± 5.5 | |
6-OHDA lesioned mouse model
All methods in the animal model conformed to the Australian National Health and Medical Research Council published code of practice for animal research and were approved by the Howard Florey Institute Animal Ethics Committee. C57Bl/6 mice were lesioned as previously described.17,18 Briefly, a mouse was injected with 0.5 mg/kg atropine (Pharmacia, Wayville, South Australia, Australia) and 10 mg/kg xylazine (Troy Laboratories, Smithfield, NSW, Australia) as premedication. Anaesthesia was then induced with 4% isofluorane (Forthane, Abbott, Australia) carried by oxygen and then maintained at 2% isofluorane. The head was then secured in a stereotaxic frame with the bite bar 3 mm above horizontal. A solution of 6-OHDA (1.65 mg/mL, Sigma) in ascorbic acid (0.2 mg/mL) was prepared and kept on ice until the time of injection. A 26 gauge needle attached via tubing to a 500 μl syringe (SGE, Australia) mounted in a syringe pump (Bioanalytical Systems Inc., USA) was inserted into the right SN through a small hole drilled through the top of the skull. The needle was left to settle for 2 min before 2 μL (3.3 μg, 1.65 mg/mL) of 6-OHDA was slowly injected (0.5 μL/min). Twenty-one days after surgery the animal was anaesthetised with sodium pentobarbitone (Lethobarb; 100 mg/kg) and perfused with 30 ml of warmed (37 °C) 0.1 M phosphate buffered saline (PBS), pH 7.4, followed by 30 ml of chilled 4% (w/v) paraformaldehyde (Sigma Aldrich) and 0.1 M PBS (4 °C), pH 7.4. The brain was then removed and left at 4 °C overnight in 30% (w/v) sucrose in PBS before being frozen and sectioned at 30 μm on a cryostat. Each consecutive section was collected into three parallel series, producing an interval of 90 μm between sections in each series. Sections were thawed onto glass microscope slides, dried at room temperature and then stored frozen in a desiccator until used. Alternate sections were stained by various histopathological techniques, including Nissl staining and tyrosine hydroxylase immunostaining for identification of regions of interest. This study focused on the development of a three-dimensional modelling method for LA-ICP-MS, hence data from a single lesioned animal is presented.Ablation procedure
A laser beam diameter of 100 μm was used for all experiments, with a scan speed of 100 μm/s and frequency of 20 Hz. Laser energy density was set at 0.28 J/cm2 to reduce fracturing of the tissue during ablation. The distance between each line was 100 μm so as to ensure total ablation of all available tissue. The total analysis time for the 12 sections was approximately 36 h.Imaging procedure
Each line of ablation generated a single data file for all selected elements. Interactive Spectral Imaging Data Analysis Software (ISIDAS), an in-house developed suite written in the Python programming language was used to amalgamate all data files into a single image containing 12 sections from a single animal. Data reduction took less than 30 s. Boxes of equivalent pixel dimensions were placed over each section, allowing the image to be broken into 12 stackable units. Adjacent sections were registered by moving and rotating the two images about the x- and y-axes. Registration was performed by aligning adjacent sections with prominent structural features located along the edge of the cerebral cortex. All alignment procedures were performed using the 13C signal. This procedure was repeated until the 12 sections were all aligned (see Fig. 1). | ||
| Fig. 1 Schematic showing the registration of each section in ISIDAS for 3D rendering. | ||
The 12 registered images were then exported as Visualisation Toolkit (.vtk) files into the MayaVi2 (Enthought, Inc., Austin, Texas, USA) open source three-dimensional data visualization suite. Three-dimensional images were produced using the ‘glyph’ imaging module. Constructed images had dimensions of approximately 15 × 7 × 1 mm (width × height × depth).
The images were quantified by ablating a representative sample of the prepared tissue standards, constructing calibration curves and applying to the images. Each pixel represented a concentration within an approximately 100 μm3 area. Calibration curves for each measured element had correlation coefficients greater than 0.99. Regions of interest were extracted from the images for statistical analysis by careful freehand drawing with the aid of a mouse brain atlas.19
Results and discussion
This paper presents a new method to quantify the spatial and regional distribution of Mn, Fe, Cu and Zn reconstructed from tissue cut into serial consecutive sections though the SN to obtain three-dimensional elemental maps of a mouse brain that had been lesioned on one side of the animal with 6-OHDA. Anatomical nuclei can readily be distinguished as can the trauma caused by the injection needle and the changes in Fe in the SN. Quantitative data extracted from the image in selected regions of interest are displayed in Table 3.| Fe | Zn | Cu | Mn | ||||
|---|---|---|---|---|---|---|---|
| SN – lesioned side | 40.7 ± 3.2 | SN – lesioned side | 15.2 ± 1.3 | SN – lesioned side | 7.20 ± 1.4 | SN – lesioned side | 0.454 ± 0.03 |
| SN – non lesioned side | 33.9 ± 1.8 | SN – non lesioned side | 15.3 ± 1.1 | SN – non lesioned side | 7.40 ± 0.7 | SN – non lesioned side | 0.423 ± 0.02 |
| APT – lesioned side | 36.5 ± 4.2 | Amyg – lesioned side | 26.2 ± 4.6 | DG – lesioned side | 10.2 ± 1.2 | PAG | 0.424 ± 0.07 |
| APT – non lesioned side | 34.0 ± 1.7 | Amyg – non lesioned side | 26.1 ± 3.5 | DG – non-lesioned side | 8.51 ± 0.5 | APT – lesioned side | 0.418 ± 0.06 |
| MM | 37.3 ± 5.6 | CTX | 22.1 ± 3.8 | CA3 – lesioned side | 10.1 ± 2.2 | APT – non lesioned side | 0.469 ± 0.04 |
| Needle track | 229 ± 206 | DG – lesioned side | 26.7 ± 5.0 | CA3 – non lesioned side | 11.5 ± 2.9 | Midbrain – lesioned | 0.497 ± 0.05 |
| DG – non lesioned side | 25.3 ± 4.4 | VTM – lesioned side | 23.1 ± 8.3 | Midbrain – non lesioned | 0.448 ± 0.04 | ||
| CA3 – lesioned side | 21.0 ± 0.8 | VTM – non lesioned side | 21.0 ± 6.6 | Needle track | 0.443 ± 0.17 | ||
| CA3 – non lesioned side | 18.8 ± 1.4 | PAG | 15.4 ± 12.6 |
Fig. 2 shows the 12 single 2D images and the 3D reconstruction of Fe in the lesioned mouse brain. A large increase in Fe (greater than 229 mg kg−1) was observed around the needle track (indicated by the black arrow), consistent with published data in similar models.1 The damage was not visible in the Nissl stained section, but likely resulted from the trauma of inserting the needle into the brain. The source of the Fe could have been from extracellular heme, which is metabolised by heme oxygenase-1 (HO-1) after haemorrhage from the traumatic brain injury (TBI). In support of this, there have been reports of increased HO-1 expression for up to six months following TBI in humans.20 Alternately, TBI would also result in an increase in local glial cell numbers, increasing the concentration of iron-laden ferritin within the region of injury.
 | ||
| Fig. 2 Two-dimensional images (top) and 3D reconstruction of Fe in 6-OHDA lesioned mouse brain (middle). The needle track is marked by the dashed arrow and the letter ‘i’ Coloured arrows: Red = lateral, blue = anterior and green = ventral. Regions of mouse brain corresponding to increased trace element concentration are shown in a Nissl stain (bottom left) and 56Fe 2D image (bottom right) of section 6. APT = anterior pretectal nucleus, SN = substantia nigra, DG = dentate gyrus, CA1 and CA3 fields of hippocampus and MM = medial mammilary nucleus. Dashed arrow denotes needle track. | ||
In our mouse lesion model, high concentrations of Fe were also observed within the SN in both hemispheres. The concentration of Fe was higher in the SN ipsilateral to the lesion (40.7 mg kg−1, as compared to 33.9 mg kg−1 in the contralateral side), which is a biological consequence of the neurotoxin.21 The 20% increase in nigral Fe falls within the same range of values previously reported, including a 35% increase by Oestreicher et al.22 and a 50% increase in a larger sample set by this laboratory.1 The source of this increased Fe is unclear, however the unilateral data supports the theory of intraneuronal transport of Fe from Fe-rich areas into the SN.23 LA-ICP-MS cannot differentiate between oxidation states of Fe; therefore this technique is unsuitable for determining if Fe increase in 6-OHDA lesioned mice is related to release of Fe3+ from ferritin.10,11 High Fe concentrations were also observed in many regions of the hippocampus, including the medial mamillary nucleus and the anterior pretectal nucleus. In this series of sections the SN was approximately 180 μm from the needle track, suggesting that the increase in Fe in the SN is likely to be an independent biological event that is not dependent on or related to the increase in Fe around the needle track.
Fig. 3 shows a 3D reconstruction of Zn concentration within the same sections of the lesioned brain. There was no obvious impact of the needle trauma or neurotoxin on regional Zn concentrations. Areas of increased Zn concentration (ca. 20 mg kg−1) were observed throughout the hippocampus.1 Zn was also observed in the cortex and amygdala.
 | ||
| Fig. 3 Two-dimensional images (top) and 3D reconstruction of Zn in 6-OHDA lesioned mouse brain (middle). Coloured arrows: Red = lateral, blue = anterior and green = ventral. Regions of mouse brain corresponding to increased trace element concentration are shown in a Nissl stain (bottom left) and 66Zn 2D image (bottom right) of section 6. Amyg = amygdala, DG = dentate gyrus, CA1 and CA3 fields of hippocampus and CTX = cortex. | ||
2D images and reconstructed 3D maps of Cu concentration are shown in Fig. 4. Cu was more concentrated around the periaqueductal grey matter (15.4 mg kg−1) and the hippocampus (10–11.5 mg kg−1), as well as in the ventral tuberomammillary nucleus (21–23 mg kg−1), an area rich in the Cu-binding dipeptide carnosine.24,25 Again, no obvious impact (7.2 vs. 7.4 mg kg−1, lesioned vs. unlesioned) of the 6-OHDA toxin was observed.
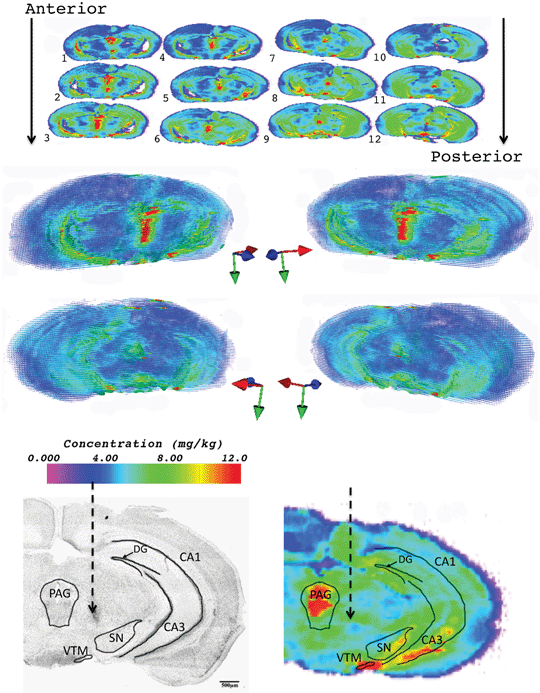 | ||
| Fig. 4 Two-dimensional images (top) and 3D reconstruction of Cu in 6-OHDA lesioned mouse brain (middle). Coloured arrows: Red = lateral, blue = anterior and green = ventral. Regions of mouse brain corresponding to increased trace element concentration are shown in a Nissl stain (bottom left) and 63Cu 2D image (bottom right) of section 6. SN = substantia nigra, DG = dentate gyrus, CA1 and CA3 fields of hippocampus, PAG = periaqueductal grey and VTM = ventral tuberomammillary nucleus. | ||
Fig. 5 shows Mn 2D images and 3D reconstructions. High concentrations of Mn were observed within the anterior pretectal nucleus as well as many regions in the centre of the brain such as the deep mesencephalic nucleus and the medial geniculate nucleus. A higher concentration of Mn was observed within the region of the needle track. This increase in Mn suggests that it is caused by a cellular response to trauma, rather than blood infiltration following the needle. Accumulation of nitrated and inactivated Mn-superoxide dismutase at the site of the traumatic brain injury may be a potential source of elevated Mn concentrations.26 This contention is supported by there being no increase in other trace elements (Cu, Zn) that are just as likely to be introduced into the wound with blood. Additionally, a significant difference (as measured by a two-tailed t-test) in Mn concentration within the SN was observed, with a mean concentration of 0.45 mg/kg on the lesioned side versus 0.42 mg/kg in the unlesioned hemisphere through the entire volume of the SN. Mn-superoxide dismutase has been shown to offer protection against 6-OHDA injury in the mouse brain27 and may be the source of elevated Mn levels. Further study is required to confirm this hypothesis.
 | ||
| Fig. 5 Two-dimensional images (top) and 3D reconstruction of Mn in 6-OHDA lesioned mouse brain (middle). Coloured arrows: Red = lateral, blue = anterior and green = ventral. Regions of mouse brain corresponding to increased trace element concentration are shown in a Nissl stain (bottom left) and 55Mn 2D image (bottom right) of section 6. PAG = periaqueductal grey, APT = anterior pretectal nucleus, DpMe—deep mesencepahlic nucleus, MGM = medial geniculate nucleus and SN = substantia nigra. | ||
Aside from the increased Fe observed in the SN and increased Zn in the cortex, higher metal concentration through the areas scanned appeared to be mostly confined to the regions surrounding the hippocampus bilaterally. There were no obvious areas of metal overlap, with higher Mn, Cu and Zn concentrations apparently confined to separate areas.
3D P images are shown in Fig. 6. No obvious structural features were observed at this resolution. Slightly higher signal intensity was observed within the midbrain, which could reflect the high cellular density. Conversely no obvious deficit is observed in the area of the needle track suggesting that the trauma was relatively minor.
 | ||
| Fig. 6 Two-dimensional images (top) and 3D reconstruction of P in 6-OHDA lesioned mouse brain (bottom). | ||
Refinement of the method is possible to produce resolutions as high as 10 to 20 μm. Simultaneous elemental bio-imaging and matrix assisted laser desorption ionisation—imaging mass spectrometry would provide unprecedented information and new knowledge about protein–metal interactions, and possibly discover new bio-markers for many diseases in which metal-protein interactions are involved. Our laboratory is currently constructing a whole brain atlas of trace metals in the mouse brain, with improvements to the 3D modelling procedure.
Conclusion
This paper has demonstrated the feasibility of the quantitative determination of trace elements in thin tissue sections presented as 3D maps. Software developed in-house enabled the rapid production of images that were easily converted into three-dimensional volume reconstructions using open source software. The 6-OHDA neurotoxin increased the concentration of Fe throughout the substantia nigra in three dimensions, supporting existing data our laboratories have produced. Furthermore, the method simultaneously determined the regional distribution of Zn, Cu, and Mn. Imaging the needle track showed persistent disruption of Fe and Mn homeostasis that was not visible in the Nissl stained sections.Acknowledgements
The developement of the ISIDAS program by Grigory Dorokhov and Michael Lake from the Computational Research Centre of Expertise, Science Faculty, UTS is gratefully acknowledged.References
- D. Hare, B. Reedy, R. Grimm, S. Wilkins, I. Volitakis, J. George, R. A. Cherny, A. I. Bush, D. I. Finkelstein and P. Doble, Metallomics, 2009, 1, 53 RSC.
- D. Hare, F. Burger, C. Austin, F. Fryer, R. Grimm, B. Reedy, A. Scolyer Richard, F. Thompson John and P. Doble, Analyst, 2009, 134, 450–453 RSC.
- C. Austin, D. Hare, A. R. Rozelle, W. H. Robinson, R. Grimm and P. Doble, Metallomics, 2009, 1, 142–147 RSC.
- A. Matusch, C. Depboylu, C. Palm, B. Wu, G. U. Hoglinger, M. K. H. Schafer and J. S. Becker, J. Am. Soc. Mass Spectrom., 2010, 21, 161–171 CrossRef CAS.
- R. McRae, P. Bagchi, S. Sumalekshmy and C. J. Fahrni, Chem. Rev., 2009, 109, 4780–4827 CrossRef CAS.
- S. Bohic, K. Murphy, W. Paulus, P. Cloetens, M. Salome, J. Susini and K. Double, Anal. Chem., 2008, 80, 9557–9566 CrossRef CAS.
- R. Ortega, G. Deves and A. Carmona, J. R. Soc. Interface, 2009, 6, S649–S658 CrossRef CAS.
- J. L. George, S. Mok, D. Moses, S. Wilkins, A. I. Bush, R. A. Cherny and D. I. Finkelstein, Curr. Neuropharmacol., 2009, 7, 9–36 Search PubMed.
- A. Schober, Cell Tissue Res., 2004, 318, 215–224 CrossRef.
- K. L. Double, M. Maywald, M. Schmittel, P. Riederer and M. Gerlach, J. Neurochem., 1998, 70, 2492–2499 CAS.
- H. P. Monteiro and C. C. Winterbourn, Biochem. Pharmacol., 1989, 38, 4177–4182 CrossRef CAS.
- D. Blum, S. Torch, N. Lambeng, M. F. Nissou, A. L. Benabid, R. Sadoul and J. M. Verna, Prog. Neurobiol., 2001, 65, 135–172 CrossRef CAS.
- S. Wilkins, L. Masters Colin, I. Bush Ashley, A. Cherny Robert and I. Finkelstein David, in The Basal Ganglia IX, Advances in Behavioral Biology, ed. H. J. Groenewegen, Springer, New York, Editon edn, 2009, vol. 58, pp. 431–442 Search PubMed.
- M. Andersson, M. R. Groseclose, A. Y. Deutch and R. M. Caprioli, Nat. Methods, 2008, 5, 101–108 CrossRef CAS.
- T. Tarohda, M. Yamamoto and R. Amamo, Anal. Bioanal. Chem., 2004, 380, 240–246 CrossRef CAS.
- T. Tarohda, Y. Ishida, K. Kawai, M. Yamamoto and R. Amano, Anal. Bioanal. Chem., 2005, 383, 224–234 CrossRef CAS.
- D. I. Finkelstein, D. Stanic, C. L. Parish, J. Drago and M. K. Horne, Curr. Prot. Neurosci., 2004, ch. 1, Unit 1 13 Search PubMed.
- D. I. Finkelstein, D. Stanic, C. L. Parish, D. Tomas, K. Dickson and M. K. Horne, Neuroscience, 2000, 97, 99–112 CrossRef CAS.
- K. B. J. Franklin and G. Paxinos, The Mouse Brain in Stereotaxic Coordinates, Academic Press, San Diego, 1997 Search PubMed.
- R. Beschorner, D. Adjodah, J. M. Schwab, M. Mittelbronn, I. Pedal, R. Mattern, H. J. Schluesener and R. Meyermann, Acta Neuropathol., 2000, 100, 377–384 CrossRef CAS.
- Y. He, P. S. P. Thong, T. Lee, S. K. Leong, C. Y. Shi, P. T. H. Wong, S. Y. Yuan and F. Watt, Brain Res., 1996, 735, 149–153 CrossRef CAS.
- E. Oestreicher, G. J. Sengstock, P. Riederer, C. W. Olanow, A. J. Dunn and G. W. Arendash, Brain Res., 1994, 660, 8–18 CrossRef CAS.
- M. Gerlach, K. L. Double, M. B. H. Youdim and P. Riederer, Journal of Neural Transmission Supplementa, 2006, 70, 133–142 Search PubMed.
- P. Q. Trombley, M. S. Horning and L. J. Blakemore, Biochemistry-Moscow, 2000, 65, 807–816 CAS.
- H. Otani, A. Okumura, K. Nagai and N. Okumura, Neurosci. Lett., 2008, 445, 166–169 CrossRef CAS.
- H. Bayir, V. E. Kagan, R. S. B. Clark, K. Janesko-Feldman, R. Rafikov, Z. Huang, X. Zhang, V. Vagni, T. R. Billiar and P. M. Kochanek, J. Neurochem., 2007, 101, 168–181 CAS.
- J. Callio, T. D. Oury and C. T. Chu, J. Biol. Chem., 2005, 280, 18536–18542 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Supplementary videos. See DOI: 10.1039/c0mt00039f |
| ‡ Current address: Monash Institute of Medical Research, Monash University, Victoria, Australia. |
| § Equal Senior Author. |
| This journal is © The Royal Society of Chemistry 2010 |
