Nicotianamine forms complexes with Zn(II)in vivo†
Aleksandra
Trampczynska
a,
Hendrik
Küpper
bc,
Wolfram
Meyer-Klaucke
d,
Holger
Schmidt
a and
Stephan
Clemens
*a
aUniversität Bayreuth, Lehrstuhl Pflanzenphysiologie, D-95440 Bayreuth, Germany. E-mail: stephan.clemens@uni-bayreuth.de; Fax: +49-921-552642; Tel: +49-921-552630
bUniversität Konstanz, Mathematisch-Naturwissenschaftliche Sektion, Fachbereich Biologie, D-78457 Konstanz, Germany
cUniversity of South Bohemia, Biological Faculty, CZ-370 05 České Budějovice, Czech Republic
dEMBL Outstation Hamburg, c/o DESY, D-22603 Hamburg, Germany
First published on 16th October 2009
Abstract
The non-proteinogenic amino acid nicotianamine (NA) is a major player in plant metal homeostasis. It is known to form complexes with different transition metalsin vitro. Available evidence associates NA with translocation of Fe, and possibly other micronutrients, to and between different plant cells and tissues. To date, however, it is still extremely challenging to detect metal–ligand complexes in vivo because tissue disruption immediately changes the chemical environment and thereby the availability of binding partners. In order to overcome this limitation we used various Schizosaccharomyces pombe strains expressing a plant NASgene to study formation of metal–NA complexes in vivo. Tolerance, accumulation and competition data clearly indicated formation of Zn(II)–NA but not of Cu(II)–NA complexes. Zn(II)–NA was then identified by X-ray absorption spectroscopy (XAS). About half of the cellular Zn was found to be bound by NA in NAS-expressing cells while no NA-like ligands were detected by XAS in control cells not expressing NAS. Given the high conservation of eukaryotic metal homeostasis components, these results strongly suggest the possible existence of Zn(II)–NA complexes also in planta. Reported observations implicating NA in plant Zn homeostasis would then indeed be attributable to direct interaction of Zn(II) with NA rather than only indirectly to perturbations in Fe metabolism. Re-evaluation of extended X-ray absorption fine structure (EXAFS) spectra for the Zn hyperaccumulator Thlaspi caerulescens showed that NA is as expected not a major storage ligand for Zn. Instead it is hypothesized to be involved in efficient translocation of Zn to above-ground tissues in hyperaccumulators.
Introduction
Delivery of essential transition metals such as Cu, Zn, and Fe to the countless target sites inside a cell requires intracellular trafficking. In multicellular organisms there is, in addition, the need for cell-to-cell and long distance transport. Stable metal–chelator complexes are essential to suppress uncontrolled binding of metal ions to proteins and other biological molecules, for instance in plants during symplastic passage and during transport via xylem and phloem.1,2Candidate low-molecular weight metal chelators in plants are nicotianamine (NA), a non-proteinogenic amino acid synthesized through condensation of three molecules of S-adenosylmethionine by nicotianamine synthases (NAS),3glutathione (GSH) and phytochelatins (PCs), glutathione-derived metal-binding peptides,4 and amino acids such as histidine.5 Organic acids have also repeatedly been discussed as binding partners for transition metals.6 However, metal–citrate or metal–malate complexes display low stability constants and are therefore likely to act as binding partners only in compartments such as vacuoles and the xylem which contain fewer competing molecules.7,8
NA binds several transition metals with very high affinity. Logarithms of the stability constants are 18.8 for Cu(II), 16.1 for Ni(II), 14.8 for Co(II), 14.7 for Zn(II), 12.1 for Fe(II), and 8.8 for Mn(II).9 Because the stability of complexes with most essential transition metals is much higher at pH values above 6.5,10NA is considered a symplastic chelator.11 The exceptions are Cu(II)–NA complexes which are also stable in an acidic environment down to pH 3. Numerous observations implicate NA in Fe and Cu homeostasis in plants.8 More indirect evidence suggests a role in Zn and Mn homeostasis as well. NA deficiency causes intercostal chlorosis especially in young leaves of the tomato mutant chloronerva,3,12 indicating defective Fe transport and possibly Zn transport to developing tissues. While long-distance transport of Fe is not affected in chloronerva, Cu content of shoots is strongly reduced,13 consistent with complexation of Cu(II) with NA in the acidic xylem sap as also suggested by a study of copper complexes in the xylem sap of Brassica carinata.14 In transgenic tobacco plants, made NA deficient through the overexpression of an NAaminotransferase, Fe, Zn, Mn, and Cu contents of leaves are reduced. An A. thaliana NAS quadruple mutant develops severe symptoms of Fe deficiency15 and shows reduced levels of Zn in aboveground tissues. NAS overexpression in tobacco results in 2.5 fold higher Zn levels and 1.9 fold higher Fe levels in young leaves.16 Other studies with NAS overexpressing tobacco and A. thaliana plants reported elevated Ni2+ tolerance as the major phenotype .17,18 Seeds of transgenic tobacco grown in a serpentine soil again contained more Fe, Zn, Cu, and Mn.19
NA has also been implicated in metal hyperaccumulation in plants. Certain plant species are capable of accumulating metals such as Zn in leaves up to a level of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ppm, which is >100 fold more than in the tissues of non-hyperaccumulating plants.20 The two most widely studied hyperaccumulation model systems are Thlaspi caerulescens and Arabidopsis halleri. In extracts of the former, Ni(II)–NA complexes were detected by size exclusion chromatography coupled to ICP-MS.21 Comparative transcriptome studies with A. thaliana and A. halleri found that total NAS message and protein levels are strongly elevated in A. halleri roots and shoots relative to A. thaliana.22,23 Similar observations were made for T. caerulescens.24 Thus, elevated NAS expression is part of the “hyperaccumulation syndrome”, i.e. the constitutive strong expression of metal homeostasisgenes.25 The expression difference in A. halleri roots resulted in constitutively higher NA levels. Formation of NA in S. pombe cells rescued Zn2+hypersensitivity of a mutant strain unable to store excess Zn in the endoplasmic reticulum.23 Based on these observations and the above-mentioned phenotypes of transgenic plants with reduced and elevated levels of NA16 it was proposed that (i) NA plays a role in Zn homeostasis as an intracellular Zn chelator and that (ii) higher NA levels contribute to Zn hypertolerance and hyperaccumulation in A. halleri and T. caerulescens.
000 ppm, which is >100 fold more than in the tissues of non-hyperaccumulating plants.20 The two most widely studied hyperaccumulation model systems are Thlaspi caerulescens and Arabidopsis halleri. In extracts of the former, Ni(II)–NA complexes were detected by size exclusion chromatography coupled to ICP-MS.21 Comparative transcriptome studies with A. thaliana and A. halleri found that total NAS message and protein levels are strongly elevated in A. halleri roots and shoots relative to A. thaliana.22,23 Similar observations were made for T. caerulescens.24 Thus, elevated NAS expression is part of the “hyperaccumulation syndrome”, i.e. the constitutive strong expression of metal homeostasisgenes.25 The expression difference in A. halleri roots resulted in constitutively higher NA levels. Formation of NA in S. pombe cells rescued Zn2+hypersensitivity of a mutant strain unable to store excess Zn in the endoplasmic reticulum.23 Based on these observations and the above-mentioned phenotypes of transgenic plants with reduced and elevated levels of NA16 it was proposed that (i) NA plays a role in Zn homeostasis as an intracellular Zn chelator and that (ii) higher NA levels contribute to Zn hypertolerance and hyperaccumulation in A. halleri and T. caerulescens.
Until recently, NA had been considered a plant-specific metabolite. However, filamentous fungi such as Neurospora crassa, Magnaporthe grisea and Podospora anserina carry putative NASgenes as apparent from their genome sequences. For N. crassa it was indeed demonstrated that it synthesizes NA and expresses a functional NASgene which is massively up-regulated under conditions of Zn deficiency.26
Most of the available evidence for the functions of NA in metal homeostasis is indirect. Detection and quantification of metal–ligand complexes in vivo, i.e.metal ion speciation analysis, is still extremely difficult because tissue disruption immediately changes the chemical environment and thereby the availability of binding partners.8 Non invasive techniques, such as X-ray absorption spectroscopy (XAS), have been applied to trace element accumulations in plants.27 Here the weak association of metal ions with organic acids in vacuoles, and not binding to strong ligands, was found to be the main storage form of metals and the metalloid arsenic.28–34 Recently, the focus of the studies has been expanded to cell compartments.35 Here we analyze S. pombe cells, in which we modulated metal ligand production to learn more about the in vivo formation of metal–NA complexes. A plant NASgene was expressed in various mutant strains. The question of Zn(II)–NA complex formation was addressed by subjecting such S. pombe cells to analysis by X-ray absorption spectroscopy (EXAFS and XANES) in a frozen-hydrated state, thus avoiding artefactual Zn(II)–NA complex formation during sample preparation. Further, by analyzing previously measured32 Zn–EXAFSspectra of T. caerulescens we tested whether, in hyperaccumulators, NA binds a major proportion of the accumulated Zn, i.e. is involved in Zn storage, or rather acts as a transport ligand, in which case only a small proportion of the total Zn would be bound to NA.
Results and discussion
Yeast growth, metal uptake and nicotianamine synthesis
Expression of AtNAS2 and NcNAS in the Zn(II) hypersensitive mutant zhfΔ, which lacks efficient Zn sequestration,36 has already shown an NA-dependent increase in Zn(II) tolerance.23,26 In control medium AtNAS2-expressing cells grew like wild type cells indicating that NA synthesis does not cause Zn deficiency. We extended the analysis of tolerance effects to other transition metals and included Cd(II) and Cu(II) hypersensitive pcsΔ cells37 as well as wild type cells. AtNAS2 expression partially rescued Co(II)hypersensitivity of zhfΔ cells. NA formation led to an increase in growth from 28.1% (± 2.0) to 40.5% (± 5.2) of control level (Fig. 1a). Also, it conferred a significant increase in Ni(II) tolerance to S. pombe wild type and pcsΔ cells (29.3% ± 11.6% growth inhibition instead of 60.0% ± 4.8% in pcsΔ cells)(Fig. 1b). Surprisingly, the opposite effect was seen for Cd(II) (1 μM). The pcsΔ strain became even more Cd(II) sensitive through formation of NA in the cytosol (76.1% ± 4.2% growth inhibition for pcsΔ, 33.9% ± 11.4% growth inhibition for empty vector control cells) (Fig. 1c). This effect was independent of the formation of phytochelatins. S. pombe hmt1Δ cells still form low molecular weight complexes38 yet are equally Cd(II) sensitive.39 AtNAS2 expression resulted in an increase in Cd(II)hypersensitivity of hmt1Δ cells that was comparable to the effect observed in pcsΔ cells (data not shown). The mild Cu(II)hypersensitivity of pcsΔ and hmt1Δ cells on the other hand was not affected by NA formation. Growth inhibition was not significantly different in AtNAS2 expressing and empty vector cells (shown in Fig. 1d for hmt1Δ).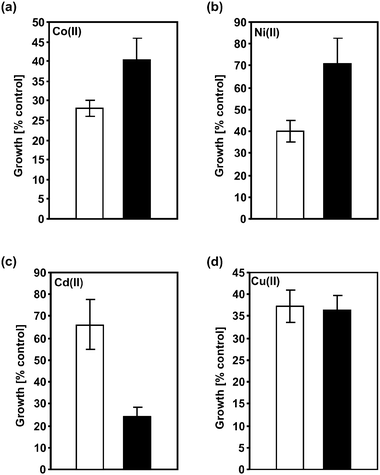 | ||
| Fig. 1 NA formation differentially affects transition metal sensitivity of S. pombe cells. AtNAS2 was expressed in various S. pombe wild type and mutant strains. Empty vector control cells (white bars) and AtNAS2-expressing cells (black bars) were cultivated in the presence of growth-inhibitory concentrations of different transition metal ions. After 20 to 24 h OD600 was determined. (a) Co(II) hypersensitive zhfΔ cells grown in the presence of 750 μM CoCl2; (b) S. pombe wild type cells in medium containing 150 μM NiCl2; (c) Cd(II) hypersensitive pcsΔ cells grown in the presence of 1 μM CdCl2; (d) hmt1Δ cells cultivated in medium with elevated CuCl2 concentration (20 μM). Error bars represent S.D., n = 4–7. | ||
These metal tolerance assays with cells heterologously synthesizing NA indicated cytoplasmic in vivo complex formation of NA with Ni(II) and Co(II) in addition to Zn(II), but not with Cu(II) even though the latter is by far the best binding partner among divalent transition metal ions for NAin vitro. This is interpreted as a confirmation of the very high affinity of dedicated Cu trafficking and sequestration pathways that are effectively suppressing the existence of hydrated Cu ions in the cytosol.40 Thus, NA is unlikely to interact with Cu ions in the cytoplasm. The essentiality of NA for copper metabolism as demonstrated in plants (e.g.ref. 3 and 13) may be based on copper transport by NA in the xylem.14 Conversely, the results for ZnCl2-, NiCl2- and CoCl2-treated cells suggest the presence of a non-negligible fraction of cytoplasmic Zn(II), Ni(II) and Co(II) available for chelation by NA. Cd(II)hypersensitivity of NA-producing cells was surprising. Possibly, NA makes intracellular binding sites available for Cd(II). Interaction with Cd(II) then causes damage. To date we have no indication as to the nature of these binding sites. Phytochelatins as the main Cd(II)chelators did not interfere with this process. The phenotype was also observed in hmt1Δ cells which still produce phytochelatins.
Metal–NA interaction was investigated further by testing interference of other metals with the suspected Zn(II)–NA complex formation in competition experiments. zhfΔ cells were exposed to growth-inhibiting Zn(II) concentrations plus an excess of other metal ions. Concentrations of NiCl2, CoCl2, CdCl2, CuCl2, and MnCl2 were chosen so that they did not affect the growth of zhfΔ cells when applied at normal Zn(II) levels. Assays showed that in the presence of non-inhibitory NiCl2 (200 μM) and CoCl2 (250 μM) concentrations, the protective effect of AtNAS2 expression against ZnCl2 toxicity (150 μM) was diminished while CdCl2 addition (10 μM) had no effect and extra CuCl2 (2.5 μM) as well as extra MnCl2 (400 μM) slightly enhanced growth under excess Zn2+ conditions (Fig. 2). These effects were also observed in drop assays on agar plates under analogous conditions (Fig. 2). Thus, Co(II) and Ni(II) appeared to be able to out-compete Zn(II) for NA binding. Fe was not studied because it was not possible to establish stable conditions of Fe overload in S. pombe cultures.
 | ||
| Fig. 2 Transition metals differentially interfere with the protective effect of NA formation against Zn(II) toxicity. zhfΔ cells expressing AtNAS2 were grown either in liquid culture (shown on the left) or on plates in drop assays (shown on the right) in the presence of an excess of ZnCl2 (150 μM) (white bars), in 150 μM ZnCl2 plus a non-inhibitory excess of another transition metal ion (black bars) or in medium containing only the tolerable excess of another transition metal ion (grey bars). Growth is shown in % of growth in normal medium. Empty vector control cells in medium containing 150 μM ZnCl2 showed a growth reduction to about 23% of growth in normal medium while AtNAS2-expressing cells reached a level of about 60%. (a) NiCl2 (200 μM); (b) CdCl2 (10 μM); (c) CoCl2 (250 μM); (d) CuCl2 (2.5 μM); (e) MnCl2 (400 μM). Error bars represent S.D., n = 3. Respective conditions on plates were 150 μM ZnCl2 (+Zn(II)), 225 μM NiCl2 (a), 225 μM CoCl2 (b), 2.5 μM CdCl2 (c), 2.5 μM CuCl2 (d), and 250 μM MnCl2 (e). The concentrations of metal ions other than Zn(II) did not affect the growth of zhfΔ cells as apparent from a comparison of the respective metal excess with control plates which contained only regular micronutrient concentrations (please note: for clarity the control plate is shown in (a) through (e)). zhfΔ cells expressing AtNAS2 were grown to an OD600 of 1. Serial dilutions were spotted onto EMM plates. From top to bottom the ODs were 1.0, 0.2, 0.04, 0.008, 0.0016. | ||
Overall, competition experiments yielded results that fit the tolerance data and—with the discussed exception of Cu(II)—are in agreement with the in vitro stability constants for NA–metal complexes.9Ni(II) and Co(II) excess reduced the protective effect of NA formation against Zn toxicity. Their binding constants are higher or very similar to those for Zn(II), respectively. Mn(II)–NA complexes on the other hand are much less stable than Zn(II)–NA complexes. Correspondingly, MnCl2 excess did not interfere with AtNAS2-dependent Zn(II) tolerance (Fig. 2) and NAS expression did not confer any protection against MnCl2 toxicity. Along these lines, the lack of interference by Cd(II) would suggest lower stability of Cd(II)–NA complexes as compared to Zn(II)–NA complexes. Thus, these phenotypic observations indicate the suitability of ligand synthesis modulation in a heterologous system to obtain knowledge on the formation of metal–ligand complexes in a cellular environment.
Next we analyzed the effect of NA synthesis on elemental profiles of zhfΔ cells. AtNAS2 expressing cells as well as control cells were transferred either to normal medium or to medium containing an excess of ZnCl2, CuCl2, NiCl2, FeCl2, MnCl2, CdCl2, or CoCl2. After different incubation times cells were harvested and subjected to elemental profiling by ICP-OES. AtNAS2-expressing cells contained significantly more Zn than cells carrying the empty vector both under control and ZnCl2 excess (50 μM) conditions. For instance, after 1 h of incubation in control medium empty vector cells contained 106.9 ± 10.1 μg/g Zn and AtNAS2-expressing cells 185.3 ± 18.4 μg/g Zn (n = 5). Fe levels were also found to be slightly elevated (33.5 ± 7.8 μg/g vs. 52.6 ± 9.8 μg/g, n = 5) while no other metal showed significant AtNAS2-dependent changes in control medium (data not shown). Under all tested metal excess conditions Zn accumulation was significantly higher in NA-producing cells whereas no other metal was accumulated more strongly in AtNAS2-expressing cells when present in the medium at elevated levels (data not shown).
The observed effect of NA formation on Zn accumulation was investigated more closely in time-course experiments. Cells were first grown in the presence of thiamine which suppresses the nmt1 promoter of pSGP72. AtNAS2transcription was then triggered by transfer into thiamine-free medium. After different incubation times, cell aliquots were taken and incubated in high Zn medium for 1 h. Elemental profiles were determined by ICP-OES. Also, AtNAS2 protein levels and NA concentrations were monitored (Fig. 3). AtNAS2 protein and NA were clearly detectable 6 h after transfer. This was paralleled by an increase in the accumulation of Zn. AtNAS2-expressing cells taken from the culture 6 h after activating transcription accumulated about 30% more Zn than control cells. Maximum AtNAS2-dependent Zn accumulation was observed after 12 h when AtNAS2-expressing cells consistently accumulated more than twice as much as control cells (please note that in Fig. 3 data for one representative experiment are shown; the exact onset of NAS expression and NA production can vary). Taken together, the experiments with NA-producing S. pombe cells suggested intracellularZn(II)–NA formation. In order to obtain direct evidence for such complexes in vivo, we subjected AtNAS2-expressing zhfΔ cells to EXAFS analysis.
 | ||
| Fig. 3 Time-course analysis of NAS expression, NA formation and Zn accumulation. zhfΔ cells carrying the empty vector or pSGP72-AtNAS2 were transferred from thiamine-containing medium (1 μM) to thiamine-free medium to de-repress the nmt1 promoter and activate AtNAS2transcription. Following transfer AtNAS2-HA protein level, NA concentration and Zn accumulation were monitored for 12 h. Aliquots were taken in 3 h intervals. (a) Western analysis using a monoclonal anti-HA antibody. Results are shown for zhfΔ cells carrying pSGP72-AtNAS2. Amido black staining as loading control is shown in the bottom panel. (b) Quantification of NA in zhfΔ cells carrying pSGP72-AtNAS2. Please note: no NA was detectable in zhfΔ cells carrying the empty vector. (c) Zn accumulation of zhfΔ cells carrying empty vector (white bars) or pSGP72-AtNAS2 (black bars). Aliquots were taken at the indicated time points and cultivated for 1 h in the presence of 400 μM ZnCl2. The results of a typical experiment are shown. Three biological replicates were performed with comparable results. | ||
EXAFS measurements
Quantification of the elemental composition of an organism (=ionomics) has been well-established in recent years and has already led to important insights into the physiology of plants.27 A major bottleneck in unravelling metal homeostasis mechanisms, however, is our lack of knowledge about in vivo metal–ligand complexes. Particularly suited for speciation analysis are synchrotron radiation techniques such as EXAFS and XANES.41 They can evaluate the ligand environment without destroying intracellular compartmentation.35 When frozen-hydrated rather than dried or fixed samples are analyzed under suitable measuring conditions, artefacts of element re-complexation are minimal.32EXAFS and XANES have been successfully applied to the in vivo characterization of ligand environments of accumulated metals or metalloids in hyperaccumulator plants.29,32,42–45 Such analyses become difficult, however, when complexes are of low abundance in a complicated biological matrix, because they require high metal concentrations and ideally dominance of one ligand in a complex matrix. Ligands binding only a few percent of the total metal cannot be determined. This is most likely the case for metals bound to low-molecular weight chelators such as NA that are present only in submicromolar to micromolar concentrations in plant cells.15 In T. caerulescens, for instance, NA concentrations are around 0.5 mM in leaf dry weight according to recent data of Callahan et al.46 (corrigendum), i.e. around 50–100 μM in fresh weight. This is not much compared to the enormous Zn concentrations in T. caerulescens or A. halleri leaves, which are in the millimolar range. In order to overcome this limitation and to be able to study the formation of metal–NA complexes in a meaningful matrix, i.e. in the cytosol of a biological system, and at sufficiently high concentrations, we exploited NAS expression in a heterologous system. This approach provided conditions suitable at least for Zn. It was not possible to load S. pombe cells adequately with Cu or Fe. Thus, the synchrotron-based analysis was restricted to ZnCl2-treated cells.First, several Zn model complexes were measured as “fingerprints” of the most important potential types of ligands in fission yeast to facilitate the evaluation of the in vivoEXAFS data. Some of these model compounds were described by Küpper et al.,32 reduced Zn(II)–glutathione (Zn(II)–GSH) and Zn(II)–NA were added. The measurements showed that these ligands yield clearly distinguishable XAS signals (see ESI File S1† ). Especially the white line intensity and position of maxima and minima were different for reduced Zn(II)–glutathione compared to all other models, reflecting the change in coordination number, ligand type and individual bond lengths. The differences between Zn(II)–NA and the other ligands were, however, much more visible and quantifiable (see below) in the k3-weighted EXAFS (Fig. 4b) than in the XAS data (Fig. 4a), because the main difference to other oxygen ligands was caused by the backscattering of the C of the carboxyl groups of NA, visible as the peak at 2.8 ± 0.1 Å in the Fourier transform of the EXAFS (Fig. 4c). The visibility of this feature in NA, but not in smaller organic acid ligands like citrate, shows that the binding of Zn(II) makes the NA molecule rather rigid. This is in line with the high Zn affinity of NA, which is much higher than for Zn(II)–GSH. For Zn(II)–GSH, no scattering contribution of the carboxyl carbon was found, indicating a less rigid structure of the complex. Therefore, the EXAFS feature at 2.8 ± 0.1 Å could be used for distinguishing between NA and other ligands, incl. other organic acids.
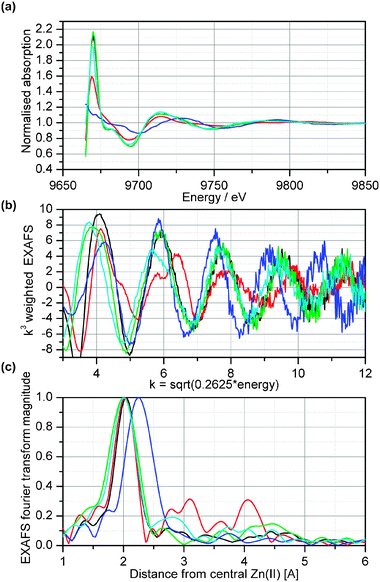 | ||
| Fig. 4 Comparison of Zn-XAS of different model compounds. The data for Zn(II)–NA and Zn(II)–GSH are from the current project, the other model compound spectra were recorded by Küpper et al.32 Black: Zn(II) with oxygen ligands (average of aqueous, citrate and malate model); red: Zn(II)–histidine; green:Zn(II) bound to oxygen of oxidized glutathione, blue: Zn(II) bound to thiol group of reduced glutathione, cyan: Zn(II)–NA. (a) Raw data after normalisation. Note that X-ray absorbance was measured as an excitation spectrum of X-ray fluorescence. (b) Normalised EXAFS. (c) Fourier transform of the EXAFS. | ||
In all samples of S. pombe cells grown on ZnCl2, the metal concentration was high enough (see Table 1) to obtain EXAFS data with a good signal to noise ratio, as shown in Fig. 5. Inspection of the XASspectra already showed that Zn was bound in a very different way in the AtNAS2 overexpressing cells as compared to the empty vector controls (Fig. 5). Fitting the measured EXAFSspectra of the S. pombe samples with a linear combination of EXAFSspectra of the model compounds revealed that in the vector control, as expected, NA (or similar ligands) did not contribute any significant proportion to the zinc ligation. In contrast, in the AtNAS2 overexpressing cells more than half of the total zinc was bound by NA (Fig. 6). These proportions were almost identical in cells treated with 100 or 400 μM ZnCl2, leading to the very small standard errors in the average of both treatments shown in Fig. 6. Fitting the XANES instead of the EXAFS region yielded the same tendencies in ligand binding, i.e. an increase of NA ligation of Zn in the AtNAS2 overexpressing cells (not shown). However, since the main difference between the XAS of NA compared to other oxygen ligands is the feature at 2.8 ± 0.1 Å (see above), which hardly influences the XANES region, the standard errors of the XANES fits were much larger than those of the EXAFS fits. The ligand proportions were also confirmed by the refinement of the EXAFSspectra, i.e. by fitting them with a theoretical model (Table 2, Fig. 6C, ESI† ). This refinement additionally yielded information about the ligand distances. A very good agreement was found between the distances of the ligands in the model compounds and the S. pombe cells. This did not only apply to the oxygen and sulfur ligands in the first shell but also to the carbon of the NAcarboxyl groups in the multiple scattering (Table 2). Furthermore, hardly any deviation in any of the parameters (ligand numbers, distances, Debye–Waller factors and Fermi energy) was found between the S. pombe cells treated with 100 or 400 μM ZnCl2, respectively, demonstrating the reliability of the results (Table 2).
 | ||
| Fig. 5 Examples of data obtained by in situ X-ray absorbance measurements (Zn-edge) of frozen-hydrated S. pombe cells. (a) Raw data after normalization and fit with datasets of model compounds (see Table 2). Note that X-ray absorbance was measured as an excitation spectrum of X-ray fluorescence. Dark green: zhfΔ-pSGP72 with 400 μM ZnCl2; light green: zhfΔ-pSGP72 with 100 μM ZnCl2; red: zhfΔ-AtNAS2 with 400 μM ZnCl2; dotted red: refinement of zhfΔ-AtNAS2 with 400 μM ZnCl2; orange: zhfΔ-AtNAS2 with 100 μM ZnCl2. (b) Normalized EXAFS and fit with theoretical model. (c) Fourier transform of the EXAFS and fits with theoretical model (done on normalised EXAFS) as well as fit with datasets of model compounds. | ||
 | ||
| Fig. 6 Proportions of ligands detected in S. pombe cells as determined by fitting EXAFSspectra of ZnCl2-treated cells with a linear combination of EXAFSspectra of model compounds. The data represent average and standard error of experiments with cells treated with 100 and 400 μM ZnCl2. | ||
| Sample | Ligand atom: number of ligands (±se) | Distance (± se) [Å] | DW (± se) | EF (± se) [eV] | FI |
|---|---|---|---|---|---|
| Zn(II)–NA | O: 6.3 (0.3) | 2.093 (0.003) | 0.014 (0.001) | −7.8 (0.5) | 0.322 |
| C: 3.5 (0.9) | 2.917 (0.013) | 0.013 (0.005) | |||
| Zn(II)–GSH (reduced) | O: 1.0 (0.5) | 2.058 (0.039) | 0.018 (0.012) | −9.8 (1.0) | 0.160 |
| S: 3.2 (0.3) | 2.338 (0.004) | 0.008 (0.001) | |||
| Vector control, | O: 3.5 (0.3) | 1.983 (0.008) | 0.017 (0.002) | −5.9 (0.7) | 0.246 |
| 100 μM ZnCl2 | S: 0.9 (0.2) | 2.309 (0.006) | 0.007 (0.003) | ||
| C: 0.5 (0.3) | 2.991 (0.050) | 0.002 (0.002) | |||
| Vector control, | O: 3.5 (0.3) | 1.985 (0.010) | 0.019 (0.002) | −5.5 (0.7) | 0.239 |
| 400 μM ZnCl2 | S: 1.2 (0.3) | 2.306 (0.006) | 0.008 (0.003) | ||
| C: 0.5 (0.2) | 2.986 (0.031) | 0.002 (0.003) | |||
| AtNAS2 expressing cells, | O: 5.0 (0.2) | 2.084 (0.010) | 0.025 (fixed) | −7.5 (0.8) | 0.305 |
| 100 μM ZnCl2 | S: 0.8 (0.2) | 2.310 (0.006) | 0.006 (0.003) | ||
| C: 1.8 (0.5) | 2.906 (0.014) | 0.006 (0.005) | |||
| AtNAS2 expressing cells, | O: 5.3 (0.2) | 2.074 (0.008) | 0.025 (fixed) | −7.6 (0.7) | 0.278 |
| 400 μM ZnCl2 | S: 0.5 (0.2) | 2.305 (0.007) | 0.003 (0.004) | ||
| C: 2.1 (0.6) | 2.921 (0.012) | 0.007 (0.005) |
In the NAS overexpressing cells, the EXAFSspectra could be very well explained by the fits with a linear combination of the model compound spectra. In contrast, the fits of the EXAFSspectra of the empty vector controls yielded rather large residuals. This indicates that in these cells further ligands, rather different from our model compounds, contribute to Zn(II) complexation. These could be proteins with structural Zn(II)-binding sites where mainly cysteines ligate the metal ion.
Time-course experiments (Fig. 3) demonstrated a clear positive correlation between NA levels and Zn accumulation. The size of the fraction of NA-chelated Zn(II) as determined by EXAFS analysis was in good agreement with the accumulation data. Zn levels were up to 2 fold higher in NA-producing cells. About half of the cellular Zn was chelated by NA in AtNAS2-expressing cells (Fig. 6), while almost no chelation by NA-like ligands was found in the vector controls, which also showed that yeast cells do not contain any other ligands that could produce a Zn(II)–NA like EXAFSspectrum. Thus, EXAFS data unequivocally showed that a major proportion of cellular Zn was indeed chelated by NA. Like in the model compound Zn(II)–NA in aqueous solution, the cellular EXAFSspectra showed the typical multiple scattering pattern of NA originating from the C atom of the carboxyl groups. This shows that both in vitro and in vivoNA strongly binds Zn with participation of several of its carboxyl groups.
Analysis of Thlaspi caerulescens samples
The results for AtNAS2-expressing S. pombe cells prompted us to revisit the question as to whether NA contributes to Zn storage in Zn hyperaccumulators. Fitting of the previously published Zn-EXAFS spectra of T. caerulescens leaves grown under different Cd/Zn regimes32 with this new set of model compounds did not show any significant amount of NA binding to Zn (data not shown). Instead, it was confirmed that most of the Zn in T. caerulescens is stored in association with weak oxygen ligands such as organic acids, as they are commonly found in plant vacuoles.Biological interpretation
Most components of eukaryotic metal homeostasis, e.g. metal-requiring proteins, metal transporter families, or major potential ligands such as glutathione, are conserved across kingdoms.47,48 Thus, it is both viable and informative to study the functions and activities of metal homeostasis factors, including ligands, in heterologous systems. This approach has been extensively utilized for the identification and molecular characterization of metal transporters and metal sequestration genes.4,49 Effectiveness of yeast systems for chelator studies is exemplified by experiments with the amyloid precursor protein (APP), which is implicated in Alzheimer’s disease. Modulation of Cu ion transport across the plasma membrane by APP and the effects of clioquinol, a synthetic metal chelator, were demonstrated in S. cerevisiae cells expressing APP.50 We have been following a similar reasoning to study in vivo binding of transition metal cations by NA.Published evidence for in vivo metal–NA complexes is so far based on invasive methods and confined to Ni(II)–NA in T. caerulescens,21 and in latex of Sebertia acuminata.51 as well as most recently the detection of Cu(II)–NA in T. caerulescens by EXAFS.45 In order to analyze suspected Zn binding by NA intracellularly we used a system where sequestration into the vacuole is not as pronounced as in hyperaccumulator plants, but nevertheless sufficiently high metal and ligand concentrations are reached inside the cell without causing lethality. Both phenotypic and EXAFS data demonstrated the existence of a Zn(II)–NA complex in vivo. Formation of Zn(II)–NA occurs in the presence of myriad potential Zn(II) binding sites that are conserved between S. pombe and cells of other eukaryotes including plants. Thus, despite the fact that NAS-overexpressing S. pombe cells clearly accumulate more NA and accordingly Zn(II)–NA than plant cells, this first in vivo proof of such a complex strongly suggests that the reported observations implicating NA in plant Zn homeostasis are indeed attributable to interaction of Zn(II) with NA and not just indirect consequences of perturbations in Fe metabolism.
Obviously, our data do not allow conclusions to be drawn about the exact proportions of different micronutrients chelated by NA in various plant tissues. Our conclusion is, that Zn(II)–NA complexes can be formed. Also, we are not proposing that NA functions as the final binding partner for Zn in hyperaccumulators. One has to distinguish translocation to above-ground tissue from final storage. The fits of the T. caerulescens Zn-EXAFSspectra clearly showed that NA does not act as Zn storage ligand, which is in agreement with recently published data on various Thlaspi species.46NA concentrations are in any case much too low for this. Rather, NA might contribute to Zn hyperaccumulation by enhancing the mobility of Zn(II), required for transport to the storage sites in the epidermal vacuoles, where it is only weakly associated with organic acids.29,32 Potential metal transport ligands or compartments that are not involved in storage, but in transport can hardly be determined by macroscopic (whole-tissue) EXAFS. High resolution μ-EXAFS, however, is still facing major limitations as any change in photon energy requires an adjustment of the X-ray optics to keep the focal spot unchanged. Other techniques for the in vivo detection of low abundance metal–ligand complexes are not available yet.
Experimental
Organisms, culture media and culture conditions
S. pombe strains used in this study were FY261 (h+ ade6-M216 leu1-32 ura4-Δ18 can 1-1) as a wild type strain, zhfΔ,36pcsΔ37 and hmt1Δ.39 Cells were grown in the standard media YE5S (Yeast extract + 225 mg/l of adenine, histidine, leucine, uracil, and lysine) and EMM (Edinburgh minimal medium: 14.7 mM potassium hydrogen phthalate, 15.5 mM Na2HPO4, 93.5 mM NH4Cl, 1.3 mM MgCl2, 100 μM CaCl2, 13.4 mM KCl, 0.28 mM Na2SO4, 4.2 μM panthotenic acid, 81.2 μM nicotoinic acid, 55.5 μM inositol , 40.8 nM biotin, 8.1 μM H3BO4, 2.4 μM MnSO4, 1.4 μM ZnSO4, 0.74 μM FeCl2, 0.25 μM H2MoO4, 0.6 μM KI, 0.16 μM CuSO4, 4.76 μM citric acid, 2% (w/v) glucose) supplemented appropriately. Expression of AtNAS2 in S. pombe cells under control of the thiamine-repressible nmt1 promoter, monitoring of expression by Western analysis, and growth assays were carried out as described.23Preparation of S. pombe samples for EXAFS measurements
Cells for the EXAFS analysis were grown in two ways: (i) 6 hours in medium containing 400 μM ZnCl2, (ii) cells were diluted into fresh medium containing 100 μM ZnCl2 three times during a 24 h incubation. Following incubation cells were harvested by centrifugation and washed twice with 1 μM EDTA (pH 8.0) and twice with Milli-Q water. The pellets, consisting of intact cells, were filled into EXAFS cuvettes, which were afterwards sealed with Kapton® tape. To eliminate problems of element re-distribution during sample preparation, the filled cuvettes were frozen in supercooled isopentane at about −140 °C and afterwards immediately transferred to liquid nitrogen, where they were stored until measurement.Preparation of model complexes for EXAFS measurements
The model compounds were chosen to be representative for known potential Zn(II) ligands in cells. 5 mM solutions of ZnSO4 were used as reference for the aquo complexes. The Zn(II)–NA complex was made by adding 1.2 mM NA to 1 mM of ZnSO4 solution; histidine and glutathione complexes were made by adding 50 mM ligand to a 5 mM solution of ZnSO4. The NA and GSH complexes were adjusted to pH 7 with NaOH. Zn(II)–citrate was obtained from Merck (Darmstadt, Germany) and dissolved in 50 mM citric acid. All solutions of the model complexes contained 30% (v/v) glycerol to minimize the formation of ice crystals during freezing. The samples were transferred into EXAFS cuvettes and frozen in liquid nitrogen.EXAFS measurements and data analysis
As described by Küpper et al.,32 the measurements were performed at the EMBL bending magnet beamline D2 (DESY, Hamburg, Germany) using a Si(111) double crystal monochromator, a focusing mirror and a 13-element Ge solid-state fluorescence detector. All samples were mounted in a top-loading closed-cycle cryostat (modified Oxford instruments) and kept at about 30 K. The transmitted beam was used for energy calibration by means of the Bragg reflections of a static Si(220) crystal.52 At least 1 million fluorescence counts per data point above the absorption edge were accumulated for each measurement. No photo reduction was observable in subsequent scans.The Kemp program package53 was employed for data reduction and conversion into ASCII files. Analysis of the spectra was done with DL-Excurve using constrained parameters54 with a data range from k = 2.1 Å−1 to k = 13.5 Å−1. The set of potential ligands coordinating Zn was limited to S, O, N and Zn for the first shell. EXAFS data analysis generally cannot discriminate between N and O ligation. However, rigid ligands, such as the imidazole ring of histidine or the nicotianamine structure stabilized by its central ion, can be identified via the specific multiple scattering pattern they generate. For NA, the most dominant multiple scattering feature (as determined by the model compound) was the peak at about 2.8 Å, which is attributed to the carbons of the carboxyl groups coordinating the zinc. This was included in the refinement and allowed for distinction between NA and other oxygen ligands. For the imidazole ring (with one N at about 2 Å, two C at about 3 Å, and one N and C each at about 4 Å) multiple scattering contributions up to 5th order were considered in the simulations. Sulfur ligands were calculated in the single scattering approximation.55 For all spectra the Fermi energy, the ligand distances, the Debye–Waller parameters and the coordination numbers of the potential ligands were refined. The Fermi energy defines the energy threshold of each EXAFSspectrum.56 The Debye–Waller parameter is proportional to static and dynamic disorder of each shell.
In addition to the refinement described above, the measured EXAFS data of all S. pombe and plant samples were fitted with a linear combination of the measured curves of all model complexes. This fit, subsequently called “component analysis” (CA), was done in order to have a second method to estimate the proportions of the different types of ligands around the metals. The fitting of measured XAS sample data by a linear combination of measured model data has also successfully been used by Salt et al.29 for Zn in samples of T. caerulescens and by Küpper et al.32 for Cd and Zn in the same species. In the present work the approach of Salt and co-workers was modified as described by Küpper et al.32
Elemental profiling of S. pombe cells
Washed, frozen-hydrated S. pombe cells were lyophilized and subsequently digested in 4 ml 65% HNO3 and 2 ml 30% H2O2 and analyzed by ICP-OES using an iCAP 6000 (Thermo, USA). Elements were measured at the following wavelengths: Zn, 213.8 nm and 334.5 nm; Fe, 238.2 nm and 259.9 nm; Cd, 226.5 nm; Cu, 324.7 nm; Mn, 257.6 nm; Ni, 231.6 nm; Co, 228.6 nm.NA detection and quantification
NA was analyzed through a newly established protocol using a 15N-labelled NA standard (H. Schmidt, C. Böttcher, A. Trampczynska, S. Clemens, in preparation). S. pombe cell pellets were disrupted with glass beads in H2O. Extraction was performed at 80 °C and with 5 min sonication. Following centrifugation supernatants were harvested and stored at −20 °C. Extracts were derivatized with 9-fluorenylmethoxycarbonyl (FMOC) in 250 mM sodium borate, pH 8.0, at room temperature for 1 min and subsequently analyzed by UPLC-ESI-QTOF-MS (Waters, Milford, USA). Two μl of sample were separated on a BEH C18 column (2.1 × 100 mm, 1.7 μm) at 40 °C. The flow was set to 0.5 ml per minute, solvents were water (A) and acetonitrile (B), both acidified with 0.1% formic acid. The gradient was as follows: 0.5 min 80% A, from 0.5 to 4.5 min a linear gradient to 95% B, 95% B for 0.9 min, a linear gradient to initial conditions in 1.6 min and re-equilibration (95% A) for 1 min. The mass spectrometer was operated in the ESI V+ mode. Source settings were as follows: capillary: 2.5 kV, sampling cone 30, extraction cone 30, ion guide 3.3, source temperature 120 °C, cone gas flow 10 l/h, desolvation gas flow 1000 l/h. The MS acquisition was performed from 400–600 Th with a scan time of 0.3 s and an inter-scan delay of 0.05 s. Samples were measured in duplicate. Runs were recorded and reviewed with the MassLynx 4.1 software. Quantification of the mono-FMOC derivatized NA was performed using the QuanLynx module of the software package, the 15N-labelled NA standard and a calibration curve for the authentic NA standard obtained from Toronto Research Chemicals (Toronto, Canada). The [M + H]+ ion trace (526.2189 ± 0.1 Th) was used for quantification. The retention time was set to 1.96 ± 0.05 min.Conclusion
We utilized modulation of metal ligand synthesis in yeast cells in combination with physiological characterization and analysis by EXAFS as a powerful way of elucidating Zn speciation in vivo. Using this approach we obtained several lines of evidence for the formation of Zn(II)–NA complexes in vivo. Given the high degree of conservation in eukaryotic cellular metal homeostasis this suggests the existence of such complexes also in planta and would help explain numerous observations reported in recent years for mutant plants altered in NA levels and/or YSL transporter activities.Acknowledgements
We are very grateful to Silke Haala for excellent technical assistance. This work was financially supported by the Deutsche Forschungsgemeinschaft (Cl 152/3) and in part by the EU through its Sixth Framework Program for RTD (contract no. FOOD-CT-2006-016253).References
- U. Krämer and S. Clemens, Top. Curr. Genet., 2005, 14, 216–271 Search PubMed.
- E. P. Colangelo and M. L. Guerinot, Curr. Opin. Plant Biol., 2006, 9, 322–330 CrossRef CAS.
- H. Q. Ling, G. Koch, H. Bäumlein and M. W. Ganal, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 7098–7103 CrossRef CAS.
- C. Cobbett and P. Goldsbrough, Annu. Rev. Plant Physiol. Plant Mol. Biol., 2002, 53, 159–182 CrossRef CAS.
- U. Krämer, J. D. Cotter-Howells, J. M. Charnock, A. J. M. Baker and J. A. C. Smith, Nature, 1996, 379, 635–638 CrossRef CAS.
- W. E. Rauser, Cell Biochem. Biophys., 1999, 31, 19–48 CrossRef CAS.
- H. Küpper and P. M. Kroneck, in Metal Ions in Biological Systems, ed. A. Sigel, H. Sigel and R. K. O. Sigel, Marcel Dekker Inc., 2005, vol. 44, pp. 97–144 Search PubMed.
- D. L. Callahan, A. J. Baker, S. D. Kolev and A. G. Wedd, J. Biol. Inorg. Chem., 2006, 11, 2–12 CrossRef CAS.
- I. Benes, K. Schreiber, H. Ripperger and A. Kircheiss, Cell. Mol. Life Sci., 1983, 39, 261–262 CrossRef CAS.
- R. Rellan-Alvarez, J. Abadia and A. Alvarez-Fernandez, Rapid Commun. Mass Spectrom., 2008, 22, 1553–1562 CrossRef CAS.
- C. Curie, G. Cassin, D. Couch, F. Divol, K. Higuchi, M. Jean, J. Misson, A. Schikora, P. Czernic and S. Mari, Ann. Bot., 2009, 103, 1–11 CAS.
- N. von Wiren, S. Klair, S. Bansal, J. F. Briat, H. Khodr, T. Shioiri, R. A. Leigh and R. C. Hider, Plant Physiol., 1999, 119, 1107–1114 CrossRef CAS.
- A. Pich and G. Scholz, J. Exp. Bot., 1996, 47, 41–47 CrossRef CAS.
- B. Irtelli, W. A. Petrucci and F. Navari-Izzo, J. Exp. Bot., 2009, 60, 269–277 CAS.
- M. Klatte, M. Schuler, M. Wirtz, C. Fink-Straube, R. Hell and P. Bauer, Plant Physiol., 2009, 150, 257–271 CrossRef CAS.
- M. Takahashi, Y. Terada, I. Nakai, H. Nakanishi, E. Yoshimura, S. Mori and N. K. Nishizawa, Plant Cell, 2003, 15, 1263–1280 CrossRef CAS.
- D. Douchkov, C. Gryczka, U. W. Stephan, R. Hell and H. Bäumlein, Plant, Cell Environ., 2005, 28, 365–374 CrossRef CAS.
- K. Pianelli, S. Mari, L. Marques, M. Lebrun and P. Czernic, Transgenic Res., 2005, 14, 739–748 CrossRef CAS.
- S. Kim, M. Takahashi, K. Higuchi, K. Tsunoda, H. Nakanishi, E. Yoshimura, S. Mori and N. K. Nishizawa, Plant Cell Physiol., 2005, 46, 1809–1818 CrossRef CAS.
- A. J. M. Baker and R. R. Brooks, Biorecovery, 1989, 1, 81–126 Search PubMed.
- V. Vacchina, S. Mari, P. Czernic, L. Marques, K. Pianelli, D. Schaumlöffel, M. Lebrun and R. Lobinski, Anal. Chem., 2003, 75, 2740–2745 CrossRef CAS.
- M. Becher, I. N. Talke, L. Krall and U. Krämer, Plant J., 2004, 37, 251–268 CAS.
- M. Weber, E. Harada, C. Vess, E. von Roepenack-Lahaye and S. Clemens, Plant J., 2004, 37, 269–281 CAS.
- J. E. van de Mortel, L. Almar Villanueva, H. Schat, J. Kwekkeboom, S. Coughlan, P. Moerland, E. Ver Loren van Themaat, M. Koornneef and M. G. M. Aarts, Plant Physiol., 2006, 142, 1127–1147 CrossRef CAS.
- U. Krämer, I. N. Talke and M. Hanikenne, FEBS Lett., 2007, 581, 2263–2272 CrossRef.
- A. Trampczynska, C. Böttcher and S. Clemens, FEBS Lett., 2006, 580, 3173–3178 CrossRef CAS.
- D. E. Salt, I. Baxter and B. Lahner, Annu. Rev. Plant Biol., 2008, 59, 709–733 CrossRef CAS.
- H. Küpper, F. Jie Zhao and S. P. McGrath, Plant Physiol., 1999, 119, 305–312 CrossRef CAS.
- D. E. Salt, R. C. Prince, A. J. M. Baker, I. Raskin and I. J. Pickering, Environ. Sci. Technol., 1999, 33, 713–717 CrossRef CAS.
- H. Küpper, E. Lombi, F.-J. Zhao and S. P. McGrath, Planta, 2000, 212, 75–84 CrossRef CAS.
- H. Küpper, E. Lombi, F.-J. Zhao, G. Wieshammer and S. P. McGrath, J. Exp. Bot., 2001, 52, 2291–2300 CrossRef CAS.
- H. Küpper, A. Mijovilovich, W. Meyer-Klaucke and P. M. Kroneck, Plant Physiol., 2004, 134, 748–757 CrossRef.
- J. R. Wang, F. J. Zhao, A. A. Meharg, A. Raab, J. Feldmann and S. P. McGrath, Plant Physiol., 2002, 130, 1552–1561 CrossRef CAS.
- S. M. Webb, J. F. Gaillard, L. Q. Ma and C. Tu, Environ. Sci. Technol., 2003, 37, 754–760 CrossRef CAS.
- G. Wellenreuther, M. Cianci, R. Tucoulou, W. Meyer-Klaucke and H. Haase, Biochem. Biophys. Res. Commun., 2009, 380, 198–203 CrossRef CAS.
- S. Clemens, T. Bloss, C. Vess, D. Neumann, D. Nies and U. Zur Nieden, J. Biol. Chem., 2002, 277, 18215–18221 CrossRef CAS.
- S. Clemens, E. J. Kim, D. Neumann and J. I. Schroeder, EMBO J., 1999, 18, 3325–3333 CrossRef CAS.
- D. F. Ortiz, L. Kreppel, D. M. Speiser, G. Scheel, G. McDonald and D. W. Ow, EMBO J., 1992, 11, 3491–3499 CAS.
- S. Clemens, Biochimie, 2006, 88, 1707–1719 CrossRef CAS.
- T. O'Halloran and V. Culotta, J. Biol. Chem., 2000, 275, 25057–25060 CrossRef CAS.
- R. Lobinski, C. Moulin and R. Ortega, Biochimie, 2006, 88, 1591–1604 CrossRef CAS.
- I. J. Pickering, C. Wright, B. Bubner, D. Ellis, M. W. Persans, E. Y. Yu, G. N. George, R. C. Prince and D. E. Salt, Plant Physiol., 2003, 131, 1460–1467 CrossRef CAS.
- I. J. Pickering, L. Gumaelius, H. H. Harris, R. C. Prince, G. Hirsch, J. A. Banks, D. E. Salt and G. N. George, Environ. Sci. Technol., 2006, 40, 5010–5014 CrossRef CAS.
- H. Küpper, A. Mijovilovich, B. Götz, F. C. Küpper and W. Meyer-Klaucke, Plant Physiol., 2009, 151, 702, DOI:10.1104/pp.109.139717.
- A. Mijovilovich, B. Leitenmaier, W. Meyer-Klaucke, P. M. H. Kroneck, B. Götz and H. Küpper, Plant Physiol., 2009, 151, 715, DOI:10.1104/pp.109.144675.
- D. L. Callahan, S. D. Kolev, R. A. J. O’Hair, D. E. Salt and A. J. M. Baker, New Phytol., 2007, 176, 836–848 Search PubMed.
- D. J. Eide, Biochim. Biophys. Acta, Mol. Cell Res., 2006, 1763, 711–722 CrossRef CAS.
- B. E. Kim, T. Nevitt and D. J. Thiele, Nat. Chem. Biol., 2008, 4, 176–185 CrossRef CAS.
- T. C. Fox and M. L. Guerinot, Annu. Rev. Plant Physiol. Plant Mol. Biol., 1998, 49, 669–696 CrossRef CAS.
- C. Treiber, A. Simons, M. Strauss, M. Hafner, R. Cappai, T. A. Bayer and G. Multhaup, J. Biol. Chem., 2004, 279, 51958–51964 CrossRef CAS.
- D. Schaumlöffel, L. Ouerdane, B. Bouyssiere and R. Lobinski, J. Anal. At. Spectrom., 2003, 18, 120–127 RSC.
- R. F. Pettifer and C. Hermes, J. Appl. Crystallogr., 1985, 18, 404–412 CrossRef CAS.
- M. Korbas, D. F. Marsa and W. Meyer-Klaucke, Rev. Scient. Instr., 2006, 77, 063105-1–063105-5 Search PubMed.
- N. Binstead, R. W. Strange and S. S. Hasnain, Biochemistry, 1992, 31, 12117–12125 CrossRef CAS.
- J. J. Rehr and R. C. Albers, Phys. Rev. B: Condens. Matter Mater. Phys., 1990, 41, 8139–8149 CrossRef.
- J. J. Rehr and R. C. Albers, Rev. Mod. Phys., 2000, 72, 621–654 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary file S1: EXAFS refinements of all model compounds, Supplementary file S2: EXAFS refinements of all S. pombe samples. See DOI: 10.1039/b913299f |
| This journal is © The Royal Society of Chemistry 2010 |
