DOI:
10.1039/C0MT00028K
(Paper)
Metallomics, 2010,
2, 754-765
Received
10th July 2010
, Accepted 14th September 2010
First published on
21st October 2010
Abstract
Ternary cobalt(III) complexes [CoL(B)] (1–3) of a trianionic tetradentate phenolate-based ligand (L) and phenanthroline bases (B), viz. 1,10-phenanthroline (phen in 1), dipyridoquinoxaline (dpq in 2) and dipyridophenazine (dppz in 3) are synthesized, characterized from X-ray crystallographic, analytical and spectral techniques, and their utility in photodynamic therapy (PDT) of thyroid diseases caused by TSH receptor dysfunction is probed. The complexes display a visible spectral band within the PDT spectral window at ∼690 nm. Photodynamic potential was estimated through DNA cleavage activity of the dpq and dppz complexes in UV-A light of 365 nm and red light of 676 nm. The reactions proceed via the hydroxyl radical pathway. The complexes retain their DNA photocleavage activity in red light under anaerobic conditions, a situation normally prevails in hypoxic tumor core. Investigation into the photocytotoxic potential of these complexes showed that the dppz complex 3 is approximately 4-fold more active in the HEK293 cells expressing human thyrotropin receptor (HEK293-hTSHR) than in the parental cell line and has an insignificant effect on an unrelated human cervical carcinoma cell line (HeLa). Photoexcitation of complex 3 in HEK293-hTSHR cells leads to damage hTSHR as evidenced from the decrease in cAMP formation both in absence and presence of hTSH and decrease in the TSHR immunofluorescence with a concomitant cytoplasmic translocation of the membrane protein, cadherin. The involvement of hTSHR is evidenced from the ability of complex 3 to bind to the extracellular domain of hTSHR (hTSHR-ECD) with a Kd value of 81 nM and from the photocleavage of hTSHR-ECD.
Introduction
G-protein coupled receptors (GPCRs) represent a large family of cell-surface receptors which control key cellular functions such as proliferation, survival and motility.1 Mutations in the GPCRs have been observed in tumor progression, angiogenesis and metastasis.1 The thyroid stimulating hormone receptor (hTSHR) belongs to the rhodopsin subclass of GPCRs with a characteristic 7-transmembrane serpentine domain (TMD) and a large complex extracellular domain (hTSHR-ECD). Its primary action is initiated on binding of thyroid stimulating hormone (TSH) to hTSHR-ECD with subsequent activation of adenylyl cyclases to produce cAMP. This cascade is the central feature in the growth and development of the thyroid gland along with production and release of thyroid hormones.
Owing to its physiological relevance, any perturbation in TSH receptor could result in wide varieties of abnormalities such as auto-immune condition as in Graves' thyrotoxicosis or somatic constitutively activating mutations in the hTSHR gene causing autonomous toxic adenomas, a form of thyroid cancer.1 Dysregulated signal transduction in non-canonical TSH receptor signal transduction pathways such as PKC and MAP kinase pathways may give rise to benign thyroid tumors (also hot nodules).2 The slow growth of thyroid tumors has been considered as the primary reason for their resistance to chemotherapeutic drugs like cisplatin, doxorubicin, carboplatin and presently the treatment relies solely on the use of radioiodine (131I), thyroidectomy or usage of a combination of cisplatin, doxorubicin, etoposide, and peplomycin along with the treatment of granulocyte colony-stimulating factor for support of the bone marrow. However, remissions are a common event.1,2 While targeted radiotherapy using [90Yttrium-DOTA]-TOC has been successfully applied in the treatment of iodine refractory and metastatic thyroid cancer, the use of photodynamic therapy (PDT) in thyroid adenomas and thyroid diseases is not well-documented in the literature.3 Photodynamic therapy involves highly selective activation of a photosensitizing drug in tumor with a laser light of appropriate wavelength which leads to the production of cytotoxic reactive oxygen species (ROS) resulting in the oxidation and damage to the biomolecules in the close proximity depending on its localization.4–6
Photofrin® is the currently used PDT drug which on photoactivation with 633 nm light generates 1(π–π*) state with subsequent formation of 3(π–π*) state that activates molecular oxygen to form cytotoxic singlet oxygen (1O2) species by energy transfer.4,7,8 Porphyrin derived photosensitizing agents in PDT are known for hepatotoxicity and skin sensitivity.9 These limitations have led to the advent of non-porphyrinic organic dyes like phthalocyanines, texaphyrins, squaranines and boradiazaindacenes, etc. as potential photosensitizers for PDT applications.10–15 Transition metal complexes with photoactive ligands provide an alternative platform for metal-based PDT agents.16 The problem associated with the organic dyes is that they generally follow a mechanistic pathway that forms cytotoxic singlet oxygen. Their ability to generate singlet oxygen on photoactivation within the PDT spectral window and their activity under hypoxic conditions are of paramount importance for drug activity.4,14,17 In contrast, transition metal complexes with their tunable coordination geometry and varied spectral and redox properties in the presence of low-energy d–d and/or charge transfer band(s) could offer better scope of developing metal-based PDT agents following alternate mechanistic pathway like photo-redox and Type-I pathways.15 The metal-based compounds could as well be designed to observe visible light-induced DNA cleavage activity under anoxic reaction conditions.18,19
Among the metal-based drugs, cisplatin and its analogues have been successfully used as non-PDT chemotherapeutic agents but their toxic effects on the normal cells together with the development of drug-resistance related problems have invoked keen interests to develop the chemistry of metal-based photoactivated chemotherapeutic agents.20–22 Recent reports have shown that platinum(IV) complexes upon intracellular reduction of the metal by chemical or photochemical means could release the cisplatin analogue drug in cancer cells.16,21 Sadler and co-workers have shown that a six-coordinate platinum(IV) complex with two trans azide ligands is stable in dark but generates trans diguanidine platinum(II) adduct on photoactivation killing human cancer cells in vitro.16 Additionally, dirhodium(II,II) complexes showing photocytotoxic effects in visible light using different cancer cell lines have been reported.19 Ruthenium nitrosyls have been used for site-specific delivery of cytotoxic nitric oxide (NO) on exposure to visible light as a new modality in PDT.23 Guo and coworkers have reported “molecular combo” species as red light PDT agents in which a platinum(II) based chemotherapeutic drug is conjugated to a silicon(IV) phthalocyanine moiety.10 While the focus on metal-based drugs in PDT has primarily been on the platinum group metals, the chemistry of bio-essential 3d-transition metal complexes as PDT agents is relatively unexplored.24 We have recently shown that a ternary iron(III) complex of dipyridophenazine and a tetradentate phenolate-based ligand becomes significantly cytotoxic upon photo-excitation in visible light in human cervical carcinoma cells (HeLa) and human keratinocytes (HaCaT).25 We have also reported that vanadium(IV) complex of dipyridophenazine exhibits photocytotoxicity in HeLa cells in visible light with low dark toxicity.26
The present work stems from our interest to probe the photocytotoxic potential of photoactive cobalt(III) complexes on human embryonic kidney cell line expressing the full length thyrotropin (hTSH) receptor (HEK293-hTSHR) as compared to the normal HEK293 cells. Cobalt(III) complexes are known to show UV light-induced DNA cleavage activity and have been used as highly potent prodrugs to release cytotoxic ligands like mustards upon intracellular reduction or photo-irradiation.27–29 Unlike high-spin iron(III) complexes, cobalt(III) complexes with low energy visible band within the PDT spectral window of 620–850 nm could be suitably designed for red light-induced DNA cleavage and photocytotoxicity studies. Herein, we report the synthesis, structure, DNA binding, DNA photocleavage and photocytotoxic property of three cobalt(III) complexes [CoL(B)], where H3L is a tetradentate phenolate-based ligand having lipophilic tert-butyl groups, namely, 2-bis-[3,5-di(tert-butyl)-2-hydroxybenzyl]aminoacetic acid and B is a phenanthroline base, viz. 1,10-phenanthroline (phen in 1), dipyrido[3,2-d:2′,3′-f]quinoxaline (dpq in 2) and dipyrido[3,2-a:2′,3′-c]phenazine (dppz in 3) (Fig. 1). The phenanthroline bases having quinoxaline and phenazine moieties that are present in the antitumor antibiotics like echinomycin or triostin are potent photosensitizers for their ability to generate photo-excited 3(n–π*) and/or 3(π–π*) state suitable for activation of molecular oxygen to generate ROS on photo-irradiation in UV light.30 The significant results of the present study are anaerobic DNA cleavage by the cobalt(III) complexes 2 and 3 in red-light of 676 nm and preferential photocytotoxicity of complex 3 in HEK293-hTSHR cells as compared to HEK293 and HeLa cells. TSH receptor activity has been monitored through its ability to transduce signal in the presence and absence of its cognate ligand as well as studying the changes in the basal cAMP synthesizing machinery on photo-treatment with complex 3. The work focuses on the preferential action of 3 on TSH receptor and draws a correlation between the observed photocytotoxicity of HEK293-hTSHR with damage of the hTSHR in cells as well as photocleavage of hTSH receptor extracellular domain expressed in Pichia pastoris.
![(a) Ternary cobalt(iii) complexes and the phenanthroline bases used. (b) An ORTEP view of [CoL(phen)] in 1·0.5EtOH·H2O showing 50% probability thermal ellipsoids and atom numbering for the metal and heteroatoms. The hydrogen atoms are omitted for clarity.](/image/article/2010/MT/c0mt00028k/c0mt00028k-f1.gif) |
| | Fig. 1 (a) Ternary cobalt(III) complexes and the phenanthroline bases used. (b) An ORTEP view of [CoL(phen)] in 1·0.5EtOH·H2O showing 50% probability thermal ellipsoids and atom numbering for the metal and heteroatoms. The hydrogen atoms are omitted for clarity. | |
Experimental
Materials and measurements
The reagents and chemicals were obtained from commercial sources (Sigma-Aldrich, USA; SD Fine Chemicals, India). The solvents used were purified by standard procedures.31 Supercoiled (SC) pUC19 DNA (caesium chloride purified) was purchased from the Bangalore Genei (India). All other reagents were purchased from Sigma (USA). Tris(hydroxymethyl)aminomethane–HCl (Tris–HCl) buffer was prepared using deionized and sonicated triple distilled water. The N,N-donor heterocyclic bases, dipyrido[3,2-d:2′,3′-f]quinoxaline (dpq) and dipyrido[3,2-a:2′,3′-c]phenazine (dppz), were prepared by reported procedures using 1,10-phenanthroline-5,6-dione as a precursor reacted with ethylenediamine for dpq and 1,2-phenylenediamine for dppz.32,33 The ligand H3L was prepared following a literature procedure.34
The elemental analysis was carried out using a Thermo Finnigan FLASH EA 1112 CHNS analyzer. The infrared and electronic spectra were recorded on PerkinElmer Lambda 35 and PerkinElmer spectrum one 55 spectrophotometers, respectively. Magnetic susceptibility data for polycrystalline samples of the complexes were obtained using Model 300 Lewis-coil-force magnetometer (George Associates Inc., Berkeley, USA). Hg[Co(NCS)4] was used as a standard. Experimental susceptibility data were corrected for diamagnetic contributions.35 Molar conductivity measurements were done using a Control Dynamics (India) conductivity meter. Cyclic voltammetric measurements were made at 25 °C using EG&G PAR 253 VersaStat potentiostat/galvanostat with a three-electrode configuration that consisted of a glassy carbon electrode, a platinum wire auxiliary and a saturated calomel reference electrode (SCE). Ferrocene (Ef = 0.42 V) was used as a standard in MeCN-0.1 M [Bun4N](ClO4) (TBAP). Electrospray ionization mass and 1H NMR spectral measurements were made using Bruker Daltonics make (Esquire 300 Plus ESI Model) and Bruker 400 MHz NMR spectrometers, respectively.
Syntheses
Preparation of [CoL(B)] (B = phen, 1; dpq, 2; dppz, 3).
The complexes were prepared by a general procedure in which the ligand H3L (0.26 g, 0.5 mmol) taken in ethanol (20 ml) was reacted with Et3N (0.2 ml, 1.5 mmol) followed by addition of Co(NO3)2·6H2O (0.15 g, 0.5 mmol). The colour of the solution changed from purple to dark brown. The solution was stirred for 1 h followed by addition of the phenanthroline base B (0.10 g, phen; 0.12 g, dpq; 0.14 g, dppz; 0.5 mmol). The solution was filtered after 10 min. The filtrate on slow evaporation gave dark brown microcrystalline product in analytically pure form.
[CoL(phen)] (1).
Yield, 51% (0.19 g). Anal. calcd for C44H54CoN3O4: C, 70.67; H, 7.28; N, 5.62. Found: C, 70.44; H, 7.34; N 5.58. Molar conductance in DMF (ΛM): 17 S m2 M−1. FT-IR (KBr phase): 2942s, 1633vs, 1472s, 1432s, 1303m, 1266m, 1203m, 1036m, 825m, 722s, 643w, 608w, 514w, 451w cm−1 (vs, very strong; s, strong; m, medium; w, weak). ESI-MS in MeOH: m/z 748.3 (M+H)+. UV-visible in 6% DMF/Tris HCl buffer [λmax, nm (ε, M−1 cm−1)]: 685 (320), 448 (4000), 290 (33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 450), 266 (48
450), 266 (48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 110). χM (298 K): −502.8 × 10−6 cm3 M−1. 1H NMR (DMSO-d6) δ (ppm): 9.75 (d, J = 4 Hz, 1H), 9.07 (d, J = 8 Hz, 1H), 8.70 (d, J = 8 Hz, 1H), 8.58 (d, J = 4 Hz, 1H), 8.32–8.38 (m, 2H), 8.20 (d, J = 8 Hz, 1H), 7.68 (m, 1H), 7.02 (d, J = 2 Hz, 2H), 6.70 (d, J = 2 Hz, 2H), 4.89 (d, J = 12 Hz, 2H), 1.25 (s, 21H), 0.55 (s, 15H).
110). χM (298 K): −502.8 × 10−6 cm3 M−1. 1H NMR (DMSO-d6) δ (ppm): 9.75 (d, J = 4 Hz, 1H), 9.07 (d, J = 8 Hz, 1H), 8.70 (d, J = 8 Hz, 1H), 8.58 (d, J = 4 Hz, 1H), 8.32–8.38 (m, 2H), 8.20 (d, J = 8 Hz, 1H), 7.68 (m, 1H), 7.02 (d, J = 2 Hz, 2H), 6.70 (d, J = 2 Hz, 2H), 4.89 (d, J = 12 Hz, 2H), 1.25 (s, 21H), 0.55 (s, 15H).
[CoL(dpq)] (2).
Yield, 48% (0.19 g). Anal. calcd for C46H54CoN5O4: C, 69.07; H, 6.80; N, 8.76. Found: C, 68.93; H, 6.88; N, 8.73. ΛM in DMF: 18 S m2 M−1. FT-IR (KBr phase): 2949s, 1626vs, 1469s, 1435s, 1233m, 1200w, 1166s, 1122m, 1082s, 1035m, 876w, 832m, 737m, 514w, 435w cm−1. ESI-MS in MeOH: m/z 800.4 (M + H)+. UV-visible in 6% DMF/Tris-HCl buffer [λmax, nm (ε, M−1 cm−1)]: 693 (415), 445 (5150), 340 (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 450), 282 (32
450), 282 (32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 550), 266 (44
550), 266 (44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 270). χM (298 K): −549.3 × 10−6 cm3 M−1. 1H NMR (DMSO-d6) δ (ppm): 9.88 (d, J = 4 Hz, 1H), 9.83 (d, J = 8 Hz, 1H), 9.48 (d, J = 8 Hz, 1H), 9.36 (s, 1H), 9.32 (s, 1H), 8.78 (d, J = 4 Hz, 1H), 8.52–8.55 (m, 1H), 7.88–7.85 (m, 1H), 7.01 (s, 2H), 6.69 (s, 2H), 4.92 (d, J = 12 Hz, 2H), 1.19 (s, 21H), 0.57 (s, 15H).
270). χM (298 K): −549.3 × 10−6 cm3 M−1. 1H NMR (DMSO-d6) δ (ppm): 9.88 (d, J = 4 Hz, 1H), 9.83 (d, J = 8 Hz, 1H), 9.48 (d, J = 8 Hz, 1H), 9.36 (s, 1H), 9.32 (s, 1H), 8.78 (d, J = 4 Hz, 1H), 8.52–8.55 (m, 1H), 7.88–7.85 (m, 1H), 7.01 (s, 2H), 6.69 (s, 2H), 4.92 (d, J = 12 Hz, 2H), 1.19 (s, 21H), 0.57 (s, 15H).
[CoL(dppz)] (3).
Yield, 45% (0.19 g). Anal. calcd for C50H56CoN5O4: C, 70.66; H, 6.64; N, 8.24. Found: C, 70.43; H, 6.77; N, 8.20. ΛM in DMF: 11 S m2 M−1. FT-IR (KBr phase): 2954s, 1625vs, 1443m, 1303m, 1299m, 1233s, 1200m, 1166s, 1035s, 832s, 731m, 514w, 450w cm−1. ESI-MS in MeOH: m/z 850.3 (M + H)+. UV-visible in 6% DMF/Tris HCl buffer [λmax, nm (ε, M−1 cm−1) 690 (450), 450 (4680), 380 (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 550), 362 (19
550), 362 (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 710), 274 (73
710), 274 (73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970). χM (298 K): −517.6 × 10−6 cm3 M−1. 1H-NMR (DMSO-d6) δ (ppm): 9.94 (d, J = 8 Hz, 1H), 9.90 (d, J = 4 Hz, 1H), 8.81 (d, J = 4 Hz,1H), 8.49–8.61 (m, 4H), 8.25 (s, 2H), 7.89 (d, J = 8 Hz, 1H), 7.04 (s 2H), 6.74 (s, 2H), 4.96 (d, J = 12 Hz, 2H), 1.26 (m, 21H), 0.66 (s, 15H).
970). χM (298 K): −517.6 × 10−6 cm3 M−1. 1H-NMR (DMSO-d6) δ (ppm): 9.94 (d, J = 8 Hz, 1H), 9.90 (d, J = 4 Hz, 1H), 8.81 (d, J = 4 Hz,1H), 8.49–8.61 (m, 4H), 8.25 (s, 2H), 7.89 (d, J = 8 Hz, 1H), 7.04 (s 2H), 6.74 (s, 2H), 4.96 (d, J = 12 Hz, 2H), 1.26 (m, 21H), 0.66 (s, 15H).
Solubility and stability
The complexes showed good solubility in CH2Cl2, MeOH, EtOH, MeCN, DMF and DMSO. They were stable in the solution phase.
X-ray crystallographic procedure
Dark brown block shaped crystals of 1·0.5EtOH·H2O were grown from an ethanol–DMF solution of the complex and crystal mounting was done on glass fibers with epoxy glue. X-ray diffraction data were measured in frames with increasing ω (width of 0.3° per frame) with a scan speed at 15 s per frame using a Bruker SMART APEX CCD diffractometer, equipped with a fine focus 1.75 kW sealed tube X-ray source. Empirical absorption corrections were made using a multi-scan program.36 The structure was solved by heavy-atom method and refined by full matrix least-squares using SHELX system of programs.37,38 All non-hydrogen atoms were refined anisotropically. The hydrogen atoms belonging to the complex were at their calculated positions and were refined by a riding model. The relatively high R index value could be due to high thermal vibrations associated with the tert-butyl groups present in the tetradentate ligand (L). The ethanol molecule showed positional disorder with half occupancy in each asymmetric unit. Two restraints (EADP and DFIX) were used during refinement for the thermal and positional disorders of the tert-butyl groups and ethanol molecule, respectively, using provisions available in the SHELX program. This disorder did not have any apparent effect on the overall structure of the complex. The lattice water molecule Ow(1) showed bifurcated hydrogen bonding interactions with O(2) of the phenolate group and O(3) of the carboxylate group with D⋯A bond distances of 2.832 Å and 2.997 Å, respectively. The lattice ethanol molecule did not show any chemically significant hydrogen bonding interactions. Detailed crystallographic data along with bonding parameters are given in Tables S1 and S2 (vide ESI†).
Selected crystal data for 1·0.5EtOH·H2O: C45H59N3O5.5Co, M = 788.88, monoclinic, space group P 21/c, a = 13.205(4) Å, b = 16.068(4) Å, c = 21.477(6) Å, α = γ = 90°, β = 105.504(10)°, U = 4391(2) Å3, Z = 4, ρ = 1.193 g cm−3, T = 293(2) K, μ = 0.437 mm−1, data/restraints/parameters: 7922/2/460, F(000) = 1684, GOOF = 1.03, R/wR = 0.1025/0.2441 for 2954 reflections [I > 2σ(I)], R/wR [(all data)] = 0.2535/0.3310.
DNA binding and photocleavage experiments
The experiments were done in Tris-HCl/NaCl buffer (5 mM Tris-HCl, 5 mM NaCl, pH 7.2) or phosphate buffer of pH 7.4 using DMF solution of the complexes 1–3 using procedures as described earlier (vide ESI†).25,39–43 DNA photocleavage studies were carried out using DMF solutions of the complexes 1–3 with supercoiled pUC 19 DNA in 50 mM Tris-HCl buffer containing 50 mM NaCl. The photocleavage experiments were carried out at 365 nm (UV-A light) and 676 nm (Ar–Kr mixed gas ion laser) using appropriate controls and inhibitors as mentioned earlier (vide ESI†).44
Photocytotoxicity assay
The photocytotoxicity of the complexes was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay which is based on the ability of mitochondrial dehydrogenases in the viable cells to cleave the tetrazolium rings of MTT forming dark blue membrane-impermeable crystals of formazan that can be measured at 595 nm making it soluble in the detergent.45 The level of the formazan product formed gave us a measure of the number of viable cells. About 8000 cells of human cervical carcinoma (HeLa) or 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells of Human Embryonic Kidney stably transfected with full length hTSH receptor (HEK293-hTSHR) or untransfected Human Embryonic Kidney cell line (HEK293) were plated in 96 well culture plates in DMEM containing 10% FBS and after 24 h of incubation at 37 °C in CO2 incubator (70% confluency), various concentrations of the cobalt(III) complexes dissolved in DMSO were added to the cells and incubation was continued for 4 h in the dark. After incubation, the medium was replaced with PBS and photo-irradiated with a 400–700 nm broadband visible light (Luzchem Photoreactor Model LZC-1, 10 J cm−2). Post irradiation, PBS was replaced with DMEM-FBS and incubation was continued for 12 h in the dark. Post incubation, a 20 μL volume of 5 mg mL−1 of MTT was added to each well and incubation was carried on for an additional period of 3 h in dark. The culture medium was discarded and 100 μL of 10% SDS/0.01 M HCl was added to dissolve the formazan crystals formed and the absorbance at 595 nm was determined using a BIORAD ELISA plate reader. Cytotoxicity of the test compounds was measured as the percentage ratio of the absorbance of the treated cells to the untreated controls. The IC50 values were determined by nonlinear regression analysis (GraphPad Prism).
The assay was carried out as reported earlier.46 Approximately, 5 × 104 HEK293-hTSHR or HEK293 cells were plated in 48 well culture plate in DMEM containing 10% FBS and after 24 h of incubation at 37 °C in CO2 incubator (70% confluency), various concentrations of the cobalt(III) complexes dissolved in 1% DMSO were added to the cells and incubation was continued for 4 h in the dark. Light irradiation was carried out as described previously after which the cells were allowed to recover for 12 h. The medium in each well was replaced by 200 μL fresh serum containing medium along with 0.5 mM of general phosphodiesterase inhibitor isobutyl methyl xanthine (IBMX, Sigma) for 2 h at 37 °C to achieve steady state activity of adenylyl cyclase in the cells. For analysis of hTSH stimulated cAMP production, recovery post photo-irradiation was reduced to 1 h. Incubation in this case was carried out with 100 μL IBMX containing medium for 30 min and subsequently induced with 100 μL of hTSH (5 nM) dissolved in IBMX containing medium for 15 min. The termination of the cyclase reaction was carried out by adding 0.2 M HCl to the reaction medium.
000 cells of Human Embryonic Kidney stably transfected with full length hTSH receptor (HEK293-hTSHR) or untransfected Human Embryonic Kidney cell line (HEK293) were plated in 96 well culture plates in DMEM containing 10% FBS and after 24 h of incubation at 37 °C in CO2 incubator (70% confluency), various concentrations of the cobalt(III) complexes dissolved in DMSO were added to the cells and incubation was continued for 4 h in the dark. After incubation, the medium was replaced with PBS and photo-irradiated with a 400–700 nm broadband visible light (Luzchem Photoreactor Model LZC-1, 10 J cm−2). Post irradiation, PBS was replaced with DMEM-FBS and incubation was continued for 12 h in the dark. Post incubation, a 20 μL volume of 5 mg mL−1 of MTT was added to each well and incubation was carried on for an additional period of 3 h in dark. The culture medium was discarded and 100 μL of 10% SDS/0.01 M HCl was added to dissolve the formazan crystals formed and the absorbance at 595 nm was determined using a BIORAD ELISA plate reader. Cytotoxicity of the test compounds was measured as the percentage ratio of the absorbance of the treated cells to the untreated controls. The IC50 values were determined by nonlinear regression analysis (GraphPad Prism).
The assay was carried out as reported earlier.46 Approximately, 5 × 104 HEK293-hTSHR or HEK293 cells were plated in 48 well culture plate in DMEM containing 10% FBS and after 24 h of incubation at 37 °C in CO2 incubator (70% confluency), various concentrations of the cobalt(III) complexes dissolved in 1% DMSO were added to the cells and incubation was continued for 4 h in the dark. Light irradiation was carried out as described previously after which the cells were allowed to recover for 12 h. The medium in each well was replaced by 200 μL fresh serum containing medium along with 0.5 mM of general phosphodiesterase inhibitor isobutyl methyl xanthine (IBMX, Sigma) for 2 h at 37 °C to achieve steady state activity of adenylyl cyclase in the cells. For analysis of hTSH stimulated cAMP production, recovery post photo-irradiation was reduced to 1 h. Incubation in this case was carried out with 100 μL IBMX containing medium for 30 min and subsequently induced with 100 μL of hTSH (5 nM) dissolved in IBMX containing medium for 15 min. The termination of the cyclase reaction was carried out by adding 0.2 M HCl to the reaction medium.
cAMP measurement
The amount of cAMP produced by the cells were quantified using radioimmunoassay (RIA).47 The reaction mixture (50 μL) from each well were incubated overnight with 50 μL of cAMP antiserum (NHPP) and 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cpm of 125I-cAMP in a total volume of 200 μL at 4 °C. A standard curve for cAMP was constructed using increasing concentrations of cAMP standard or ranging from 0.2 to 400 pM. The unbound adsorbed by 1% activated charcoal in 0.05 M potassium phosphate buffer (pH 6.3) containing 1.0 mg mL−1 of BSA was separated by centrifugation at 3000 g, and counted in a γ-counter (Perkin-Elmer). The bound 125I-cAMP was calculated by subtracting the free counts from the total counts added and analyzed by a non-linear regression after interpolating the unknowns from the standard curve.
000 cpm of 125I-cAMP in a total volume of 200 μL at 4 °C. A standard curve for cAMP was constructed using increasing concentrations of cAMP standard or ranging from 0.2 to 400 pM. The unbound adsorbed by 1% activated charcoal in 0.05 M potassium phosphate buffer (pH 6.3) containing 1.0 mg mL−1 of BSA was separated by centrifugation at 3000 g, and counted in a γ-counter (Perkin-Elmer). The bound 125I-cAMP was calculated by subtracting the free counts from the total counts added and analyzed by a non-linear regression after interpolating the unknowns from the standard curve.
Confocal immunofluorescence
HEK293-hTSHR cells were treated with various concentrations of complex 3 for 4 h in the dark followed by light treatment as mentioned above and were allowed to recover for 1 h in the dark. The cells were then fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at 37 °C followed by two rinses in DPBS containing 1.0 mM CaCl2. No permeabilization step was carried out in this procedure to differentiate between the plasma membrane bound receptors and the intracellular immature forms. The cells were blocked using DPBS containing 10% heat inactivated FBS, 0.1% fish skin gelatine and 7.5% sucrose in DPBS for 10 min and then incubated with 2.5 μg monoclonal antibody (311/62) against TSH receptor (raised against aa 200-413 of TSH receptor ectodomain, by a modified protocol as developed by Dighe and Agrawal for the FSH work) and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution of Anti-Pan Cadherin antibody (C3678, Sigma-Aldrich) for 1 h.46 Immunoreactivity was detected with FITC-conjugated anti-mouse secondary antibodies (Sigma-Aldrich) for detection of 311/62 and TRITC-conjugated anti rabbit secondary antibody for detecting Anti-Pan Cadherin antibody. The specificity was determined by omitting the primary antibody or using normal rabbit/mouse antibody as controls. The immunolabelled cells were imaged in a Leica SP5-AOBS confocal laser scanning microscope (Leica Microsystems, GmbH Wetzlar, Germany) and analyzed using LAS AF Lite software.
100 dilution of Anti-Pan Cadherin antibody (C3678, Sigma-Aldrich) for 1 h.46 Immunoreactivity was detected with FITC-conjugated anti-mouse secondary antibodies (Sigma-Aldrich) for detection of 311/62 and TRITC-conjugated anti rabbit secondary antibody for detecting Anti-Pan Cadherin antibody. The specificity was determined by omitting the primary antibody or using normal rabbit/mouse antibody as controls. The immunolabelled cells were imaged in a Leica SP5-AOBS confocal laser scanning microscope (Leica Microsystems, GmbH Wetzlar, Germany) and analyzed using LAS AF Lite software.
Binding of hTSHR-ECD to complex 3
The ectodomain of TSH receptor (hTSHR-ECD, amino acid 21-413) was cloned and expressed using the Pichia pastoris expression system and purified using hydrophobic interaction chromatography followed by size exclusion chromatography (by a modified protocol as developed by Dighe and co-workers for the FSH work).46 The ability of complex 3 to bind to hTSHR-ECD was studied by absorption spectral titration. Briefly, increasing concentrations of hTSHR-ECD (stock solution: 5 mg ml−1 in 0.2 M phosphate buffer, pH 7.4) was added to a fixed 30 μM solution of complex 3 (6% DMF/5 mM phosphate buffer pH 7.4), to both reference and sample channels and the absorption spectra were recorded after each addition till no change in the absorption spectra of complex 3 was observed. The dissociation constant (Kd) value was determined by plotting (εa − εf)/(εb − εf) vs. ECD concentration, where εf, εa and εb are, respectively, the molar extinction coefficients of the free complex in solution, complex bound to hTSHR-ECD at a definite concentration and the complex in completely bound form with ECD. The data were fitted using the non-linear regression analysis hyperbolic saturation binding (Graph Pad Prism 5.0). As a control, binding of complex 3 to bovine serum albumin (BSA) was investigated and the binding constant was determined as mentioned above.
Protein profile and immunoblot of photocleaved hTSHR-ECD
The photocleavage potential of complex 3 on hTSH receptor ectodomain was investigated using SDS PAGE analysis and subsequent immunoblot analysis of the protein profile. hTSHR ECD (20 μM) was diluted in a 50 mM Tris–HCl buffer (pH, 7.2) and photoirradiated in the presence of different concentrations of 3 at 400–700 nm broadband light (10 J cm−2) for 1 h, after which the samples were analyzed on a 12.5% SDS-PAGE.48,49 For immunoblot analysis, the reaction mixtures were TCA precipitated to remove salts, solubilized in Lammeli buffer and probed using a hTSHR-ECD specific polyclonal antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 104) after 12.5% SDS-PAGE and electroblotting.
104) after 12.5% SDS-PAGE and electroblotting.
Results and discussion
Synthesis and general aspects
Cobalt(III) complexes [CoL(B)] (1–3) are prepared from the reaction of Co(NO3)2·6H2O with a trianionic tetradentate ligand (H3L) and the phenanthroline base (B: phen in 1; dpq in 2; dppz in 3) (Fig. 1). Complexes 1–3 are characterized from elemental analytical and spectral data (Table 1). The tetradentate ligand H3L with two phenolates, one amine and one acetate binding site is chosen to stabilize the metal in its +3 oxidation state. The presence of tert-butyl groups in L could impart higher lipophilic character needed for better cellular activity.50 Moreover, the phenolate ligands are known to show low energy ligand-to-cobalt(III) charge transfer electronic transitions.51 Planar phenanthroline bases are used in the ternary structure as DNA binders. Moreover, the dpq and dppz ligands with their quinoxaline and phenazine moieties could serve as efficient photosensitizers generating 3(n–π*) and/or 3(π–π*) excited state.30 Although cobalt complexes of trianionic ligands are reported in the literature, they have been studied for their varied structural, redox and magnetostructural studies.52 Cobalt(III) complexes that have been studied for their nuclease activity primarily have tris-diimine ligands or mono/dianionic ligands.53 We are not aware of any cobalt(III) complex having a trianionic ligand and phenanthroline bases exhibiting photonuclease activity.
Table 1 Selected physicochemical and DNA binding data for the complexes
| Complex |
[CoL(phen)] (1) |
[CoL(dpq)] (2) |
[CoL(dppz)] (3) |
|
In KBr phase.
Visible electronic band in 6% DMF-Tris-HCl buffer within PDT window.
ΛM, molar conductance in DMF at 25 °C.
Scan rate of 50 mV s−1 in DMF-0.1 M TBAP. Potentials are vs. saturated calomel electrode (SCE). Ef = 0.5(Epa + Epc). ΔEp = (Epa − Epc), where Ef, Epa and Epc are formal, anodic and cathodic peak potentials, respectively.
Anodic peak without any cathodic peak.
K
b, DNA binding constant [s, fitting constant].
K
app, apparent DNA binding constant.
Change in the DNA melting temperature.
|
IRa/cm−1 [ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)] O)] |
1633 |
1626 |
1625 |
|
λ/nm (ε/M−1 cm−1)b |
685 (320) |
693 (415) |
690 (450) |
| ΛMc /S m2 M−1 |
17 |
18 |
11 |
|
E
f/V (ΔEp, mV)d |
|
|
|
| CoIII–CoII couple |
−0.28 (560) |
−0.24 (475) |
−0.22 (445) |
| CoIV–CoIII couple |
0.80 (80) |
0.83 (75) |
0.85e |
|
K
b/M−1 [s/ b.p.]f |
5.6(±1.7) × 104 [0.1] |
8.6(±0.6) × 104 [0.4] |
1.9(±0.5) × 105 [0.3] |
|
K
app
/M−1 |
8.3 × 104 |
1.3 × 105 |
2.7 × 105 |
| ΔTmh/°C |
0.3 |
1.7 |
2.6 |
The monomeric complexes are non-electrolytic in solution. They are stable in the solution phase as evidenced from the mass spectral data essentially showing the molecular ion peak (ESI Fig. S1–S3†). Magnetic susceptibility data on the solid samples at 25 °C show diamagnetic nature of the complexes. 1H-NMR spectra of the complexes in DMSO-d6 reveal the presence of the phenolate-based ligand and the phenanthroline base in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio (ESI Fig. S4–S6†). The electronic spectra of the complexes in 50 mM Tris-HCl buffer containing 6% DMF (pH 7.2) display a broad band near 425 nm and a weak band near 690 nm (Fig. 2, ESI Fig. S7†). The low-energy band falls within the PDT spectral window of 620–850 nm. This is important considering the fact that the human tissue is transparent to this region of the electromagnetic spectrum as most biomolecules and intracellular chromophores do not absorb in this region. The electronic spectral properties of the low-spin 3d6-cobalt(III) complexes differ considerably from their 3d5 high-spin iron(III) analogues showing only the ligand-to-metal charge transfer (LMCT) band near 500 nm.25 The higher-energy intense visible band in 1–3 is assignable to the phenolate (O−) pπ → cobalt(III) dσ* charge transfer transition whereas the lower-energy band could be attributed to a d–d transition (1A1g → 1T1g).51 The complexes show poor redox activity. A voltammogram near −0.24 V (ΔEp, 430 mV at 50 mV s−1) vs. SCE in DMF-0.1 M TBAP with poor reversibility is assignable to the Co(III)/Co(II) couple (ESI Fig. S8†). The quasi-reversible nature of the Co(III)/Co(II) couple with large peak-to-peak separation suggests possible structural changes associated with the electrochemical reduction of the metal from 3d6-Co(III) to Jahn–Teller active 3d7-Co(II). The redox property of the cobalt(III) complexes differ significantly from their 3d5-iron(III) analogues that show significantly better reversibility of the cyclic voltammetric responses near −0.6 V vs. SCE.25 Complex 3, in addition, displays a voltammogram at −1.09 V vs. SCE (ΔEp = 193 mV) due to dppz ligand reduction. The complexes also exhibit a voltammogram near 0.8 V vs. SCE and this redox process could be assignable to the Co(IV)/Co(III) couple. While this oxidative process is quasi-reversible in nature for the phen and dpq complexes, the dppz complex shows only the anodic response without having any cathodic counterpart.
1 molar ratio (ESI Fig. S4–S6†). The electronic spectra of the complexes in 50 mM Tris-HCl buffer containing 6% DMF (pH 7.2) display a broad band near 425 nm and a weak band near 690 nm (Fig. 2, ESI Fig. S7†). The low-energy band falls within the PDT spectral window of 620–850 nm. This is important considering the fact that the human tissue is transparent to this region of the electromagnetic spectrum as most biomolecules and intracellular chromophores do not absorb in this region. The electronic spectral properties of the low-spin 3d6-cobalt(III) complexes differ considerably from their 3d5 high-spin iron(III) analogues showing only the ligand-to-metal charge transfer (LMCT) band near 500 nm.25 The higher-energy intense visible band in 1–3 is assignable to the phenolate (O−) pπ → cobalt(III) dσ* charge transfer transition whereas the lower-energy band could be attributed to a d–d transition (1A1g → 1T1g).51 The complexes show poor redox activity. A voltammogram near −0.24 V (ΔEp, 430 mV at 50 mV s−1) vs. SCE in DMF-0.1 M TBAP with poor reversibility is assignable to the Co(III)/Co(II) couple (ESI Fig. S8†). The quasi-reversible nature of the Co(III)/Co(II) couple with large peak-to-peak separation suggests possible structural changes associated with the electrochemical reduction of the metal from 3d6-Co(III) to Jahn–Teller active 3d7-Co(II). The redox property of the cobalt(III) complexes differ significantly from their 3d5-iron(III) analogues that show significantly better reversibility of the cyclic voltammetric responses near −0.6 V vs. SCE.25 Complex 3, in addition, displays a voltammogram at −1.09 V vs. SCE (ΔEp = 193 mV) due to dppz ligand reduction. The complexes also exhibit a voltammogram near 0.8 V vs. SCE and this redox process could be assignable to the Co(IV)/Co(III) couple. While this oxidative process is quasi-reversible in nature for the phen and dpq complexes, the dppz complex shows only the anodic response without having any cathodic counterpart.
 |
| | Fig. 2 UV-visible absorption spectrum of complex 3 in 6% DMF-Tris HCl buffer with arrows showing selected wavelengths used for photo-induced DNA cleavage study. The inset shows the d–d band (as shoulder) of complex 3. | |
The phen complex as 1·0.5EtOH·H2O is structurally characterized by single crystal X-ray diffraction technique. The complex crystallizes in the monoclinic space group P21/c (Z = 4) with 0.5 molecule of ethanol and one water per formula unit as the solvents of crystallization. An ORTEP view of the complex is shown in Fig. 1. Selected bond distances and angles are given in Table S1 (vide ESI†). The unit cell packing diagram is shown in Fig. S9 (vide ESI†). The complex has the cobalt(III) center in a six-coordinate CoN3O3 coordination geometry bound to the trianionic ligand L that shows tetradentate mode of bonding to the metal center, while the N,N-donor 1,10-phenanthroline ligand binds in a bidentate fashion. The Co–O and Co–N distances are in the range of 1.896(6) to 1.984(7) Å. The structure of the cobalt(III) complex is significantly different from its iron(III) analogue.25 Two phenolate oxygen atoms are found to be trans to each other in the cobalt(III) complex whereas in the iron(III) complex one of the phenolate oxygen atom is trans to one nitrogen atoms of the phenanthroline base. This structural difference has a significant effect on the steric enclosure of the phenanthroline ligand. The phenanthroline base is sterically encumbered by the tertiary butyl groups in the cobalt(III) structure, while no such steric effect is exerted by the tetradentate ligand in the reported iron(III) structure.
DNA binding and photocleavage property
The binding property of the cobalt(III) complexes to calf thymus DNA has been studied using different spectral techniques. Selected DNA binding data are given in Table 1. We have carried out absorption spectral measurements to determine the equilibrium binding constant (Kb) of the complexes to CT-DNA by measuring the change in the absorption intensity of the spectral band at 266 nm for complexes 1, 2 and at 271 nm for complex 3. The complexes show minor bathochromic shift upon addition of CT-DNA (Fig. 3, ESI Fig. S10†). The Kb and s (fitting parameter) values range within 5.6 × 104 to 1.9 × 105 M−1 and 0.1 to 0.4, respectively. Complex 3 having dipyridophenazine ligand shows better binding propensity to CT-DNA. The low s values obtained from the theoretical fitting of the spectral data are indicative to the DNA groove-binding propensities of the complexes in preference to intercalation due to the presence of steric bulk of the ancillary tetradentate ligand having bulky tert-butyl groups.54 A low value of s (<1) could also result from an aggregation of the hydrophobic molecules on the DNA surface by π-stacking. The DNA binding has also been studied by a competitive ethidium bromide displacement assay using fluorescence spectral method to determine the apparent binding constant (Kapp) values that gave the order: 3 (dppz) > 2 (dpq) > 1 (phen) (ESI Fig. S11†). The thermal denaturation data show only a minor change in the DNA melting temperature (Tm) giving ΔTm values of 0.3–2.6 °C on addition of the complexes to CT-DNA indicating primarily groove binding nature of the complexes (Fig. 3).55
![(a) Absorption spectral traces of complex 3 (30 μM) in 5 mM Tris-HCl buffer (pH 7.2) (6% DMF) on increasing the quantity of CT DNA (241 μM). The inset shows the least-squares fit of Δεaf/Δεbfvs. [DNA] for complex 3 (■) using the MvH equation. (b) Thermal denaturation plots of CT-DNA (150 μM) in the presence and absence of the complexes 1–3 (10 μM) in 5 mM phosphate buffer (pH 6.8).](/image/article/2010/MT/c0mt00028k/c0mt00028k-f3.gif) |
| | Fig. 3 (a) Absorption spectral traces of complex 3 (30 μM) in 5 mM Tris-HCl buffer (pH 7.2) (6% DMF) on increasing the quantity of CT DNA (241 μM). The inset shows the least-squares fit of Δεaf/Δεbfvs. [DNA] for complex 3 (■) using the MvH equation. (b) Thermal denaturation plots of CT-DNA (150 μM) in the presence and absence of the complexes 1–3 (10 μM) in 5 mM phosphate buffer (pH 6.8). | |
Photo-induced DNA cleavage activity of the cobalt(III) complexes has been studied using SC pUC19 DNA (30 μM, 0.2 μg) in a medium of Tris-HCl/NaCl (50 mM, pH, 7.2) buffer on irradiation with UV-A light of 365 nm and red light of 676 nm. The extent of DNA cleavage from SC to the nicked circular (NC) form has been measured from the gel electrophoresis study (Fig. 4 and 5, ESI Fig. S12–S14†). Selected photo-induced DNA cleavage data are given in Table 2. The DNA cleavage activity of the complexes at 365 nm follows the order: 3 (dppz) ≥ 2 (dpq) ≫ 1 (phen) (ESI Fig. S12†). A 5.0 μM concentration of 2 and 3 shows essentially complete cleavage of SC DNA for an photoexposure time of 2 h (lanes 9,10 in ESI Fig. S12†). The phen complex 1 does not show any apparent DNA photocleavage activity at this wavelength. The results suggest that the UV-A light-induced DNA cleavage reaction possibly involves quinoxaline ring of dpq and phenazine moiety of dppz towards generating the photo-excited 3(n–π*) and/or 3(π–π*) state(s).30 While complex 2 has a spectral band at 340 nm, complex 3 has bands at 380 and 362 nm that are close to this photo-excitation wavelength. Control experiments with Co(NO3)2·6H2O (30 μM) and the ligands H3L, phen, dpq or dppz (30 μM) alone do not show any apparent DNA cleavage activity at 365 nm. The complexes are cleavage inactive in the dark thus ruling out any hydrolytic DNA cleavage pathway. We have studied the groove binding preference of the complexes using DNA minor groove binder distamycin-A which alone shows ∼25% cleavage of the SC DNA at 365 nm for an exposure time of 2 h (ESI Fig. S12†). Addition of complex 2 (5 μM) to distamycin-A bound DNA shows no apparent increase in the DNA cleavage activity suggesting minor groove-binding propensity of complex 2. The DNA cleavage activity of complex 3 remains unaffected in the presence of distamycin-A indicating major groove binding preference of the dppz complex. The dppz complex shows significant inhibition in DNA cleavage in the presence DNA major groove blocker methyl green.
![Photocleavage of SC pUC19 DNA (0.2 μg, 30 μM) by [CoL(B)] (30 μM; B = phen, 1; dpq, 2; dppz, 3) in Tris-HCl/NaCl buffer (pH 7.2) containing 10% DMF on photo-exposure to red light of 676 nm (50 mW) for 2 h: lane 1, DNA control; lane 2, DNA + dpq (100 μM); lane 3, DNA + dppz (100 μM); lane 4, DNA + Co(NO3)2·6H2O (30 μM); lane 5, DNA + 2 (in dark); lane 6, DNA + 3 (in dark); lane 7, DNA + 1; lane 8, DNA + 2; lane 9, DNA + 3; lane 10, DNA + 2 (under argon); lane 11, DNA + 3 (under argon).](/image/article/2010/MT/c0mt00028k/c0mt00028k-f4.gif) |
| | Fig. 4 Photocleavage of SC pUC19 DNA (0.2 μg, 30 μM) by [CoL(B)] (30 μM; B = phen, 1; dpq, 2; dppz, 3) in Tris-HCl/NaCl buffer (pH 7.2) containing 10% DMF on photo-exposure to red light of 676 nm (50 mW) for 2 h: lane 1, DNA control; lane 2, DNA + dpq (100 μM); lane 3, DNA + dppz (100 μM); lane 4, DNA + Co(NO3)2·6H2O (30 μM); lane 5, DNA + 2 (in dark); lane 6, DNA + 3 (in dark); lane 7, DNA + 1; lane 8, DNA + 2; lane 9, DNA + 3; lane 10, DNA + 2 (under argon); lane 11, DNA + 3 (under argon). | |
 |
| | Fig. 5 The bar diagram showing mechanistic aspects of UV-A (365 nm) and red light-induced (676 nm) SC pUC19 DNA (0.2 μg, 30 μM bp) cleavage activity of complex 3 in the presence of external reagents (NaN3, TEMP, DABCO and KI of 500 μM; DMSO, 4 μL; catalase and SOD of 4 units) under aerobic and argon atmosphere. The blue bars represent the photocleavage activity (expressed as %NC form) in UV-A light and the red bars represent the activity in red light. | |
Table 2 Photo-induced SC DNA (0.2 μg, 30 μM bp) cleavage dataa
| Sl. No. |
Reaction condition |
[Complex/ligand]: 365 nm [676 nm]/μM |
t/min |
%NC365nm [%NC676nm] |
|
SC and NC are supercoiled and nicked circular forms of pUC19 DNA. The cleavage reactions are under aerobic conditions except for serial numbers 5 and 7 that are done under argon atmosphere. The error in measuring %DNA is 3–5%. t, exposure time. Light sources: UV-A light (365 nm, 6 W) and visible light [Ar–Kr CW Laser (676 nm, 50 mW)]. Co(NO3)2·6H2O (30 μM) alone converts SC DNA to NC DNA by 3% and 4% in UV-A and 676 nm light respectively.
|
| 1 |
DNA control |
— |
2 h |
2 [2] |
| 2 |
DNA + [CoL(dppz)] (3) (in dark) |
5 [30] |
2 h |
3 [11] |
| 3 |
DNA + dpq ligand |
30 [100] |
2 h |
13 [4] |
| 4 |
DNA + dppz ligand |
30 [100] |
2 h |
15 [7] |
| 3 |
DNA + [CoL(phen)] (1) |
5 [30] |
2 h |
7 [15] |
| 4 |
DNA + [CoL(dpq)] (2) |
5 [30] |
2 h |
97 [85] |
| 5 |
DNA + [CoL(dpq)] (2) (in argon) |
5 [30] |
2 h |
3 [36] |
| 6 |
DNA + [CoL(dppz)] (3) |
5 [30] |
2 h |
98 [99] |
| 7 |
DNA + [CoL(dppz)] (3) (in argon) |
5 [30] |
2 h |
6 [42] |
The cobalt(III) complexes show a visible band within the PDT window near 690 nm. We have carried out photolysis of SC DNA in the presence of the complexes (30 μM) in red light of 676 nm using a continuous-wave (CW) Ar–Kr mixed-gas ion laser (50 mW laser power). While the dpq and dppz complexes show photo-induced oxidative DNA cleavage activity, the phen complex is photo-inactive (Fig. 4, lanes 7–9). The cleavage data reveal the necessity of photo-active phenanthroline bases like dpq or dppz in the ternary cobalt(III) structure to observe DNA photocleavage activity. Similar DNA cleavage by copper(II) complexes on photo-irradiation at the low-energy metal-based d–d band has been reported.56,57 The compounds showing photocleavage of DNA at ∼700 nm are of interest in the PDT chemistry considering greater skin penetration of red light at near-IR wavelengths. The PDT drug Photofrin® is active at 630 nm and barring the phthalocyanines, other organic dyes showing DNA cleavage at near-IR wavelength are rare in the literature.4 The presence of a low-energy visible band in the 3d6-cobalt(III) complexes makes these complexes as potent PDT agents at near-IR light.
The mechanistic aspects of the photo-induced DNA cleavage reaction of complex 3 have been investigated using various external reagents in UV-A light of 365 nm and red light of 676 nm (Fig. 5, ESI Fig. S13–S14†). Addition of singlet oxygen quenchers, viz. DABCO, TEMP, sodium azide and L-histidine show no apparent effect on the DNA photocleavage activity. No changes in the DNA cleavage activity is observed in D2O thus ruling out the formation of singlet oxygen in a type-II pathway.58 Hydroxyl radical scavengers like DMSO and KI, however, have shown significant inhibition in the DNA cleavage activity at these wavelengths. Hydrogen peroxide and superoxide scavengers, viz. catalase and SOD, respectively, show significant inhibition in the DNA cleavage activity. The results suggest the involvement of peroxo species and superoxide radicals (O2˙−) in the photocleavage reactions in both UV-A and red light possibly involving metal-assisted photo-activation of the dpq or dppz ligand in a buffer medium. The peroxo species is likely to generate reactive hydroxyl radicals. Interestingly, both dpq and dppz ligands alone are known to photocleave DNA in high energy UV light via only type-II singlet oxygen pathway.30 The presence of a redox active metal like cobalt(III) could make the alternate photo-redox pathway more viable than the type-II process that requires transfer of energy from the excited triplet of the complex to the molecular oxygen in its triplet state requiring high singlet oxygen quantum yield. We have studied the DNA photocleavage activity under anaerobic medium. The complexes do not show any DNA photocleavage activity under argon at 365 nm (Fig. 5). The DNA photocleavage reaction in red light under argon, however, shows partial inhibition thus indicating an alternate DNA cleavage pathway operating under anoxic conditions (Fig. 4, lanes 10, 11). Photo-excitation of the d–d band at 676 nm could possibly lead to DNA cleavage via oxygen-independent type-I pathway.15,59
Cytotoxicity study
The photocytotoxic potential of the dppz complex 3 has been tested in visible light using HeLa, HEK293 and HEK293-hTSHR cells by MTT assay (Fig. 6). Complex 2 does not show photocytotoxicity in HEK293 or HEK293-hTSHR cells (ESI Fig. S15†). Visible light irradiation after treatment with complex 3 for 4 h in dark reduces the viability of the HEK293 hTSHR cells in a dose-dependent manner giving an IC50 value of 7.3 μM. Interestingly, both untransfected HEK 293 parent cell lines as well as unrelated cervical carcinoma cell lines (HeLa) show a lesser degree of photocytotoxicity with nearly 4-fold higher IC50 value of 29.9 μM for HEK293 and >100 μM for HeLa. Moreover, no cell death has been observed in HeLa, HEK293 or HEK293-hTSHR cells when treated with complex 3 in the dark indicating its non-toxic nature in absence of light-irradiation. The dppz ligand alone, on the other hand, exhibits moderate photocytotoxicity on HeLa, HEK293 and HEK293-hTSHR cells indicating a different mode of action for the dppz base in effecting photocytotoxicity. The IC50 values of complex 3 for the HeLa, HEK293 and HEK293-hTSHR cells are given in Table 3. The fact that the three cell line types that differ only by the presence of hTSH receptor would behave marked differently in presence of 3 on treatment with light point to a selective role of 3 on the thyrotropin receptor. One possible reason could be the presence of an extremely large extracellular domain in hTSHR as compared to other receptors which could provide potential binding sites on the leucine rich repeat (LRR) folds in the extracellular domain. The second more deterministic effect would be interference of complex 3 with the signaling machinery specific to TSH receptor itself.
![Photocytotoxicity of [CoL(dppz)] (3) determined by MTT assay 12 h post-photoirradiation: (a) Effect of complex 3 upon visible light exposure (400–700 nm, 10 J cm−2) on HEK293 (●) and HEK293-hTSHR (■) cells; (b) and (c) Controls with dppz (●), H3L (■) and Co(NO3)3·6H2O (□) in HEK293 and HEK293-hTSHR, respectively; (d) Effect of complex 3 (■) and dppz (●) on HeLa cells upon exposure to visible light. The red and black lines represent visible light treatment and non-irradiated sample data, respectively. The error bars correspond to the standard deviation of the mean of three individual experiments.](/image/article/2010/MT/c0mt00028k/c0mt00028k-f6.gif) |
| | Fig. 6 Photocytotoxicity of [CoL(dppz)] (3) determined by MTT assay 12 h post-photoirradiation: (a) Effect of complex 3 upon visible light exposure (400–700 nm, 10 J cm−2) on HEK293 (●) and HEK293-hTSHR (■) cells; (b) and (c) Controls with dppz (●), H3L (■) and Co(NO3)3·6H2O (□) in HEK293 and HEK293-hTSHR, respectively; (d) Effect of complex 3 (■) and dppz (●) on HeLa cells upon exposure to visible light. The red and black lines represent visible light treatment and non-irradiated sample data, respectively. The error bars correspond to the standard deviation of the mean of three individual experiments. | |
Table 3 IC50 values of complex 3 in HeLa, HEK293 and HEK293-hTSHR Cellsa
| Compound (method) |
HEK293 |
HEK293-hTSHR |
HeLa |
| IC50 (error)/μM |
IC50 (error)/μM |
IC50 (error)/μM |
| Dark |
Lightb |
Dark |
Lightb |
Dark |
Lightb |
|
The dpq complex 2 did not show any cytotoxicity in the dark as well as on photo-irradiation with visible light in all the cell lines studied.
The light source was 400–700 nm broadband light from Luzchem Photoreactor (LZC-1). The total light dose provided was 10 J cm−2.
|
| [CoL(dppz)] (3) (MTT assay) |
>100 |
29.9 (±3.2) |
>100 |
7.3 (±0.9) |
>100 |
>100 |
| [CoL(dppz)] (3) (cAMP RIA) |
>100 |
26.5 (±2.5) |
>100 |
8.6 (±1.3) |
— |
— |
| dppz ligand (MTT assay) |
>100 |
41.1 (±1.8) |
>100 |
38.2 (±2.6) |
>100 |
95.1(±4.3) |
Cyclic AMP determination
Cyclic AMP produced by activation of hTSH receptor by hTSH controls import of iodine into the thyroid follicular cells, secretion of thyroid hormone and most importantly, mediates survival of thyrocytes by prevention of apoptosis.60 To examine whether complex 3 mediates its function through inhibition of basal cAMP production machinery of HEK293-hTSHR cells, adenylyl cyclase assay was carried out for HEK293-hTSHR cells and HEK293 cells as control, 12 h post photo irradiation. The HEK293 cells showed a decrease in cAMP level production with an IC50 value of 26.5 μM whereas the HEK293-hTSHR cells showed a marked dose dependant decrease in cAMP levels with an IC50 value of 8.6 μM (Fig. 7). Whether this decrease in the rate of cAMP synthesis directly affects the survivability of cells, either by reducing to the levels below the minimum threshold for cell survival or interfering with other linked pathways is a matter yet to be resolved. This decrease is, however, consistent with the observed reduction of cell viability noticed in cell cytotoxicity assay. It is also worth noting that the basal level of cAMP in HEK293-hTSHR (8 pM well−1) is one log order higher than HEK293 cells (0.8 pM well−1) which indicates the importance of cAMP in this cell type, a situation also encountered in primary thyrocytes isolated from tumors.61
 |
| | Fig. 7 Cyclic AMP measurement using radioimmunoassay in (a) HEK293 cells and (b) HEK293-hTSHR cells 12 h after treatment with complex 3 and photo-irradiation with visible light (400–700 nm, 10 J cm−2). The error bars correspond to mean ± s.d. (n = 3). | |
One might also speculate that this observed reduction of cAMP levels is due to the cell death and not directly related to TSH receptor per se. To negate this possibility cyclase assay was performed immediately after light treatment in presence and absence of 5 nM hTSH when cell death is minimal. We observed that there was a dose-dependent decrease in hTSH stimulated response with 50% inhibition at 50 μM of complex 3. However, no such decrease was observed in the basal cAMP level which goes on to suggest that the treatment had no effect on adenylyl cyclases activity (Fig. 8). Thus complex 3 exerts its photocytotoxic effects in HEK293-hTSHR cells by affecting the hTSH receptor.
 |
| | Fig. 8 hTSH stimulation assay to appraise thyrotropin (hTSH) receptor damage in HEK293-hTSHR cells through cyclic AMP measurement using radioimmunoassay 1 h post-irradiation with visible light. The error bars correspond to mean ± s.d. (n = 3). | |
Immunofluorescence
To further validate the decrease in levels of cAMP to the state of TSH receptor in HEK293-hTSHR, immunofluorescence study was conducted using hTSHR specific monoclonal antibody and a polyclonal antibody against a cell surface marker such as pan-Cadherin, a protein involved in cell adhesion and cell–cell contact. The intensity of FITC fluorescence (green) would designate the levels of TSH receptor whereas TRITC fluorescence (red) demarcates the levels of Cadherin in HEK293-hTSHR cells. We observed that in untreated cells, both TSH receptor as well as Cadherin has robust expression on the cell surface. However, on photoirradiation with a progressive increase in complex 3 concentrations, the levels of TSH receptor decreased on the cell surface as shown by a decrease in green fluorescence and a progressive intracellular increase of Cadherin levels (Fig. 9). The change of cellular morphology is also marked with transition from the epithelial spindle shape to extensively damaged membrane blebbed form. The decrease in levels of TSH receptor can be correlated with the damage of cell membrane and the decrease in TSH-mediated cAMP signaling as observed in the previous section. Interestingly, the total membrane Cadherin levels do not seem to alter, but there seems to be a higher distribution of trapped intracellular Cadherin as compared to the membrane bound form. This could be explained by the fact that damage of hTSH receptor leads to loss of cell surface integrity which in turn leads to loss of surface bound Cadherins. At the same time, the synthesized Cadherin accumulates in intracellular compartments that largely escape further degradation leading to enhanced intracellular Cadherin levels in HEK293-hTSHR.62
 |
| | Fig. 9 Indirect immunofluorescence with confocal microscopy images of HEK293-hTSHR cells 1 h after PDT with complex 3. The green FITC fluorescence corresponds to hTSHR levels (left panels) and red TRITC fluorescence corresponds to the Pan Cadherin levels (central panel) in the HEK293-TSHR cells (vide text). The right panel corresponds to merged images. | |
hTSHR-ECD binding
Binding of complex 3 to the extracellular domain of hTSHR was studied by absorption spectroscopy as a more direct evidence of interaction of complex 3 with hTSHR-ECD. As a control, we also determined the binding constant of complex 3 to BSA, which is a well-known binder and transporter of hydrophobic molecules.63 Addition of hTSHR-ECD to a solution of complex 3 in DMF/5 mM phosphate buffer pH 7.4 showed decrease in the intensity of the ligand centered charge transfer bands with concomitant red shift of about 5 nm (Fig. S16, ESI†). Isosbestic points were also observed at 300 nm, 350 nm and 383 nm. The dissociation constant (Kd) of complex 3 with hTSHR-ECD was found to be 81.2 nM whereas the binding to BSA was significantly lower (Kd = 387 nM). This clearly signifies higher binding affinity (∼4.7-fold) of complex 3 to the ECD over BSA, the model protein. Although, the binding to the transmembrane domain of hTSHR could not be studied due to the insolubility of the full length TSH receptor, one cannot rule out a possible interaction of complex 3 with the transmembrane domain as well. The dppz ligand alone did not bind to the hTSHR-ECD as there was no perturbation in the absorption spectra of the ligand upon addition of increasing concentrations of hTSHR-ECD.
Protein profile and immunoblot of photocleaved TSH receptor ectodomain
The ability of complex 3 to photocleave hTSHR-ECD upon phototreatment was examined by analyzing the SDS-PAGE profile of the untreated as well as complex 3 treated hTSHR ECD. hTSHR-ECD purifies at two bands of molecular weight 53 kDa and 45 kDa, the former corresponding to the higher glycosylated form and the latter being a lower glycosylated isoform. A third band at a higher molecular weight was also observed which corresponds to higher oligomer of hTSHR-ECD. Upon light treatment with the complex 3, intensity of bands corresponding to hTSHR-ECD decreased, with almost complete disappearance at 100 μM of complex 3. To further validate that the cleaved band is indeed that of hTSHR-ECD, the protein was precipitated using TCA, electrophoresed on 12.5% SDS Gel and immune-probed with hTSHR-ECD specific polyclonal antibody. The profile obtained matched with that of the SDS-PAGE profile with a decrease of band intensity with increasing complex 3 concentrations (Fig. 10). The fact that no other band except the parent hTSHR-ECD band was observed indicates that upon photoactivation, complex 3 generates highly diffusive HO˙ radicals, as shown by the mechanistic investigation in DNA photocleavage, which could damage the peptide backbone as shown in Scheme 1 in a sequence-independent manner.
 |
| | Fig. 10 (A) Coomassie stained SDS-PAGE gel showing the reduction in hTSHR-ECD (22 μM) band intensity upon treatment with various concentrations of complex 3 and exposure to visible light (400–700, 10 J cm−2): lane 1, hTSHR-ECD control; lane 2, hTSHR-ECD + complex 3 (6 μM); lane 3, hTSHR-ECD + complex 3 (12 μM); lane 4, hTSHR-ECD + complex 3 (25 μM); lane 5, hTSHR-ECD + complex 3 (50 μM); lane 6, hTSHR-ECD + complex 3 (100 μM); lane 7, protein molecular weight marker. (B) Western Blotting to substantiate hTSHR-ECD photodamage on photoexcitation with visible light (400–700 nm, 10 J cm−2) by various concentrations of complex 3. | |
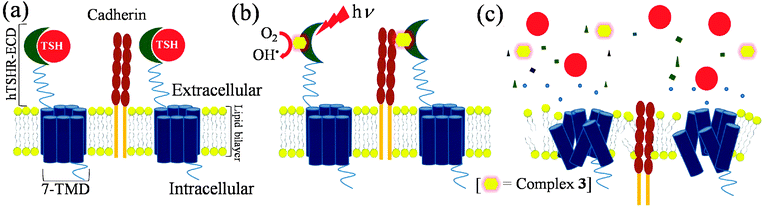 |
| | Scheme 1 Proposed scheme of TSHR damage by complex 3: (a) binding of TSH to TSH receptor activates adenylyl cyclases resulting in cAMP synthesis which activates other downstream proteins finally resulting in thyroid growth and differentiation; (b) complex 3 on incubation for 4 h in dark with HEK293-hTSHR cells binds to TSH receptor after which photoexposure is given and (c) hydroxyl radicals generated upon photo-exposure damages the receptor which results in loss of membrane integrity and subsequent translocation of Cadherin to cytoplasm, finally resulting in cell death. | |
Conclusions
We present a new class of cobalt(III) complexes with a tetradentate phenolate-based ligand and planar phenanthroline bases showing a low energy absorption band within the PDT spectral window. The complexes display moderate binding to CT-DNA due to the presence of planar phenanthroline bases within the sterically crowded tert-butyl groups. The presence of a near-IR visible band at ∼690 nm makes the dpq and dppz cobalt(III) complexes excellent model photonucleases in red light at 676 nm. The mechanism of photocleavage involves a photo-redox process which generates hydroxyl radicals as the reactive species. The non-toxic dppz complex 3 behaves as a highly potent photocytotoxin in HEK293-hTSHR cells as compared to its parent cell-line HEK293 and the HeLa cells upon photoexcitation of the complex in visible light. Direct hTSH receptor damage has been evidenced from confocal immunofluorescence, SDS-PAGE and Western blot analysis. In all, we present near-IR active photonucleolytic cobalt(III) complexes with the dipyridophenazine complex 3 showing unprecedented photocytotoxicity in HEK293-hTSHR cells arising from direct hTSH receptor damage thus providing a new platform for the treatment of thyroid pathologies using PDT as a treatment modality.
Acknowledgements
We thank the Department of Science and Technology (DST), Government of India, for financial support (SR/S5/MBD-02/2007). We are grateful to the Alexander von Humboldt Foundation, Germany, for an electrochemical system. SS thanks CSIR for a research fellowship and ARC thanks DST for J. C. Bose national fellowship.
Notes and references
-
(a) R. T. Dorsam and J. S. Gutkind, Curr. Opin. Oncol., 2008, 20, 19–24 CrossRef;
(b) E. Baudin and M. Schlumberger, Lancet Oncol., 2007, 8, 148–156 CrossRef CAS;
(c) T. Kondo, S. Ezzat and S. L. Asa, Nat. Rev. Cancer, 2006, 6, 292–306 CrossRef CAS;
(d) L. J. DeGroot and J. Quintans, Endocr. Rev., 1989, 10, 537–562 CrossRef CAS.
- S. T. Chen, H. Y. Shieh, J. D. Lin, K. S. Chang and K. H. Lin, J. Endocrinol., 2000, 165, 379–389 Search PubMed.
- F. Iten, B. Muller, C. Schindler, H. Rasch, C. Rochlitz, D. Oertli, H. R. Maecke, J. Muller-Brand and M. A. Walter, Cancer, 2009, 115, 2052–62 CrossRef CAS.
-
R. Bonnett, Chemical Aspects of Photodynamic Therapy, Gordon & Breach, London, UK, 2000 Search PubMed.
- M. R. Detty, S. L. Gibson and S. J. Wagner, J. Med. Chem., 2004, 47, 3897–3915 CrossRef CAS.
- L. K. J. Boerner and J. M. Zaleski, Curr. Opin. Chem. Biol., 2005, 9, 135–144 CrossRef CAS.
- B. W. Henderson, T. M. Busch, L. A. Vaughan, N. P. Frawley, D. Babich, T. A. Sosa, J. D. Zollo, A. S. Dee, M. T. Cooper, D. A. Bellnier, W. R. Greco and A. R. Oseroff, Cancer Res., 2000, 60, 525–529 CAS.
-
(a) M. C. DeRosa and R. J. Crutchley, Coord. Chem. Rev., 2002, 233–234, 351–371 CrossRef CAS;
(b) H. Ali and J. E. Van Lier, Chem. Rev., 1999, 99, 2379–2450 CrossRef;
(c) G. Stochel, A. Wanad, E. Kulis and Z. Stasicka, Coord. Chem. Rev., 1998, 171, 203–220 CrossRef CAS;
(d) J. L. Sessler, G. Hemmi, T. D. Mody, T. Murai, A. Burrell and S. W. Young, Acc. Chem. Res., 1994, 27, 43–50 CrossRef CAS;
(e) W.-H. Wei, Z. Wang, T. Mizuno, C. Cortez, L. Fu, M. Sirisawad, L. Naumovski, D. Magda and J. L. Sessler, Dalton Trans., 2006, 1934–1942 RSC.
-
(a) M. J. Ochsner, J. Photochem. Photobiol., B, 1996, 32, 3–9 CrossRef CAS;
(b) S. I. Moriwaki, J. Misawa, Y. Yoshinari, I. Yamada, M. Takigawa and Y. Tokura, Photodermatol., Photoimmunol. Photomed., 2001, 17, 241–243 CrossRef CAS.
- J. Mao, Y. Zhang, J. Zhu, C. Zhang and Z. Guo, Chem. Commun., 2009, 908–910 RSC.
- D. Ramaiah, I. Eckert, K. T. Arun, L. Weidenfeller and B. Epe, Photochem. Photobiol., 2004, 79, 99–104 CrossRef CAS.
- M. Kar and A. Basak, Chem. Rev., 2007, 107, 2861–2890 CrossRef CAS.
- S. Atilgan, Z. Ekmeckci, A. L. Dogan, D. Guc and E. U. Akkaya, Chem. Commun., 2006, 4398–4400 RSC.
- K. Szaciłowski, W. Macyk, A. Drzewiecka-Matuszek, M. Brindell and G. Stochel, Chem. Rev., 2005, 105, 2647–2694 CrossRef CAS.
- C. J. Burrows and J. G. Muller, Chem. Rev., 1998, 98, 1109–1151 CrossRef CAS.
-
(a) F. S. Mackay, J. A. Woods, P. Heringová, J. Kašpárková, A. M. Pizarro, S. A. Moggach, S. Parsons, V. Brabec and P. J. Sadler, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 20743–20748 CrossRef CAS;
(b) N. J. Farrer, L. Salassa and P. J. Sadler, Dalton Trans., 2009, 10690 RSC.
- B. Armitage, Chem. Rev., 1998, 98, 1171–1200 CrossRef CAS.
- D. Lahiri, T. Bhowmick, B. Pathak, O. Shameema, A. K. Patra, S. Ramakumar and A. R. Chakravarty, Inorg. Chem., 2009, 48, 339–349 CrossRef CAS.
-
(a) A. M. Angeles-Boza, H. T. Chifotides, J. D. Aguirre, A. Chouai, P. K.-L. Fu, K. R. Dunbar and C. Turro, J. Med. Chem., 2006, 49, 6841–6847 CrossRef CAS;
(b) H. T. Chifotides and K. R. Dunbar, Acc. Chem. Res., 2005, 38, 146–156 CrossRef CAS.
- B. Rosenberg, L. VanCamp, J. E. Trosko and V. H. Mansour, Nature, 1969, 222, 385–386 CrossRef CAS.
-
(a) S. Dhar, Z. Liu, J. Thomale, H. Dai and S. J. Lippard, J. Am. Chem. Soc., 2008, 130, 11467–11476 CrossRef CAS;
(b) S. Dhar, F. X. Gu, R. Langer, O. C. Farokhzad and S. J. Lippard, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17356–17361 CrossRef CAS.
-
(a) D. Wang and S. J. Lippard, Nat. Rev. Drug Discovery, 2005, 4, 307–320 CrossRef CAS;
(b) Y. Jung and S. J. Lippard, Chem. Rev., 2007, 107, 1387–1407 CrossRef CAS.
-
(a) M. J. Rose, N. L. Fry, R. Marlow, L. Hinck and P. K. Mascharak, J. Am. Chem. Soc., 2008, 130, 8834–8846 CrossRef CAS;
(b) A. D. Ostrowski and P. C. Ford, Dalton Trans., 2009, 10660 RSC.
- U. Schatzschneider, Eur. J. Inorg. Chem., 2010, 1451–1467 CAS.
-
(a) S. Saha, R. Majumdar, M. Roy, R. R. Dighe and A. R. Chakravarty, Inorg. Chem., 2009, 48, 2652–2663 CrossRef CAS;
(b) M. Roy, S. Saha, A. K. Patra, M. Nethaji and A. R. Chakravarty, Inorg. Chem., 2007, 46, 4368–4370 CrossRef CAS.
- P. K. Sasmal, S. Saha, R. Majumdar, R. R. Dighe and A. R. Chakravarty, Chem. Commun., 2009, 1703–1705 RSC.
- D. C. Ware, B. D. Palmer, W. R. Wilson and W. A. Denny, J. Med. Chem., 1993, 36, 1839–1846 CrossRef CAS.
- K. E. Erkkila, D. T. Odom and J. K. Barton, Chem. Rev., 1999, 99, 2777–2796 CrossRef CAS.
- S. Arounaguiri, D. Eswaramoorthy, A. Ashokkumar, A. Dattagupta and B. G. Maiya, Proc.–Indian Acad. Sci., Chem. Sci., 2000, 112, 1–17 Search PubMed.
- K. Toshima, R. Takano, T. Ozawa and S. Matsumura, Chem. Commun., 2002, 212–213 RSC.
-
D. D. Perrin, F. Armarego and D. R. Perrin, Purification of Laboratory Chemicals, Pergamon Press, Oxford, 1980 Search PubMed.
-
(a) J. E. Dickeson and L. A. Summers, Aust. J. Chem., 1970, 23, 1023–1027 CAS;
(b) J. G. Collins, A. D. Sleeman, J. R. Aldrich-Wright, I. Greguric and T. W. Hambley, Inorg. Chem., 1998, 37, 3133–3141 CrossRef CAS.
- E. Amouyal, A. Homsi, J.-C. Chambron and J.-P. Sauvage, J. Chem. Soc., Dalton Trans., 1990, 1841–1845 RSC.
- J. G. Wilson, Aust. J. Chem., 1990, 43, 1283–1289 CAS.
-
O. Khan, Molecular Magnetism, VCH, Weinheim, 1993 Search PubMed.
- N. Walker and D. Stuart, Acta Crystallogr., Sect. A: Found. Crystallogr., 1983, A39, 158–166 CrossRef CAS.
-
(a)
G. M. Sheldrick, SHELX-97, Programs for Crystal Structure Solution and Refinement, University of Göttingen, Göttingen, Germany, 1997 Search PubMed;
(b) G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, A64, 112–122 CrossRef CAS.
-
C. K. Johnson, ORTEP, Report ORNL-5138, Oak Ridge National Laboratory, Oak Ridge, TN, 1976 Search PubMed.
- M. E. Reichman, S. A. Rice, C. A. Thomas and P. Doty, J. Am. Chem. Soc., 1954, 76, 3047–3053 CrossRef CAS.
-
(a) J. D. McGhee and P. H. von Hippel, J. Mol. Biol., 1974, 86, 469–489 CrossRef CAS;
(b) M. T. Carter, M. Rodriguez and A. J. Bard, J. Am. Chem. Soc., 1989, 111, 8901–8911 CrossRef CAS.
- M. Waring, J. Mol. Biol., 1965, 13, 269–282 CrossRef CAS.
- J.-B. LePecq and C. Paoletti, J. Mol. Biol., 1967, 27, 87–106 CrossRef CAS.
- M. Lee, A. L. Rhodes, M. D. Wyatt, S. Forrow and J. A. Hartley, Biochemistry, 1993, 32, 4237–4245 CrossRef CAS.
- J. Bernadou, G. Pratviel, F. Bennis, M. Girardet and B. Meunier, Biochemistry, 1989, 28, 7268–7275 CrossRef CAS.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS.
- G. Agrawal and R. R. Dighe, J. Biol. Chem., 2009, 284, 2636–2647 CAS.
- W. M. Hunter and R. C. Greenwood, Nature, 1962, 194, 495–496 CAS.
- C. V. Kumar, A. Buranaprapuk, H. C. Sze, S. Jockusch and N. J. Turro, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5810–5815 CrossRef CAS.
- H. Schägger and G. von Jagow, Anal. Biochem., 1987, 166, 368–379 CrossRef CAS.
- M. S. Shongwe, C. H. Kaschula, M. S. Adsetts, E. W. Ainscough, A. M. Brodie and M. J. Morris, Inorg. Chem., 2005, 44, 3070–3079 CrossRef CAS.
- M. S. Shongwe, S. K. M. Al-Hatmi, H. M. Marques, R. Smith, R. Nukada and M. Mikuriya, J. Chem. Soc., Dalton Trans., 2002, 4064–4069 RSC.
-
(a) A. S. Rothin, H. J. Banbery, F. J. Berry, T. A. Hamor, C. J. Jones and J. A. McCleverty, Polyhedron, 1989, 8, 491–504 CrossRef CAS;
(b) C. N. Verani, S. Gallert, E. Bill, T. Weyhermüller, K. Wieghardt and P. Chaudhuri, Chem. Commun., 1999, 1747–1748 RSC;
(c) M. M. H. Khalil, E. H. Ismail, S. A. Azim and E. R. Souaya, J. Therm. Anal. Calorim., 2010, 101, 129–135 CrossRef CAS;
(d) R. E. Cowley, R. P. Bontchev, J. Sorrell, O. Sarracino, Y. Feng, H. Wang and J. M. Smith, J. Am. Chem. Soc., 2007, 129, 2424–2425 CrossRef CAS.
-
(a) S. Arounaguiri and B. G. Maiya, Inorg. Chem., 1996, 35, 4267–4270 CrossRef CAS;
(b) P. Nagababu, J. N. Lavanya Latha, Y. Prashanthi and S. Satyanarayana, J. Chem. Pharm. Res., 2009, 1, 238–249 Search PubMed;
(c) D. Lahiri, S. Roy, S. Saha, R. Majumdar, R. R. Dighe and A. R. Chakravarty, Dalton Trans., 2010, 39, 1807–1816 RSC;
(d) G. N. Novodarova and M. E. Vol'pin, Catal. Lett., 1990, 4, 265–270 CrossRef CAS.
- R. B. Nair, E. S. Teng, S. L. Kirkland and C. J. Murphy, Inorg. Chem., 1998, 37, 139–141 CrossRef CAS.
- A. Chouai, S. E. Wicke, C. Turro, J. Bacsa, K. R. Dunbar, D. Wang and R. P. Thummel, Inorg. Chem., 2005, 44, 5996–6003 CrossRef CAS.
- S. Dhar, D. Senapati, P. K. Das, P. Chattopadhyay, M. Nethaji and A. R. Chakravarty, J. Am. Chem. Soc., 2003, 125, 12118–12124 CrossRef CAS.
- B. Maity, M. Roy, S. Saha and A. R. Chakravarty, Organometallics, 2009, 28, 1495–1505 CrossRef CAS.
- A. U. Khan, J. Phys. Chem., 1976, 80, 2219–2227 CrossRef CAS.
- M. Tanaka, K. Ohkubo and S. Fukuzumi, J. Am. Chem. Soc., 2006, 128, 12372–12373 CrossRef CAS.
-
(a) M. Rivas and P. Santisteban, Mol. Cell. Endocrinol., 2003, 213, 31–45 CrossRef CAS;
(b) M. D. Michael, L. F. Michael and E. R. Simpson, Mol. Cell. Endocrinol., 1997, 134, 147–156 CrossRef CAS.
- H.-J. Breustedt, S. Nolde, M. Schumacher and H. Hilz, Acta Endocrinol., 1977, 86, 99–111 Search PubMed.
- F. Larsson, H. Fagman and M. Nilsson, Cell. Mol. Life Sci., 2004, 61, 1834–1842 CAS.
- B. Sarkar, Biol. Trace Elem. Res., 1989, 21, 137–144 CrossRef CAS.
|
| This journal is © The Royal Society of Chemistry 2010 |
Click here to see how this site uses Cookies. View our privacy policy here. ![(a) Ternary cobalt(iii) complexes and the phenanthroline bases used. (b) An ORTEP view of [CoL(phen)] in 1·0.5EtOH·H2O showing 50% probability thermal ellipsoids and atom numbering for the metal and heteroatoms. The hydrogen atoms are omitted for clarity.](/image/article/2010/MT/c0mt00028k/c0mt00028k-f1.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 450), 266 (48
450), 266 (48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 110). χM (298 K): −502.8 × 10−6 cm3 M−1. 1H NMR (DMSO-d6) δ (ppm): 9.75 (d, J = 4 Hz, 1H), 9.07 (d, J = 8 Hz, 1H), 8.70 (d, J = 8 Hz, 1H), 8.58 (d, J = 4 Hz, 1H), 8.32–8.38 (m, 2H), 8.20 (d, J = 8 Hz, 1H), 7.68 (m, 1H), 7.02 (d, J = 2 Hz, 2H), 6.70 (d, J = 2 Hz, 2H), 4.89 (d, J = 12 Hz, 2H), 1.25 (s, 21H), 0.55 (s, 15H).
110). χM (298 K): −502.8 × 10−6 cm3 M−1. 1H NMR (DMSO-d6) δ (ppm): 9.75 (d, J = 4 Hz, 1H), 9.07 (d, J = 8 Hz, 1H), 8.70 (d, J = 8 Hz, 1H), 8.58 (d, J = 4 Hz, 1H), 8.32–8.38 (m, 2H), 8.20 (d, J = 8 Hz, 1H), 7.68 (m, 1H), 7.02 (d, J = 2 Hz, 2H), 6.70 (d, J = 2 Hz, 2H), 4.89 (d, J = 12 Hz, 2H), 1.25 (s, 21H), 0.55 (s, 15H).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 450), 282 (32
450), 282 (32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 550), 266 (44
550), 266 (44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 270). χM (298 K): −549.3 × 10−6 cm3 M−1. 1H NMR (DMSO-d6) δ (ppm): 9.88 (d, J = 4 Hz, 1H), 9.83 (d, J = 8 Hz, 1H), 9.48 (d, J = 8 Hz, 1H), 9.36 (s, 1H), 9.32 (s, 1H), 8.78 (d, J = 4 Hz, 1H), 8.52–8.55 (m, 1H), 7.88–7.85 (m, 1H), 7.01 (s, 2H), 6.69 (s, 2H), 4.92 (d, J = 12 Hz, 2H), 1.19 (s, 21H), 0.57 (s, 15H).
270). χM (298 K): −549.3 × 10−6 cm3 M−1. 1H NMR (DMSO-d6) δ (ppm): 9.88 (d, J = 4 Hz, 1H), 9.83 (d, J = 8 Hz, 1H), 9.48 (d, J = 8 Hz, 1H), 9.36 (s, 1H), 9.32 (s, 1H), 8.78 (d, J = 4 Hz, 1H), 8.52–8.55 (m, 1H), 7.88–7.85 (m, 1H), 7.01 (s, 2H), 6.69 (s, 2H), 4.92 (d, J = 12 Hz, 2H), 1.19 (s, 21H), 0.57 (s, 15H).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 550), 362 (19
550), 362 (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 710), 274 (73
710), 274 (73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970). χM (298 K): −517.6 × 10−6 cm3 M−1. 1H-NMR (DMSO-d6) δ (ppm): 9.94 (d, J = 8 Hz, 1H), 9.90 (d, J = 4 Hz, 1H), 8.81 (d, J = 4 Hz,1H), 8.49–8.61 (m, 4H), 8.25 (s, 2H), 7.89 (d, J = 8 Hz, 1H), 7.04 (s 2H), 6.74 (s, 2H), 4.96 (d, J = 12 Hz, 2H), 1.26 (m, 21H), 0.66 (s, 15H).
970). χM (298 K): −517.6 × 10−6 cm3 M−1. 1H-NMR (DMSO-d6) δ (ppm): 9.94 (d, J = 8 Hz, 1H), 9.90 (d, J = 4 Hz, 1H), 8.81 (d, J = 4 Hz,1H), 8.49–8.61 (m, 4H), 8.25 (s, 2H), 7.89 (d, J = 8 Hz, 1H), 7.04 (s 2H), 6.74 (s, 2H), 4.96 (d, J = 12 Hz, 2H), 1.26 (m, 21H), 0.66 (s, 15H).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells of Human Embryonic Kidney stably transfected with full length hTSH receptor (HEK293-hTSHR) or untransfected Human Embryonic Kidney cell line (HEK293) were plated in 96 well culture plates in DMEM containing 10% FBS and after 24 h of incubation at 37 °C in CO2 incubator (70% confluency), various concentrations of the cobalt(III) complexes dissolved in DMSO were added to the cells and incubation was continued for 4 h in the dark. After incubation, the medium was replaced with PBS and photo-irradiated with a 400–700 nm broadband visible light (Luzchem Photoreactor Model LZC-1, 10 J cm−2). Post irradiation, PBS was replaced with DMEM-FBS and incubation was continued for 12 h in the dark. Post incubation, a 20 μL volume of 5 mg mL−1 of MTT was added to each well and incubation was carried on for an additional period of 3 h in dark. The culture medium was discarded and 100 μL of 10% SDS/0.01 M HCl was added to dissolve the formazan crystals formed and the absorbance at 595 nm was determined using a BIORAD ELISA plate reader. Cytotoxicity of the test compounds was measured as the percentage ratio of the absorbance of the treated cells to the untreated controls. The IC50 values were determined by nonlinear regression analysis (GraphPad Prism).
000 cells of Human Embryonic Kidney stably transfected with full length hTSH receptor (HEK293-hTSHR) or untransfected Human Embryonic Kidney cell line (HEK293) were plated in 96 well culture plates in DMEM containing 10% FBS and after 24 h of incubation at 37 °C in CO2 incubator (70% confluency), various concentrations of the cobalt(III) complexes dissolved in DMSO were added to the cells and incubation was continued for 4 h in the dark. After incubation, the medium was replaced with PBS and photo-irradiated with a 400–700 nm broadband visible light (Luzchem Photoreactor Model LZC-1, 10 J cm−2). Post irradiation, PBS was replaced with DMEM-FBS and incubation was continued for 12 h in the dark. Post incubation, a 20 μL volume of 5 mg mL−1 of MTT was added to each well and incubation was carried on for an additional period of 3 h in dark. The culture medium was discarded and 100 μL of 10% SDS/0.01 M HCl was added to dissolve the formazan crystals formed and the absorbance at 595 nm was determined using a BIORAD ELISA plate reader. Cytotoxicity of the test compounds was measured as the percentage ratio of the absorbance of the treated cells to the untreated controls. The IC50 values were determined by nonlinear regression analysis (GraphPad Prism).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cpm of 125I-cAMP in a total volume of 200 μL at 4 °C. A standard curve for cAMP was constructed using increasing concentrations of cAMP standard or ranging from 0.2 to 400 pM. The unbound adsorbed by 1% activated charcoal in 0.05 M potassium phosphate buffer (pH 6.3) containing 1.0 mg mL−1 of BSA was separated by centrifugation at 3000 g, and counted in a γ-counter (Perkin-Elmer). The bound 125I-cAMP was calculated by subtracting the free counts from the total counts added and analyzed by a non-linear regression after interpolating the unknowns from the standard curve.
000 cpm of 125I-cAMP in a total volume of 200 μL at 4 °C. A standard curve for cAMP was constructed using increasing concentrations of cAMP standard or ranging from 0.2 to 400 pM. The unbound adsorbed by 1% activated charcoal in 0.05 M potassium phosphate buffer (pH 6.3) containing 1.0 mg mL−1 of BSA was separated by centrifugation at 3000 g, and counted in a γ-counter (Perkin-Elmer). The bound 125I-cAMP was calculated by subtracting the free counts from the total counts added and analyzed by a non-linear regression after interpolating the unknowns from the standard curve.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution of Anti-Pan Cadherin antibody (C3678, Sigma-Aldrich) for 1 h.46 Immunoreactivity was detected with FITC-conjugated anti-mouse secondary antibodies (Sigma-Aldrich) for detection of 311/62 and TRITC-conjugated anti rabbit secondary antibody for detecting Anti-Pan Cadherin antibody. The specificity was determined by omitting the primary antibody or using normal rabbit/mouse antibody as controls. The immunolabelled cells were imaged in a Leica SP5-AOBS confocal laser scanning microscope (Leica Microsystems, GmbH Wetzlar, Germany) and analyzed using LAS AF Lite software.
100 dilution of Anti-Pan Cadherin antibody (C3678, Sigma-Aldrich) for 1 h.46 Immunoreactivity was detected with FITC-conjugated anti-mouse secondary antibodies (Sigma-Aldrich) for detection of 311/62 and TRITC-conjugated anti rabbit secondary antibody for detecting Anti-Pan Cadherin antibody. The specificity was determined by omitting the primary antibody or using normal rabbit/mouse antibody as controls. The immunolabelled cells were imaged in a Leica SP5-AOBS confocal laser scanning microscope (Leica Microsystems, GmbH Wetzlar, Germany) and analyzed using LAS AF Lite software.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 104) after 12.5% SDS-PAGE and electroblotting.
104) after 12.5% SDS-PAGE and electroblotting.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)]
O)]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio (ESI Fig. S4–S6†). The electronic spectra of the complexes in 50 mM Tris-HCl buffer containing 6% DMF (pH 7.2) display a broad band near 425 nm and a weak band near 690 nm (Fig. 2, ESI Fig. S7†). The low-energy band falls within the PDT spectral window of 620–850 nm. This is important considering the fact that the human tissue is transparent to this region of the electromagnetic spectrum as most biomolecules and intracellular chromophores do not absorb in this region. The electronic spectral properties of the low-spin 3d6-cobalt(III) complexes differ considerably from their 3d5 high-spin iron(III) analogues showing only the ligand-to-metal charge transfer (LMCT) band near 500 nm.25 The higher-energy intense visible band in 1–3 is assignable to the phenolate (O−) pπ → cobalt(III) dσ* charge transfer transition whereas the lower-energy band could be attributed to a d–d transition (1A1g → 1T1g).51 The complexes show poor redox activity. A voltammogram near −0.24 V (ΔEp, 430 mV at 50 mV s−1) vs. SCE in DMF-0.1 M TBAP with poor reversibility is assignable to the Co(III)/Co(II) couple (ESI Fig. S8†). The quasi-reversible nature of the Co(III)/Co(II) couple with large peak-to-peak separation suggests possible structural changes associated with the electrochemical reduction of the metal from 3d6-Co(III) to Jahn–Teller active 3d7-Co(II). The redox property of the cobalt(III) complexes differ significantly from their 3d5-iron(III) analogues that show significantly better reversibility of the cyclic voltammetric responses near −0.6 V vs. SCE.25 Complex 3, in addition, displays a voltammogram at −1.09 V vs. SCE (ΔEp = 193 mV) due to dppz ligand reduction. The complexes also exhibit a voltammogram near 0.8 V vs. SCE and this redox process could be assignable to the Co(IV)/Co(III) couple. While this oxidative process is quasi-reversible in nature for the phen and dpq complexes, the dppz complex shows only the anodic response without having any cathodic counterpart.
1 molar ratio (ESI Fig. S4–S6†). The electronic spectra of the complexes in 50 mM Tris-HCl buffer containing 6% DMF (pH 7.2) display a broad band near 425 nm and a weak band near 690 nm (Fig. 2, ESI Fig. S7†). The low-energy band falls within the PDT spectral window of 620–850 nm. This is important considering the fact that the human tissue is transparent to this region of the electromagnetic spectrum as most biomolecules and intracellular chromophores do not absorb in this region. The electronic spectral properties of the low-spin 3d6-cobalt(III) complexes differ considerably from their 3d5 high-spin iron(III) analogues showing only the ligand-to-metal charge transfer (LMCT) band near 500 nm.25 The higher-energy intense visible band in 1–3 is assignable to the phenolate (O−) pπ → cobalt(III) dσ* charge transfer transition whereas the lower-energy band could be attributed to a d–d transition (1A1g → 1T1g).51 The complexes show poor redox activity. A voltammogram near −0.24 V (ΔEp, 430 mV at 50 mV s−1) vs. SCE in DMF-0.1 M TBAP with poor reversibility is assignable to the Co(III)/Co(II) couple (ESI Fig. S8†). The quasi-reversible nature of the Co(III)/Co(II) couple with large peak-to-peak separation suggests possible structural changes associated with the electrochemical reduction of the metal from 3d6-Co(III) to Jahn–Teller active 3d7-Co(II). The redox property of the cobalt(III) complexes differ significantly from their 3d5-iron(III) analogues that show significantly better reversibility of the cyclic voltammetric responses near −0.6 V vs. SCE.25 Complex 3, in addition, displays a voltammogram at −1.09 V vs. SCE (ΔEp = 193 mV) due to dppz ligand reduction. The complexes also exhibit a voltammogram near 0.8 V vs. SCE and this redox process could be assignable to the Co(IV)/Co(III) couple. While this oxidative process is quasi-reversible in nature for the phen and dpq complexes, the dppz complex shows only the anodic response without having any cathodic counterpart.
![(a) Absorption spectral traces of complex 3 (30 μM) in 5 mM Tris-HCl buffer (pH 7.2) (6% DMF) on increasing the quantity of CT DNA (241 μM). The inset shows the least-squares fit of Δεaf/Δεbfvs. [DNA] for complex 3 (■) using the MvH equation. (b) Thermal denaturation plots of CT-DNA (150 μM) in the presence and absence of the complexes 1–3 (10 μM) in 5 mM phosphate buffer (pH 6.8).](/image/article/2010/MT/c0mt00028k/c0mt00028k-f3.gif)
![Photocleavage of SC pUC19 DNA (0.2 μg, 30 μM) by [CoL(B)] (30 μM; B = phen, 1; dpq, 2; dppz, 3) in Tris-HCl/NaCl buffer (pH 7.2) containing 10% DMF on photo-exposure to red light of 676 nm (50 mW) for 2 h: lane 1, DNA control; lane 2, DNA + dpq (100 μM); lane 3, DNA + dppz (100 μM); lane 4, DNA + Co(NO3)2·6H2O (30 μM); lane 5, DNA + 2 (in dark); lane 6, DNA + 3 (in dark); lane 7, DNA + 1; lane 8, DNA + 2; lane 9, DNA + 3; lane 10, DNA + 2 (under argon); lane 11, DNA + 3 (under argon).](/image/article/2010/MT/c0mt00028k/c0mt00028k-f4.gif)

![Photocytotoxicity of [CoL(dppz)] (3) determined by MTT assay 12 h post-photoirradiation: (a) Effect of complex 3 upon visible light exposure (400–700 nm, 10 J cm−2) on HEK293 (●) and HEK293-hTSHR (■) cells; (b) and (c) Controls with dppz (●), H3L (■) and Co(NO3)3·6H2O (□) in HEK293 and HEK293-hTSHR, respectively; (d) Effect of complex 3 (■) and dppz (●) on HeLa cells upon exposure to visible light. The red and black lines represent visible light treatment and non-irradiated sample data, respectively. The error bars correspond to the standard deviation of the mean of three individual experiments.](/image/article/2010/MT/c0mt00028k/c0mt00028k-f6.gif)





