Copper(I) and copper(II) binding to β-amyloid 16 (Aβ16) studied by electrospray ionization mass spectrometry†
Yu
Lu
,
Michel
Prudent
,
Liang
Qiao
,
Manuel A.
Mendez
and
Hubert H.
Girault
*
Laboratoire d'Electrochimie Physique et Analytique, Station 6, Ecole Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland. E-mail: hubert.girault@epfl.ch; Fax: +41 216933667; Tel: +41 216933151
First published on 26th May 2010
Abstract
Copper-β-amyloid 16 (Aβ16) complexes were investigated by electrospray ionization mass spectrometry (ESI-MS). Copper(I) and (II) complexes were formed on-line in a microchip electrospray emitter by using a sacrificial copper electrode as the anode in positive ionization mode. In the presence of ascorbic acid in the peptide solution, the amount of Cu(I)-Aβ16 generated electrochemically was even higher. A kinetic model is proposed to account for the generation of copper complexes. The structure of Cu(I)-Aβ16 was investigated by tandem mass spectrometry (MS/MS), and the binding site of Cu(I) to Aβ16 was identified at the His13, His14 residues. Cu(II)-Aβ16 was also investigated by MS/MS and, based on the unusual observations of a-ions, the two binding residues of His13 and His14 of Aβ16 to Cu(II) were also confirmed. This approach provides direct information on Cu(I)-Aβ16 complexes generated in solution from metallic copper and gives evidence that both His13 and His14 are involved in the coordination of both Cu(I)- and Cu(II)-Aβ16 complexes.
Introduction
Beta-amyloid (Aβ) deposits in tissues are one of the main pathological characteristics of Alzheimer's disease (AD), and many studies have been devoted to understanding the aggregation mechanism.1–3 Indeed, Aβ is usually present in healthy brains in a soluble form, but amyloid plaques are often observed in AD patients. In vivo, the most usual forms of Aβ consist of 40 amino-acids (Aβ40), which at physiological pH are largely random coils and of 42 amino-acids (Aβ42), which have a strong tendency to aggregate.4 Beta-amyloid plaques have been found to contain large amounts of transition metal cations such as Zn, Cu, and Fe (mM range) and it has also been suggested that the altered homeostasis of these transition metal ions is related to degenerative diseases.5–7 For this reason, many groups have investigated the oxidative damage of proteins and peptides induced by transition metals.8–10 Considering the redox activity of metal ions such as copper and iron, these ions play a crucial role in generating reactive oxygen species (ROS) responsible for the oxidative stress in the brain and for neuronal toxicity. A basic knowledge of the coordination of these metal ions at different oxidation states with amyloid peptides is therefore essential to apprehend their reactivity and reveal their potential role in the degeneration process. Furthermore, understanding the coordination of Cu(Aβ) and Zn(Aβ) may provide a strategy to rationalize novel therapeutics.11,12It has been proposed that the metal binding site lies in the N-terminus domain, more precisely in the first 16 amino acids of Aβ (Aβ16). This peptide shows no tendency to aggregate or to form fibrils under moderate concentrations, and represents a model for soluble metallated Aβ peptides. Currently, most studies on the structure of Cu(Aβ) complexes are mainly focused on the binding sites of Cu(II) to Aβ peptides. A recently published review13 recapitulates the various propositions reported in the literature and summarized the most reasonable coordination environments for Cu(II) binding to Aβ peptides considering the experimental differences and the limitation of each method. Two likely complexations involving the N-terminus, His6 and His13 or His14, Asp1-COO− on one hand, and His6, His13, His14 and Asp1-COO− on the other hand are usually considered. By comparison, the binding of the reduced Cu(I) with Aβ has not yet been fully characterized. Extended X-ray absorption fine structure (EXAFS) spectroscopy has been used to speculate a linear two-coordinate geometry with two imidazole ligands as recently reported by two different groups.14,15 Most copper(I) compounds are known for their instability in aerobic environments. It has also been reported that Cu(I)-Aβ complexes have a limited stability in air i.e. 20 min and are only stable in inert atmospheres or in the presence of strong reducing agents.14,15 Therefore, the preparation of pure copper(I) complexes is usually an experimental challenge. Copper(I) ions are involved in oxidation damages (Fenton and Haber-Weiss reactions) and since there is some evidence that Cu(I) could play an important role in the aggregation of amyloid peptide and is biologically relevant to the oxidative stress in the brain, we report here a mass spectrometric study of copper(I) and copper(II) binding to Aβ16.
Mass spectrometry (MS) coupled to electrospray ionization (ESI) is a powerful tool widely used in studying biological molecules and their complexes. Usually, the study of metal–protein interactions is carried out by mixing the biomolecules of interest with an electrolyte solution of the metal salt.16 Alternatively, soluble anodes can be used both to apply the high voltage to the electrospray emitter and to generate metal ions in the absence of counter ions, therefore preventing the charge neutralization effect induced by counter anions when using a salt.17,18 Here a microchip emitter has been used as the sensitivity is increased by two orders of magnitude compared to the commercial ESI source. We have shown that a sacrificial copper anode coupled to an electrospray emitter can generate a mixture of both Cu(I) and Cu(II) ions.19–21 Indeed, the dissolution of copper metal proceeds first by the generation of Cu(I) ions that can be further oxidized to Cu(II). However, in the presence of adequate ligands, Cu(I) ions can be scavenged to form complexes.20 In the present work, we apply this methodology to investigate the binding of both Cu(I) and Cu(II) to Aβ16.
Experimental
Chemicals
Aβ16 peptide (DAEFRH6DSGYEVH13H14QK, M = 1955.1 g mol−1) was purchased from Bachem (Bubendorf, Switzerland). Cupric chloride dihydrate (CuCl2·2H2O) and zinc chloride (ZnCl2) were bought from Acros Organics (Geel, Belgium). Ascorbic acid was bought from AppliChem GmbH (Darmstadt Germany) and methanol from Riedel-de-Haën (Seelze, Germany). Deionized water (18.2 MΩ cm) was from a Milli-Q system from Millipore (Bedford, MA). The lyophilized Aβ16 peptide was dissolved with deionized water at a final concentration of 1 mg mL−1 as a stock solution stored at −20 °C. Aβ16 solution was diluted at a final concentration of 10 μM in 50/50 (vol/vol) MeOH/H2O before each experiment. All the solutions were prepared fresh daily.MS setup and microspray interface
Metal ion on-line complexation was carried out using a microspray interface described previously.19 In brief, it consists of a single microchannel (45 μm × 120 μm × 1 cm) polyimide microchip developed by DiagnoSwiss SA (Monthey, Switzerland). The tip of the end of microchannel was cut by a blade as shown in Scheme 1 in order to form a stable Taylor cone under a high voltage. Therefore, the nanospray was stably produced by this well-shaped emitter, which could retain the stability for half an hour at least and be reused at least tens of times. A reservoir of polycarbonate (∅ = 3 mm, h = 5 mm) was glued at the inlet of the microchannel. The sample was loaded (V = 20 μL) in the reservoir in which a metallic electrode was immersed. MeOH/H2O (50/50) and methanol were used for the cleaning and rinse of the reservoir and the microchannel for three times before new sample loading respectively. A platinum wire was used for the mixture of the peptide and Cu(II) salt solution while a copper plate electrode (S = 30 mm2) were used for the peptide solution in the absence and presence of ascorbic acid. Both electrodes were sanded and rinsed with methanol before each experiment.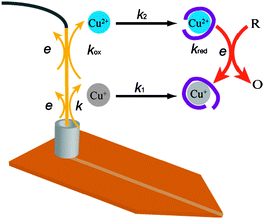 | ||
| Scheme 1 Mechanism for the generation of Cu(I)- and Cu(II)-Aβ16 complexes in the absence and presence of reducing agent. | ||
A LCQ DUO ion trap mass spectrometer (Thermo, San Jose, CA) was used in positive ion mode. The heated capillary was kept at 200 °C. The commercial ESI interface was removed and the microchip was mounted on a plate fixed on the probe slide adapter of the mass spectrometer. After the MS power supply onset (U = 3.5 kV) the chip was moved closer to the entrance of the MS. The use of high voltage is to be handled cautiously. The current was set between 30 and 50 nA by adjusting the distance between the microspray outlet and the entrance of the MS, and monitored by a nano-ammeter. The ion optics parameters were kept constant for each experiment. The MS fragments were assigned based on the calculation of a web-based software, MS-Products from UCSF (http://prospector.ucsf.edu/prospector/mshome.htm).
Results and discussion
Soluble copper anode for the formation of copper(I) complexes
The probable mechanism for the generation of copper(I) ions by using a sacrificial copper electrode as an anode during the electrospray process has been proposed and discussed previously in reference.19 Briefly, the detachment of Cu+ ions from the copper electrode into the solution is immediately followed by a complexation by proper ligands, which competes with the second oxidation on the electrode to form Cu2+ ions. Indeed, in the absence of suitable ligands, free Cu+ ions are thermodynamically unstable in aqueous solution. In the case of the presence of Aβ16 peptide in the solution, which is known to bind both Cu+ and Cu2+ ions, the formation of Aβ16 to Cu+ ions implies that the complexation reaction has a rate of the same order as that of the oxidation of Cu+ ions.Here, we have considered a simple kinetic model to predict the generation of copper complexes using a sacrificial electrode following the reaction Scheme 1. Using the steady-state approximation, we assume that the rate of production of Cu+ ions is equal to the combined rate of the complexation and oxidation of Cu+ ions. We also neglect the reverse reactions of the complexation of both Cu+ and Cu2+ ions. Furthermore, it has been reported that the reduction potential of Cu(I)-Aβ16/Cu(II)-Aβ16 is much higher than Cu(I)/Cu(II), and therefore the oxidation of Cu(I)-Aβ16 on the electrode was not considered.22 Considering the steric hindrance of the large Aβ16 peptide, we can anyway assume that the rate of oxidation of Cu(I)-Aβ16 complex on the electrode is very slow. The present kinetic model was used both in the absence and presence of reducing agents in solution. The simulated time evolution for the concentrations of Cu(I)-Aβ16, Cu(II)-Aβ16, Cu(II) and Aβ16 in the electrode reservoir are illustrated in Fig. 1. The details of the mathematical model are given in the ESI.†
![Simulated concentrations of Cu(i)-Aβ16, Cu(ii)-Aβ16, Cu(ii) and Aβ16 (a) in the absence of reducing agent and (b) in the presence of reducing agents. The assignments of the rate constants for the calculation are given as follows: k = 1.6 μM min−1, k1 = k2 = 85 μM−1 min−1, kox = 400 min−1, kred[AA] = 5 min−1.](/image/article/2010/MT/c004693k/c004693k-f1.gif) | ||
| Fig. 1 Simulated concentrations of Cu(I)-Aβ16, Cu(II)-Aβ16, Cu(II) and Aβ16 (a) in the absence of reducing agent and (b) in the presence of reducing agents. The assignments of the rate constants for the calculation are given as follows: k = 1.6 μM min−1, k1 = k2 = 85 μM−1 min−1, kox = 400 min−1, kred[AA] = 5 min−1. | ||
Fig. 1a shows that the concentrations of the complexes of both Cu+ and Cu2+ increased gradually as the electrospray went on until the depletion of Aβ16 according to the simulated curves. The concentration of free Cu2+ ions did not increase until the depletion of Aβ16 in the reservoir. The fact that about 55% ± 10% copper(I) complex was experimentally observed on-line shown below is consistent with the simulated results and corroborate the assumptions made. In this case, the ratio between the rates of the complexation and oxidation of Cu+ is found to be close to 0.5, i.e. about the same order of magnitude.
In the presence of reducing agents in excess, we have neglected the oxidation at the electrode of the reducing agent as here we have used ascorbic acid that can only be electrochemically oxidized at potentials higher than that of the formation of Cu+, and we have also neglected the reduction of bare Cu2+. Fig. 1b shows the time evolution of the different species showing that only Cu(I)-Aβ16 was produced until the depletion of Aβ16 peptide, which is also consistent with the dominant amount of copper(I) complex observed experimentally by mass spectrometry. The model indicates that the fast rate of the reduction of Cu(II)-Aβ16 by the reducing agent plays an important role to ensure a high yield of formation of copper(I) complexes.
All in all, this simple kinetic model accounts well for the trends observed experimentally, and confirms that copper(I) complexes can be formed in solution.
Copper-Aβ16 complexes
Copper-Aβ16 complexes were generated on-line by using a sacrificial copper electrode as anode in ESI-MS. After 10 min of electrospray, the doubly and triply charged complexes of Aβ16 bound with copper ions were observed. As shown in Fig. 2, the complex bound with one copper ion Cu1(Aβ16)1 was observed at m/z = 672.9 Th and m/z = 1009.4 Th, respectively, and the complex bound with two copper ions Cu2(Aβ16)1 was observed at m/z = 694.0 Th and m/z = 1040.3 Th. As the spray time increased, the number of copper ions bound to Aβ16 increased as well. Up to six copper ions were observed to bind to a single Aβ16 peptide. Several studies using Electron paramagnetic resonance (EPR), Circular Dichroism Spectroscopy (CD) and Isothermal Titration Calorimetry (ITC) showed that the Aβ peptide can bind two equivalents of Cu(II) in a sequential way where the first Cu(II) equivalent shows about a 100 times stronger affinity for the Aβ peptide than the second one.23 The observation of Cun(Aβ16)1 may stem from the extremely soft ionization provided by the microchip24 and the formation of copper complexes with the amide backbone of the peptide as in the Biuret reaction or may stem from the formation of the clusters of copper ions generated by the copper sacrificial electrode.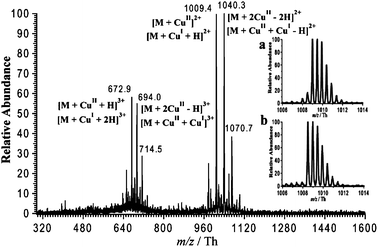 | ||
| Fig. 2 Mass spectrum of Aβ16 (10 μM in 50/50 (vol/vol MeOH/H2O) obtained using a Cu electrode as an anode for 10 min. The insert (a) shows the isotopic distribution of doubly charged copper-Aβ16 complexes generated from copper electrode in the presence of ascorbic acid and the insert (b) shows the isotopic distribution of doubly charged Cu(II)-Aβ16 complex obtained from the addition of a Cu(II) salt as a reference. | ||
Thereafter, we focus on the Cu1(Aβ16)1 complex i.e. that with the highest affinity. Given that both Cu(I) and Cu(II) ions can be produced in solution when using a sacrificial copper electrode,19,20 the isotopic distribution of each charged peak was used to identify the Aβ16 oxidation states of the copper ions involved and their concentrations as detailed previously.19 Shortly, in the present work, Cu(I)-Aβ16 complex displays its isotopic distribution (+2 charged ions are taken as an example) as follows: 1009.0 Th, 1009.5 Th, 1009.9 Th, 1010.4 Th and 1010.9 Th while the isotopic peaks of Cu(II)-Aβ16 complex ranges at 1008.5 Th, 1009.0 Th, 1009.5 Th, 1009.9 Th and 1010.4 Th. In this case, the isotopic distribution of the mixture of Cu(I)- and Cu(II)-Aβ complexes should theoretically be the sum of the isotopic distribution of each Cu(I)- and Cu(II)-Aβ complexes. Therefore, the ratio at which the calculated isotopic peaks of mixture match the experimental best in the whole isotopic distribution range is considered as the component ratio of Cu(I)- and Cu(II)-Aβ complexes. In this way, it was found that 55% ± 10% of the complexes contain Cu(I).19 To favor further the formation of Cu(I) complexes, ascorbic acid solution was added before spraying. As predicted by the kinetic model, the amount of Cu(I) complexes observed on-line increased greatly in the presence of ascorbic acid. As shown in Fig. 2a, the isotopic distribution of the doubly charged copper complex at m/z = 1009.4 Th obtained with a sacrificial copper electrode in the presence of ascorbic acid was quite similar to the theoretical isotopic distribution of a Cu(I) complex (data not shown) but different from that of a Cu(II) complex. In comparison, the peak at m/z = 1009.4 Th obtained by analyzing a solution of a Cu(II) salt mixed with Aβ16 showed an isotopic distribution corresponding to that of a Cu(II) complex (see Fig. 2b), confirming that a great amount of Cu(I) complex observed with a soluble copper anode in the presence of ascorbic acid was generated in solution and not in the gas phase. Furthermore, according to the calculation it was confirmed that 86% ± 4% of Cu(I)-Aβ16 complex was observed by using a soluble copper electrode in the presence of ascorbic acid. It was reported that Cu(I) complex could also be formed by mixing off-line the reducing agent with the Cu(II) complexes25 and we have tested this off-line reduction method using ascorbic acid to reduce Cu(II) complexes before the analysis of mass spectrometry. However, Cu(I) complexes were observed only one or two minutes in the same experimental conditions partially due to the exposition to the air and immediate re-oxidization into Cu(II) complex. However, in the case of using sacrificial copper electrode, a great proportion of Cu(I) complexes was observed and found to be stable for about half an hour. Therefore, the present system provide an easy methodology to investigate Cu(I)-Aβ16, which are otherwise difficult to generate and remain stable in aerobic conditions.25
Cu(I)-Aβ16 complex
To investigate the structure of monometallated Cu(I)-Aβ16 complex, collision-induced dissociation (CID) was applied to generate fragments of Cu(I)-Aβ16 complex to determine the binding site between copper(I) ion and Aβ16. As shown in Fig. 3, the CID spectrum of [M+CuI+H]2+ at 30% of collision energy displayed many product ions. Although there was a little of overlap of [M+CuII]2+ and [M+CuI+H]2+ in this selected m/z region, CID spectrum of [M+CuI+H]2+ displayed distinct fragments from that of [M+CuII]2+ and main fragments of tandem mass spectrum of [M+CuI+H]2+ resulted from its fragmentation. The most intense peak in CID spectrum of [M+CuII]2+ only possess 26% abundance in tandem mass spectrum of [M+CuI+H]2+ because of the high abundance of the parent ion Cu(I)-Aβ16 complex. After subtraction of all fragments shown in the CID spectrum of [M+CuII]2+ from CID spectrum of [M+CuI+H]2+, the fragments resulting from the fragmentation of [M+CuI+H]2+ were confirmed. As shown in Fig. 3, most fragments obtained are conventional b-ions and complexes of b-ions and y-ions bound to Cu+. The b-ions present are b5, b6, b72+, b11, b12, b13 while the b-ions bound to Cu+ are [b13+CuI–H]+, [b14+CuI]2+, [b15+CuI]2+. All the y-ions displayed in CID spectrum are bound to Cu+ such as [y3+CuI–H]+, [y5+CuI–H]+, [y9+CuI–H]+, [y10+CuI–H]+, [y11+CuI–H]+, [y13+CuI]2+, and [y15+CuI]2+. According to these fragments, the binding site of Cu+ to Aβ16 was deduced sequentially. Due to the presence of [y9+CuI–H]+, [y10+CuI–H]+, [y11+CuI–H]+, [y13+CuI]2+, [y15+CuI]2+ and b5, b6, b7, [b14+CuI]2+, [b15+CuI]2+, the binding site was confined to the region between Ser8 and His14. Furthermore, the presence of b11, b12, [y5+CuI–H]+ revealed the binding site of Cu+ as His13-His14. Since histidine residues are common ligands for Cu(I), these results indicated that Cu(I) should be coordinated to two imidazole ligands of both His13 and His14 of the Aβ16 peptide (see Fig. 3 insert), which is quite consistent with structural studies of Cu(I)-Aβ complex in which extended X-ray absorption fine structure (EXAFS) spectroscopy.14,15 The observation of [y5+CuI–H]+, [b13+CuI–H]+ and b12 all together proves that not only His14 but also His13 are involved in the coordination of Cu(I) further. To the best of our knowledge, these results represent the first mass spectrometric evidence that the binding site of Cu(I) to Aβ16 is located as the His13–His14 residues.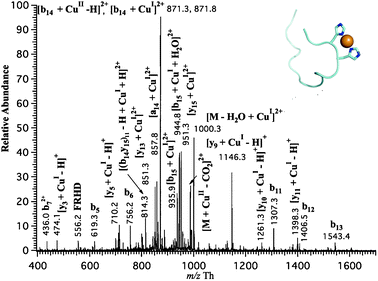 | ||
| Fig. 3 Tandem mass spectrum of copper-Aβ16 complexes obtained with a Cu electrode (t = 10 min), in which the selected parent ion is at m/z = 1009 Th ± 2.5 Th at 30% of collision energy and possible mode of Cu+ coordinating to Aβ16 schematically drawn by software Pymol.26 | ||
In order to verify the binding site of Cu(I) further, comparison of tandem mass spectrometry between [M+CuI+H]2+ and [M+ZnII]2+ was processed because it has been reported that copper and zinc possess very similar coordination environments and the binding structure of Zn(II) to Aβ16 has been well investigated. The CID spectra of [M+CuI+H]2+ and [M+ZnII]2+ obtained in the absence of copper showed the similarity to some extent and only displayed the differences in the intensities and patterns of some peaks between each other (data not shown). As shown in Fig. 4, the diagrams summarized all the fragment assignments stemming from [M+CuI+H]2+ and [M+ZnII]2+ schematically. The complexes of y-ions (y9, y10, y11, y13, y15) and b-ions (b14, b15) bound to Cu+ (or Zn2+) and bare b-ions (b5, b6, b7) indicated that the similar binding sites of Cu+ and Zn2+ were both involved in the region between Ser8 and His14. The coexisting presence of y10, [y10+ZnII–2H]+ only in the CID spectrum of [M+ZnII]2+ indicates His6 is also involved in the coordination to Zn2+ while the coordination of Cu+ is exclusive in the region of Ser8-His14. Moreover, the presence of y3 and [b12+ZnII–2H]+ only in CID spectrum of [M+ZnII]2+ instead of [y3+CuI–H]+, [y5+CuI–H]+, b11 and b12 in CID spectrum of [M+CuI+H]2+ showed essential differences of the binding sites, which indicated that the binding site of Cu+ was just located in His13–His14 while Zn2+ bound more residues besides this region. As discussed above and according to the published literature,14,15 this results directly from the tandem spectrum was consistent to two-coordinate geometry with two imidazole ligands and the possible binding mode of Cu+ to Aβ16 is shown schematically in Fig. 3 insert.
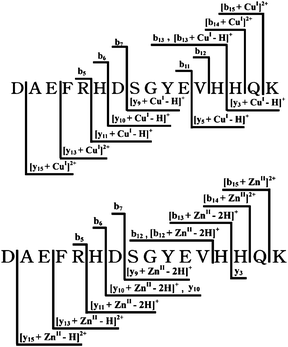 | ||
| Fig. 4 Schematic diagram summarizing the fragments ions assignments for Cu(I)- and Zn(II)-Aβ16 complexes produced by CID. | ||
Cu(II)-Aβ16 complex
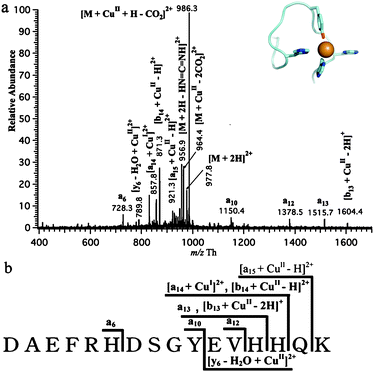
The binding site of Cu(II) to Aβ16 was also investigated by using MS/MS. As shown in Fig. 5a, the most intense peak in CID spectrum of [M+CuII]2+ was observed as [M+CuII–CO2]2+ associated to the loss of a carboxy group, as a result of the presence of Cu(II) ion. The fragment with the loss of 2 CO2 was also demonstrated in the spectrum as well as the fragment [M+2H–HN![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH]2+ with the loss of a part of guanidine moiety from arginine (HN
NH]2+ with the loss of a part of guanidine moiety from arginine (HN![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH, 42 Da) and the fragments b13, b14 and y6, were displayed to bind to Cu(II) as well. In addition, there were uncommon a-ions with a low abundance observed in CID spectrum, such as a6, a10, a12, a13 and [a14+CuI]2+ besides conventional the complexes of b-ions and y-ions.
NH, 42 Da) and the fragments b13, b14 and y6, were displayed to bind to Cu(II) as well. In addition, there were uncommon a-ions with a low abundance observed in CID spectrum, such as a6, a10, a12, a13 and [a14+CuI]2+ besides conventional the complexes of b-ions and y-ions.
 | ||
| Fig. 5 (a) Tandem mass spectrum of Cu(II)-Aβ16 complex obtained with a Pt electrode in the presence of CuCl2 (20 μM), in which the selected parent ion is also at m/z = 1009 Th ± 2.5 Th at 30% of collision energy and possible mode of Cu2+ coordinating to Aβ16 schematically drawn by software Pymol.26 (b) Schematic representation of the observed in CID spectrum. | ||
As shown in Fig. 5b, the diagram summarizes all the fragments displayed in CID spectrum of [M+CuII]2+ schematically. The presence of [b14+CuII–H]2+ and a13 in the spectrum indicated that His14 was linked to the coordination of Cu(II) to Aβ16 and it should be one of the binding residues of Cu(II) to Aβ16. In the same way, due to the presence of [b13+CuII–2H]+ and a13, it is easy to deduce that His13 is another binding residue of Cu(II) to Aβ16. As a result, we can conclude that the two most likely N ligands of the Cu(II) ion are His13 and His14. His13, His14 bind both redox states of the copper ions, meaning that the change of coordination upon redox cycling must involve a structural change of the other ligands. First of all, the present data suggest that the copper ion does not migrate along the peptide chain upon redox cycling, and that His 13 and His 14 represent strong anchoring points. Furthermore, upon oxidation from Cu(I) to Cu(II) the coordination geometry usually changes from a tetrahedral arrangement (coordination number of 4) to a square-planar coordination sphere (coordination number of 4). Upon oxidation or reduction, it is likely that His13 and His 14 remain bound to the copper ions but that the peptide chain must change its conformation to account for the change in coordination.
The highly similar coordination environment shared with Cu(I) and Cu(II) ions to Aβ16 implies a structural rearrangement of some residues of Aβ16 during the redox cycling of Cu(I) and Cu(II) ions on the basis of His13 and His14 bound to Aβ16 and this information will certainly be helpful for the explanation of the production of oxygen radicals in the etiology of AD.
The generation of unusual a-ions in CID spectrum of [M+CuII]2+ might be ascribed to the high oxidation ability of Cu2+ in the gas phase since it has been reported that the generation of a-ions are highly linked to the oxidation environment of the dissociation.19,27 The observation of [a14+CuI]2+ also corroborated that the reduction of Cu2+ occurred during the process of CID. Moreover, CO2 loss from [M+CuII]2+ also represents the crucial feature of electron detachment dissociation,28 which process results in the dissociation of Cα–C and the generation of a-ions. Therefore, the reduction of bound Cu2+ in the gas phase during the process of CID may lead to the transfer of electrons to the backbone of the peptide, which then resulted in Cα–C fragmentation and the generation of a-ions. Also, the fact that as the collision energy increased, the intensity of all a-ions increased subsequently along with the decrease of intensity of [M+CuII–CO2]2+ confirmed that these a-ions did come from the dissociation of [M+CuII–CO2]2+, which also means these a-ions were the result of high oxidation environment of CID of [M+CuII–CO2]2.
Conclusion
Copper-Aβ16 complexes with two oxidation states were generated on-line by using a sacrificial copper electrode as an anode of mass spectrometry and Cu(I)-Aβ16 complex in high abundance was formed especially in the presence of ascorbic acid. A kinetic model was further built and simulated the process of the production of both Cu(I)- and Cu(II)-Aβ16 complexes. The binding sites of Cu+ and Cu2+ to Aβ16 were well studied with the help of tandem mass spectrometry. The binding site of Cu+ was successfully identified as His13, His14 residues, which were further found to be involved in the binding site of Cu2+ to Aβ16. The same coordination environment shared with both Cu(I) and Cu(II) to Aβ16 implies the conformational change of the peptide chains on the redox cycling of copper ions bound to Aβ16.Acknowledgements
The authors thank the China Scholarship Council (CSC) for financial support. Dr Xiaofeng Liu is acknowledged for the producing of the insets of Fig. 3 and 5(a) with software Pymol. The authors also thank Swiss National Science Fundation (SNCF) for the funding for the project of Analytical tools for proteome analysis and redoxomics (grant no 200020_127142/1).Notes and references
- H. A. Lashuel, S. R. LaBrenz, L. Woo, L. C. Serpell and J. W. Kelly, J. Am. Chem. Soc., 2000, 122, 5262–5277 CrossRef CAS.
- H. A. Lashuel, D. Hartley, B. M. Petre, T. Walz and P. T. Lansbury, Nature, 2002, 418, 291–291 CrossRef CAS.
- P. T. Lansbury and H. A. Lashuel, Nature, 2006, 443, 774–779 CrossRef CAS.
- K. Takano, Curr. Alzheimer Res., 2008, 5, 540–547 CrossRef CAS.
- A. I. Bush, Curr. Opin. Chem. Biol., 2000, 4, 184–191 CrossRef CAS.
- A. I. Bush, Trends Neurosci., 2003, 26, 207–214 CrossRef CAS.
- E. Gaggelli, H. Kozlowski, D. Valensin and G. Valensin, Chem. Rev., 2006, 106, 1995–2044 CrossRef CAS.
- D. G. Smith, R. Cappai and K. J. Barnham, Biochim. Biophys. Acta, Biomembr., 2007, 1768, 1976–1990 CrossRef CAS.
- L. Guilloreau, S. Combalbert, A. Sournia-Saquet, H. Mazarguil and P. Faller, ChemBioChem, 2007, 8, 1317–1325 CrossRef CAS.
- M. Brzyska, K. Trzesniewska, A. Wieckowska, A. Szczepankiewicz and D. Elbaum, ChemBioChem, 2009, 10, 1045–1055 CrossRef CAS.
- P. Zatta, D. Drago, S. Bolognin and S. L. Sensi, Trends Pharmacol. Sci., 2009, 30, 346–355 CrossRef CAS.
- G. Meloni, V. Sonois, T. Delaine, L. Guilloreau, A. Gillet, J. Teissie, P. Faller and M. Vasak, Nat. Chem. Biol., 2008, 4, 366–372 CrossRef CAS.
- P. Faller and C. Hureau, Dalton Trans., 2009, 1080–1094 RSC.
- R. A. Himes, G. Y. Park, G. S. Siluvai, N. J. Blackburn and K. D. Karlin, Angew. Chem., Int. Ed., 2008, 47, 9084–9087 CrossRef CAS.
- J. Shearer and V. A. Szalai, J. Am. Chem. Soc., 2008, 130, 17826–17835 CrossRef CAS.
- G. Drochioiu, M. Manea, M. Dragusanu, M. Murariu, E. S. Dragan, B. A. Petre, G. Mezo and M. Przybylski, Biophys. Chem., 2009, 144, 9–20 CrossRef CAS.
- M. Prudent and H. H. Girault, Analyst, 2009, 134, 2189–2203 RSC.
- U. A. Mirza and B. T. Chait, Anal. Chem., 1994, 66, 2898–2904 CrossRef CAS.
- M. Prudent and H. H. Girault, J. Am. Soc. Mass Spectrom., 2008, 19, 560–568 CrossRef CAS.
- M. Prudent, C. Roussel and H. H. Girault, Electrochem. Commun., 2007, 9, 2067–2074 CrossRef CAS.
- M. Prudent and H. H. Girault, Metallomics, 2009, 1, 157–165 RSC.
- V. A. Streltsov and J. N. Varghese, Chem. Commun., 2008, 3169–3171 RSC.
- C. S. Atwood, R. C. Scarpa, X. D. Huang, R. D. Moir, W. D. Jones, D. P. Fairlie, R. E. Tanzi and A. I. Bush, J. Neurochem., 2000, 75, 1219–1233 CrossRef CAS.
- L. Deng, N. Sun, E. N. Kitova and J. S. Klassen, Anal. Chem., 2010, 82, 2170–2174 CrossRef CAS.
- N. C. Maiti, D. L. Jiang, A. J. Wain, S. Patel, K. L. Dinh and F. M. Zhou, J. Phys. Chem. B, 2008, 112, 8406–8411 CrossRef CAS.
- http://www.pymol.org/ .
- L. Qiao, H. Y. Bi, J. M. Busnel, J. Waser, P. Y. Yang, H. H. Girault and B. H. Liu, Chem.–Eur. J., 2009, 15, 6711–6717 CrossRef CAS.
- F. Kjeldsen, O. A. Silivra, I. A. Ivonin, K. F. Haselmann, M. Gorshkov and R. A. Zubarev, Chem.–Eur. J., 2005, 11, 1803–1812 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Details of the kinetic model and the whole simulation. See DOI: 10.1039/c004693k |
| This journal is © The Royal Society of Chemistry 2010 |
