Development of second generation amidinohydrazones, thio- and semicarbazones as Trypanosoma cruzi-inhibitors bearing benzofuroxan and benzimidazole 1,3-dioxide core scaffolds
Alicia
Merlino
a,
Diego
Benitez
a,
Santiago
Chavez
a,
Jonathan
Da Cunha
a,
Paola
Hernández
a,
Luzineide W.
Tinoco
b,
Nuria E.
Campillo
c,
Juan A.
Páez
c,
Hugo
Cerecetto
*a and
Mercedes
González
*a
aGrupo de Química Medicinal, Laboratorio de Química Orgánica, Facultad de Ciencias-Facultad de Química, Universidad de la República, Iguá 4225,
, 11400 Montevideo, Uruguay. E-mail: megonzal@fq.edu.uy; hcerecet@fq.edu.uy; Fax: +598 2 5250749; Tel: 598 2 5258618 (ext. 216)
bLaboratory of Analysis and Development of Enzyme Inhibitors - LADIE, NPPN, Federal University of Rio de Janeiro,
cInstituto de Química Médica, CSIC, Madrid, Spain
First published on 11th August 2010
Abstract
Trypanosoma cruzi is the causative agent of Chagas' disease. The thiosemicarbazone moiety as a pharmacophore has been described for inhibition of the essential cysteine protease, cruzipain, of this parasite. Our recent study identified an amidinohydrazone containing benzofuroxan as a hit compound for cruzipain inhibition with trypanosomicidal activity. Structural modification of the amidinohydrazone, thio- and semicarbazone motifs, using benzofuroxan and including a benzimidazole 1,3-dioxide system as new core scaffolds are described. These changes allowed for the identification of new structural motifs with desired antitrypanosomal activity. The new amidinohydrazone, thio-, and semicarbazone derivatives had excellent anti-trypanosomal activity without improved cruzipain-inhibitory activity compared with the parent compounds. Relevant structural features of these derivatives for further modification have also been determined.
Introduction
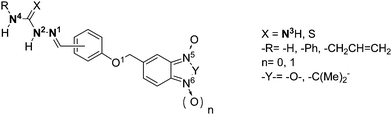
Chagas' disease is the third most neglected disease in Latin America after malaria and schistosomiasis, and affects at least 15 million people with more than 25 million at risk of infection.1 The infectious agent is the protozoan parasite Trypanosoma cruzi (T. cruzi), that causes symptoms progressing from mild swelling to intestinal disease and ultimately heart failure. Currently, there are two nitroaromatic-based drugs for treatment of this disease, Nifurtimox (Nfx, Lampit®, Scheme 1a) and Benznidazole (Bnz, Rochagan®, Scheme 1a).2,3 Treatment with these drugs is inadequate, since the parasite is often not completely eliminated despite chronic administration, and there are unacceptable side effects.4,5 Moreover, resistance to these agents has emerged.6 For these reasons the development of safer and more effective drugs against Chagas' disease is urgently needed.7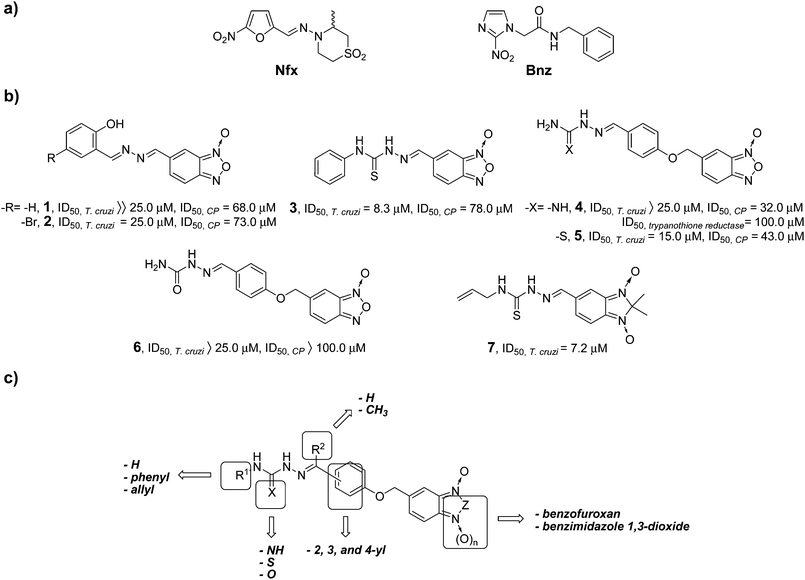 | ||
| Scheme 1 a) Drugs used clinically as anti-T. cruzi agents. b) Hybrid parent compounds with antitrypanosomal activity. c) Structural modifications described. | ||
The use of multitarget-directed ligands (MTDLs) has emerged as a strategy for the development of new drugs to treat Chagas' disease.8 The approach is based on the combination of two or more pharmacophores into a new chemical entity, also defined as a hybrid-drug, and is only just beginning to be used in drug design for treatment of this disease. A recent focus of our attention has been on developing MTDLs as anti-T. cruzi agents using free radical-releasing moieties linked to DNA-interacting, sterol-biosynthesis-inhibitor and cruzipain (CP)-inhibitor pharmacophores.9
Using this strategy, we hybridized benzofuroxan heterocycle and amidinohydrazone, thio- and semicarbazone moieties10 to generate compounds with trypanosomicidal activity involving at least two mechanism of action. In addition,11 we provided evidence that benzofuroxan derivatives were able to modify parasite dehydrogenase activity and to affect mitochondrial membrane potential, whereas amidinohydrazone and thiosemicarbazone moieties had already been described as pharmacophores for trypanothione reductase and CP inhibitors, respectively.12 In this study we identified parent compounds 1–5 (Scheme 1b) with good in vitro trypanosomicidal activity.10 Compounds 1–5 had modest CP inhibitory activity, with the amidinohydrazone 4 having some trypanothione reductase inhibitory activity. Semicarbazone analogue 6 was a poor CP inhibitor and anti-T. cruzi agent. Another heterocycle system that we used as a scaffold for the development of new trypanosomicidal agents was the benzimidazole 1,3-dioxide system (i.e.7, Scheme 1b).9 We hypothesized that this heterocycle might act as a free radical-releasing pharmacophore; however, it was recently reported that this mode of action was not observed in the parasite.11 Derivatives of this heterocycle have the advantage of being water soluble.
In our efforts to develop selective and novel trypanosomicidal compounds, we selected parent compounds 3–7 as molecular templates and the structural modifications we sought to investigate are shown in Scheme 1c. Thus, we used benzofuroxan and benzimidazole 1,3-dioxide as core scaffolds linked to amidinohydrazone, thio- and semicarbazone pharmacophores aiming to produce new anti-T. cruzi agents. The derivatives synthesized were examined for antiproliferative in vitro activity against the T. cruzi, Tulahuen 2 strain, for non-specific cytotoxicity on human red blood cells and J-774 mouse macrophages, and inhibition of CP. The ability of the compounds to interact with CP was examined by docking analyses.
Results and discussion
The starting point for the synthesis of the new hybrid derivatives was the preparation of the corresponding carbonylphenyloxymethylbenzofuroxan (8–11) as shown in Scheme 2. Aldehydes 8 and 9 were prepared according to a previous report10 with minor modification, using 5-bromomethylbenzofuroxan13 as starting material. Due to the unusual stability that the intramolecular hydrogen bond confers to salicylaldehyde and 2-hydroxyacetophenone, the alkylation reaction could not be done using the aforementioned strategy to synthesize derivatives 10 and 11. Consequently, these carbonyl derivatives, 10 and 11, were obtained using solvent-free microwave irradiation with K2CO3 and tetrabutylammonium iodide (TBAI) as phase transfer catalyst (Scheme 2). Carbonyl-containing benzimidazole 1,3-dioxide derivatives (12–14) were prepared by treatment of the corresponding benzofuroxan with 2-nitropropane and piperidine as base in THF at room temperature (Scheme 2). Amidinohydrazone, thio- and semicarbazone containing benzofuroxans 15–25 were prepared by condensation of the corresponding carbonyl derivative and hydrazones under acidic conditions as shown in Scheme 2. The same heterocycle transformation conditions used for the synthesis of derivatives 12–14 was used to prepare benzimidazole 1,3-dioxides 26–37 (Scheme 2). Compounds 4–6 (Scheme 1) were used as starting materials for the preparation of derivatives 26–28. We were unable to obtain the desired amidinohydrazones 26 and 31 because in both cases the starting material decomposed under the reaction conditions (piperidine/THF/r.t.). All the structures were determined by 1H NMR, 13C NMR, NOE-diff, COSY, HSQC, and HMBC experiments, IR and MS. The purity of products 9–25, 27–30, and 32–37 was determined by TLC and microanalysis.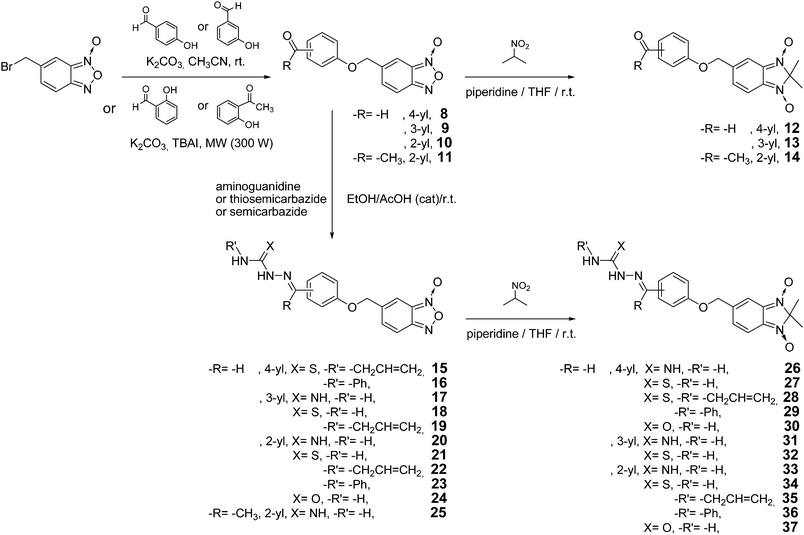 | ||
| Scheme 2 Synthetic procedure used for the preparation of hybrid compounds and precursors. | ||
The new benzofuroxan and benzimidazole 1,3-dioxide derivatives, 9–25, 27–30, and 32–37, were assessed in vitro for antiproliferative activity against the epimastigote form of the T. cruzi, Tulahuen 2 strain.9,10 The occurrence of the epimastigote form of T. cruzi as an obligate mammalian intracellular stage has been reevaluated and confirmed.14 Furthermore, it should be noted that a good correlation between antiproliferative epimastigote activity and in vivo anti-T. cruzi activity was observed with compounds from our chemical library.9,15 Compounds were incorporated into the growth medium at 25.0 μM and the ability to inhibit the growth of the parasite was evaluated by comparison with untreated controls on day 5. The ID50 doses (50% inhibitory dose) were determined for the most active compounds, and Nfx and Bnz were used as reference trypanosomicidal drugs. The starting carbonyl derivatives, 8–14, exhibited in some cases relevant anti-T. cruzi activities, the most active being the 2-substituted aldehyde 10 (Table 1). The semicarbazones and the amidinohydrazones, except for the 2-substituted amidinohydrazone 25, were not active against T. cruzi in culture. Derivative 25 showed higher activity than parent compound 4 (Table 1). The new N4-unsubstituted thiosemicarbazones, 18, 21, 27, 32 and 34, were less or as active as parent compound 5 (Table 1), whereas benzofuroxan 18 and benzimidazole 1,3-oxide derivatives 27 and 34 were the most active. However, when the thiosemicarbazone N4-position was substituted, and in particular by a phenyl moiety (i.e.16, 23 and 36, Table 1), higher anti-T. cruzi activities were observed compared with N4-unsubstituted derivatives. The semicarbazone, N4-allyl and N4-phenyl thiosemicarbazones derived from 2-formyl benzofuroxane and benzimidazole 1,3-dioxide, 35, 23 and 37, respectively, were the most active compounds obtained, having higher activities than references Nfx and Bnz.
| Seriesa | Compd | ID50/μM | Compd | ID50/μM | Compd | ID50/μM | Compd | ID50/μM | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T2b,c | rbcc,d | T2b,c | rbcc,d | T2b,c | rbcc,d | T2b,c | rbcc,d | ||||||
| a bfx: benzofuroxan; bz: benzimidazole 1,3-dioxide; ca: carbonyls; ah: amidinohydrazones; th: thiosemicarbazones; at: N4-allylthiosemicarbazones; pt: N4-phenylthiosemicarbazones. b T2: Tulahuen 2 strain. c The results are the means of three independent experiments. d rbc: Human red blood cells. e From ref. 10. f Problems with compound solubility were observed at higher doses. g “—”: compound was not obtained. h ag: the compound formed aggregates in the buffer at all doses assayed. i ns: not studied. | |||||||||||||
| bfx | ca | 8 | 8.7e | < 50 | 9 | 28 ± 2 | 50 ± 5 | 10 | 6.3 ± 0.2 | > 100 | 11 | 7.0 ± 1.2 | > 200 |
| bz | 12 | 17.0 ± 1.9 | 65 ± 3 | 13 | > 25 | < 50 | 14 | > 25 | < 50 | ||||
| bfx | ah | 4 | > 25 e | > 200 | 17 | > 25 | > 100f | 20 | > 25 | 58 ± 3 | 25 | 16.6 ± 2.3 | > 200 |
| bz | 26 | —g | — | 31 | — | — | 33 | > 25.0 | 114 ± 3 | ||||
| bfx | th | 5 | 15.0 e | > 200 | 18 | 15.0 ± 0.4 | agh | 21 | > 25 | 189 ± 1 | |||
| bz | 27 | 17.0 ± 0.8 | ag | 32 | 24 ± 2 | 141 ± 5 | 34 | 14 ± 2 | > 200 | ||||
| bfx | at | 15 | > 25.0 | ag | 19 | 13 ± 2 | ag | 22 | 23 ± 1 | 139 ± 2 | |||
| bz | 28 | 23.0 ± 1.5 | > 100h | 35 | 2.7 ± 1.4 | ag | |||||||
| bfx | pt | 16 | 7 ± 3 | ag | 23 | 3.6 ± 0.4 | > 200 | ||||||
| bz | 29 | > 25 | < 50 | 36 | 7.7 ± 0.3 | 66.0 ± 0.8 | Nfx | 7.7e | > 100f | ||||
| bfx | se | 6 | > 25.0 e | > 200 | 24 | > 25 | > 100h | Bnz | 7.4 e | nsi | |||
| bz | 30 | > 25 | > 200 | 37 | 4.7 ± 1.2 | 50.0 ± 0.4 | AmpB | 0.152 ± 0.006 | 1.5 ± 0.2 | ||||
Two different biological systems, erythrocytes and macrophages, were used to estimate the potential toxicity of the hybrid derivatives (Tables 1 and 2), 9,10,16 Erythrocytes served as a biological model for mammalian cells in direct contact with the trypomastigote, and macrophages a related model for the amastigote stage of the parasite. The compounds were added to cell suspensions at doses of 50.0–200.0 μM for erythrocytes and at doses of 100.0–400.0 μM for macrophages, toxicities were evaluated by comparison with untreated controls after 24 or 48 h and the ID50 doses (50% cytotoxic dose) were determined. Nfx was used as the reference trypanosomicidal drug and amphotericin B (AmpB), used experimentally as an anti-T. cruzi agent,17 was used as a positive control in that it is a haemolytic agent (Table 1). Only soluble compounds were included in the macrophage-cytotoxicity assays (Table 2). A general relationship between the type of substituent (carbonyl, amidinohydrazone, thio- or semicarbazone) or type of core scaffold (benzofuroxan or benzimidazole 1,3-dioxide) and cytotoxicity was not evident. However, like anti-T. cruzi activities, a correlation between cytotoxicity and the substituent's position on the phenoxy moiety was apparent. This was particularly evident in the case of 4- and 2-substituted derivatives, the latter being less cytotoxic than the former (compare rbc cytotoxicities of 8 to 10 and 11, as well as 29 to 36). One of the best anti-T. cruzi agents, 2-thiosemicarbazone 23, had excellent selectivity indexes. The selectivity indexes for derivative 23 were SIerythrocyte/T.cruzi > 55.6 and SImacrophage/T.cruzi > 111.1, and in contrast to the thiosemicarbazone parent compound 5 (SIerythrocyte/T.cruzi > 13.3 and SImacrophage/T.cruzi = 26.7) and the carbonyl parent compound 10 (SIerythrocyte/T.cruzi > 15.9 and SImacrophage/T.cruzi = 12.4), and Nfx (SIerythrocyte/T.cruzi = 13.0) and AmpB (SIerythrocyte/T.cruzi ∼ 10.0) (Tables 1 and 2). Thus, derivative 23 had far better selectivity indexes than parent derivatives 5 and 10 and reference compounds.
| Seriesa | Compd | ID50/μMb | Compd | ID50/μMb | |
|---|---|---|---|---|---|
| a bfx: benzofuroxan; bz: benzimidazole 1,3-dioxide; ca: carbonyls; ah: amidinohydrazones; th: thiosemicarbazones; at: N4-allylthiosemicarbazones; pt: N4-phenylthiosemicarbazones. b Results shown are the means of three independent experiments. c From ref. 10. | |||||
| bfx | ca | 8 | 60.0c | 10 | 78 ± 2 |
| bz | 12 | < 100 | |||
| bfx | ah | 4 | < 50 c | 20 | < 100 |
| bfx | th | 5 | 400.0c | 21 | < 100 |
| bfx | at | 22 | 138 ± 2 | ||
| bfx | pt | 23 | > 400 | ||
| bz | 36 | < 100 | |||
| bfx | se | 24 | < 100 | ||
We investigated the ability of the hybrid derivatives to inhibit CP to obtain information on their mechanism of trypanosomicidal activity. The compounds were tested at 25.0, 50.0, and 100.0 μM for the ability to inhibit CP. The compounds were assayed using Z-phenyl-arginine-7-amido-4-methylcoumarin hydrochloride (Z-Phe-Arg-AMC) as fluorogenic CP substrate, and compared with untreated control assays (Table 3). As non-specific compound aggregation could interfere with the assay or promote promiscuous CP inhibition, the assays were also done, for compounds 17 and 19, at acidic pH and in the presence of Triton X-100 (Table 4). The acidic pH is especially relevant for the protonation of the amidinohydrazone moiety18 that could change the interaction in the active site, and the use of the non-ionic surfactant is relevant to reduce aggregation and false inhibition.19 In spite of the fact that the new hybrid compounds were more active and selective against T. cruzi than the parent compounds, the CP inhibitory properties were less than the parent compounds. The best CP inhibitors were the 4- and 3-thiosemicarbazonyl containing benzofuroxans 16, 18 and 19, and the 4- and 3-thiosemicarbazonyl containing benzimidazole 1,3-dioxides 29 and 32. The thiosemicarbazone moiety was previously implicated as a CP inhibitor pharmacophore.20 None of the 2-substituted derivatives, 10, 11, 14, 20–25 and 33–37, had significant activity in this assay. Although the pH change did not improve the activity of amidinohydrazone 17 (Table 4), thiosemicarbazone 19 tested in presence of Triton X-100, and at pH 7.3, demonstrated that the effect against CP was not the result of an unrelated inhibitory activity.
| Compd. | % of inhibition at 100.0 μMa,b | ID50/μMa,b | Compd. | % of inhibition at 100.0 μMa,b | ID50/μMa,b |
|---|---|---|---|---|---|
| a In buffer, pH 7.3, and absence of Triton X-100 (see Experimental Section). b The results are the means of three independent experiments. c mbth: (m-bromophenyl)ethylketone thiosemicarbazone (reference compound). | |||||
| 8 | 6.3 ± 0.6 | >100.0 | 12 | 35.5 ± 1.5 | >100.0 |
| 9 | 43.0 ± 0.8 | ∼ 100.0 | 13 | 45.0 ± 1.0 | ∼ 100.0 |
| 10 | 0.0 | >100.0 | 14 | 0.0 | >100.0 |
| 11 | 12.0 ± 0.7 | >100.0 | |||
| 27 | 13.7 ± 1.9 | >100.0 | |||
| 15 | 38.0 ± 2.0 | >100.0 | 28 | 51.0 ± 0.7 | 100.0 ± 2.0 |
| 16 | 95 ± 2 | 75.0 ± 2.1 | 29 | 72 ± 4 | 85.0 ± 1.3 |
| 30 | 0.0 | >100.0 | |||
| 17 | 42.0 ± 1.8 | ∼ 100.0 | |||
| 18 | 80 ± 3 | 78.0 ± 1.6 | 32 | 95 ± 3 | 64.0 ± 1.9 |
| 19 | 100 ± 2 | 55.0 ± 2.0 | |||
| 20 | 39.0 ± 1.5 | >100.0 | 33 | 2.5 ± 0.5 | >100.0 |
| 21 | 49.0 ± 0.8 | 100.0 ± 2.0 | 34 | 35.0 ± 1.6 | >100.0 |
| 22 | 11.0 ± 2.1 | >100.0 | 35 | 6.6 ± 0.9 | >100.0 |
| 23 | 0.0 | >100.0 | 36 | 0.0 | >100.0 |
| 24 | 7.0 ± 1.3 | >100.0 | 37 | 0.0 | >100.0 |
| 25 | 21.0 ± 0.8 | >100.0 | |||
| mbth | 100.0 ± 0.5 | 0.10 ± 0.08 |
To gain insight into CP–ligand interactions, NMR and docking analyses were done. When a small molecule interacts with a biomolecule, a decrease in T1 is observed due to the slowing of ligand molecular motions as it binds to the macromolecule.21 Unfortunately, we were unable to obtain information using derivatives 15, 16, 18, 19 and 32 as ligands due to the low solubility of these derivatives under the assay conditions. Attempts to work with other conditions, pH and amount of DMSO were unsuccessful due to the relatively low stability of CP. Docking analysis using parent compound 4, the hybrid derivatives 16–19, 21, 23, 29 and 34, and (m-bromophenyl)ethylketone thiosemicarbazone (mbth) (Table 3) were in agreement with the experimental results (Table 5, Fig. 1). The CP–ligand complexes were analyzed using FlexiDock, a program that performs docking of conformationally flexible ligands into receptor binding sites and provides control of ligand binding characteristics, taking into account rigid, partially flexible, or fully flexible receptor side chains. FlexiDock incorporates the van der Waals, electrostatic, torsional and constraint energy terms of the Tripos force field, and uses a genetic algorithm (GA) to determine the optimum ligand geometry. GA borrows methodology and terminology from biological (or Darwinian) evolution, in that an iterative process is used in which the most fit members of a population will have the best chance of propagating themselves in future generations of analysis. The reference compound mbth showed small differences in the type of interaction compared with our derivatives (Table 6), the thiocarbonyl carbon was favorably positioned to form a covalent bond with the sulfur atom of the catalytic Cys25. Moreover, the His159 was properly oriented for protonating the resulting anion via the thiocarbonyl sulfur. So, as previously described, the potent inhibitory capacity of this compound could be explained by the possibility of forming a reversible covalent interaction with the active-site cysteine (Cys25). In the cases of our new hybrid derivatives, some relevant interactions could be identified (Table 6). Except for compound 23, all derivatives interacted by hydrogen bonding and/or hydrophobic interactions with one or more residues present in the substrate-binding cleft.
| Compd. | K d/Ma | ΔG/kcal mol−1a | ID50,CP |
|---|---|---|---|
| a Values were estimated from the most stable conformers. | |||
| 4 | 9.7 × 10−4 | −4.11 | 32.0 |
| 16 | 4.4 × 10−3 | −3.21 | 75.0 |
| 17 | 6.0 × 10−3 | −3.03 | > 100.0 |
| 18 | 1.2 × 10−3 | −3.92 | 78.0 |
| 19 | 2.9 × 10−3 | −3.46 | 55.0 |
| 21 | 5.7 × 10−3 | −3.06 | 100.0 |
| 23 | 3.6 × 10−1 | −0.61 | > 100.0 |
| 29 | 5.4 × 10−3 | −3.09 | 85.0 |
| 34 | 9.6 × 10−3 | −2.75 | > 100.0 |
| mbth | 6.2 × 10−4 | −4.37 | 0.10 |
|
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Cys25 (S) | Gly66 (N) | Gly66 (O) | Leu67 | Leu157 | Asp158 (O) | His159 (Nδ1) | Glu205 (Oε2) | |
| a HB: hydrogen bond. b According to labelled structure. c Hph: hydrophobic interactions. d Bfx: benzofuroxan ring. e no interaction. f all: allyl moiety. g The same atomic labels at the thiosemicarbazone moiety as those of the hybrid derivatives analyzed. | ||||||||
| 4 | HBa (N6-Bfx)b | HB (Y = –O–) | HB (N5-Bfx) | Hphc (Bfxd) | —e | HB/N2 | — | HB/N3 |
| 16 | — | — | — | — | Hph (Bfx) | — | — | — |
| 17 | — | — | HB/N4, HB (N2) | — | — | HB/N3 | — | — |
| 18 | — | HB (O1) | — | — | — | — | — | HB/N4 |
| 19 | — | HB/N1 | HB/N4 | Hph (allf) | Hph (Bfx) | — | — | — |
| 21 | — | — | — | — | — | HB/N2 | HB/N4 | |
| 23 | — | — | — | — | — | — | — | — |
| 29 | — | — | — | — | Hph | — | — | — |
| 34 | — | — | HB/N4 | -- | — | HB/N2 | — | — |
| mbth | — | — | HB/N2 | Hph | — | — | — | — |
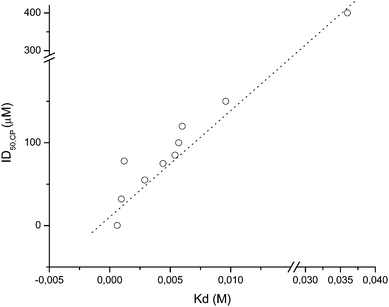 | ||
| Fig. 1 Theoretical Kdvs. experimental ID50 values for CP–ligand complexes analyzed. Notes: ID50 for derivative 23 was inferred by extrapolation as ca. 400 μM; the line shows tendency. | ||
In the CP–19 complex (Fig. 2a) Gly66 formed stable hydrogen bonds with the N1 (3.5 Å) and N4 (2.8 Å) thiosemicarbazone atoms of the ligand (Table 6), this interaction being a common stabilizing feature in CP–small molecule inhibitor complexes.9,22–26 In addition, binding was assisted by hydrophobic interactions between the benzofuroxan rings of the ligand and Leu157 and between the allyl moiety and Leu67. The stabilizing effect of these hydrophobic interactions became evident after examination of the CP–16 complex (Fig. 2b). Compound 16, with an ID50 of 75 μM, was stabilized in the active cleft only by Leu157 and the non-polar regions of Ala133 oriented toward the benzofuroxan ring of the inhibitor. This amino acid residue at the bottom of the S2 subsite was of particular importance as it was crucial in determining the substrate specificity of the enzyme.26 Together with flexible Glu205, CP has an S2 subsite that is able to accept both basic and hydrophobic residues. In fact, when comparing the complexes of the amidinohydrazones 17 and 4 (as the protonated forms), CP–17 and CP–4, respectively, it could be seen that when the molecule occupies the active-cleft with the benzofuroxan moiety directed to the bottom of the S2 subsite, in the case of 17, the Glu205 sidechain adopted a conformation oriented toward the solvent (Fig. 2c). However, when the parent ligand 4 was docked at that site the Glu205 sidechain was oriented toward the inhibitor and interacted with a guanidine nitrogen (Fig. 2d, Table 6). The ability of parent compound 4 to adopt this conformation in the active site of the enzyme could explain its high performance as a CP inhibitor. These preferential orientations also explained why there was no relative improvement in the inhibitory activities of amidinohydrazone 17 and thiosemicarbazone 19 at pH 5.3.
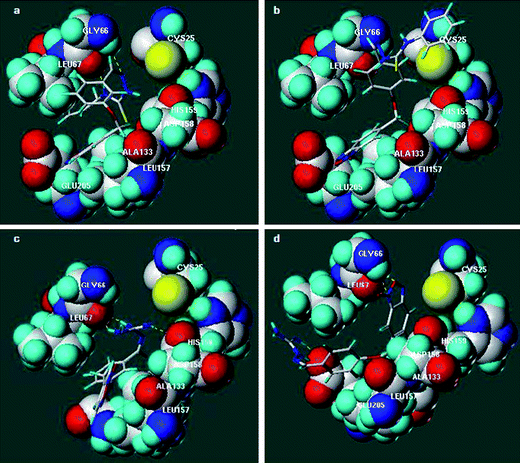 | ||
| Fig. 2 Position of some representative derivatives in the binding-cleft of CP determined by docking. a. CP–19. b. CP–16. c. CP–17. d. CP–4. | ||
Experimental section
Chemistry
5-(3-Formylphenyloxymethyl)benzo[1,2-c]1,2,5-oxadiazole N-oxide (9). 5-Bromomethylbenzofuroxan (1 eq.) was added to a solution of 3-hydroxybenzaldehyde (200 mg, 1.64 mmol) in dry CH3CN and K2CO3 (1 eq.). The reaction mixture was stirred at room temperature under nitrogen atmosphere and checked by TLC (Al2O3, petroleum ether–AcOEt, 7
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) until the disappearance of the reactants. The solvent was evaporated in vacuo and the reaction mixture was partitioned between H2O (50.0 mL) and EtOAc (3 × 50 mL). The organic phase was dried over Na2SO4, filtered and concentrated in vacuo to obtain the desired product. Yellow solid (80%), mp 145.8 °C. 1H NMR (CDCl3, 400 MHz) δ (ppm): 5.16 (s, 2H), 7.31 (m, 1H), 7.36–7.69 (bs, 3H), 7.49 (s, 1H), 7.53 (d, 1H, J = 7.6 Hz), 7.55 (t, 1H, J1 = 2.0 Hz, J2 = 1.2 Hz), 10.00 (s, 1H). 13C NMR (CDCl3, 100 MHz) δ (ppm): 68.7 (CH2), 112.4 (Ar), 122.2 (Ar), 124.9 (Ar), 130.5 (Ar), 137.9 (C–C = N-NH-), 158.4 (C-O-CH2), 191.8 (CHO). MS (EI), m/z (abundance, %): 270 (M+•, 2), 254 (10), 149 (19), 133 (76), 121 (15), 77 (27). (Found: C, 61.9; H, 3.5; N, 10.2. C14H10N2O4 requires C, 62.2; H, 3.7; N, 10.4%)
3) until the disappearance of the reactants. The solvent was evaporated in vacuo and the reaction mixture was partitioned between H2O (50.0 mL) and EtOAc (3 × 50 mL). The organic phase was dried over Na2SO4, filtered and concentrated in vacuo to obtain the desired product. Yellow solid (80%), mp 145.8 °C. 1H NMR (CDCl3, 400 MHz) δ (ppm): 5.16 (s, 2H), 7.31 (m, 1H), 7.36–7.69 (bs, 3H), 7.49 (s, 1H), 7.53 (d, 1H, J = 7.6 Hz), 7.55 (t, 1H, J1 = 2.0 Hz, J2 = 1.2 Hz), 10.00 (s, 1H). 13C NMR (CDCl3, 100 MHz) δ (ppm): 68.7 (CH2), 112.4 (Ar), 122.2 (Ar), 124.9 (Ar), 130.5 (Ar), 137.9 (C–C = N-NH-), 158.4 (C-O-CH2), 191.8 (CHO). MS (EI), m/z (abundance, %): 270 (M+•, 2), 254 (10), 149 (19), 133 (76), 121 (15), 77 (27). (Found: C, 61.9; H, 3.5; N, 10.2. C14H10N2O4 requires C, 62.2; H, 3.7; N, 10.4%)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, as eluent).
3, as eluent).
5-(2-Formylphenyloxymethyl)benzo[1,2-c]1,2,5-oxadiazole N-oxide (10). Yellow solid (65%), mp 146.0 °C (d). 1H NMR (CDCl3, 400 MHz) δ (ppm): 5.37 (s, 2H), 7.15 (t, 1H, J = 7.5 Hz), 7.33 (d, 1H, J = 8.4 Hz), 7.69 (t, 1H, J1 = 8.6 Hz, J2 = 7.4 Hz), 7.57–7.77 (bs, 3H), 7.76 (d, 1H, J = 7.5 Hz), 10.50 (s, 1H). 13C NMR (CDCl3, 100 MHz) δ (ppm): 69.5 (CH2), 114.8 (Ar), 122.2 (Ar), 125.5 (C–C = N–NH–), 129.3 (Ar), 137.2 (Ar), 160.7 (C-O-CH2), 190.2 (CHO). MS (EI), m/z (abundance, %): 270 (M+•, 2), 253 (30), 149 (100), 133 (49), 121 (38), 105 (11), 89 (47), 77 (12). (Found: C, 62.0; H, 3.3; N, 10.1. C14H10N2O4 requires C, 62.2; H, 3.7; N, 10.4%).
5-(2-Acetylphenyloxymethyl)benzo[1,2-c]1,2,5-oxadiazole N-oxide (11). Yellow solid (32%), mp 125.7–126.4 °C. 1H NMR (acetone-d6, 400 MHz) δ (ppm): 2.59 (s, 3H), 5.41 (s, 2H), 7.09 (dt, 1H, J1 = 7.2 Hz, J2 = 7.8 Hz, J1 = 0.8 Hz, J2 = 0.4 Hz), 7.29 (d, 1H, J = 8.4 Hz), 7.54 (m, 1H), 7.58–7.68 (bs, 3H), 7.71 (dd, 1H, J = 7.6 Hz, J = 1.6 Hz). 13C NMR (acetone-d6, 100 MHz) δ (ppm): 31.1 (CH3), 69.2 (CH2), 113.4 (Ar), 121.1 (Ar), 128.9 (C–C = N–NH–), 130.0 (Ar), 133.5 (Ar), 157.3 (C-O-CH2), 198.1 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). MS (ESI), m/z: 307 (M+• + Na+). (Found: C, 63.7; H, 4.2; N, 9.5. C15H12N2O4 requires C, 63.4; H, 4.2; N, 9.8%).
O). MS (ESI), m/z: 307 (M+• + Na+). (Found: C, 63.7; H, 4.2; N, 9.5. C15H12N2O4 requires C, 63.4; H, 4.2; N, 9.8%).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). The precipitate was filtered and washed with ethanol.
4). The precipitate was filtered and washed with ethanol.
5-(3-Amidinohydrazonophenyloxymethyl)benzo[1,2-c]1,2,5-oxadiazole N-oxide (17). Pale brown solid (85%), mp 155.2 °C (d). 1H NMR (DMSO-d6, 400 MHz) δ (ppm): 5.23 (s, 2H), 7.11 (s, 1H), 7.37 (m, 2H), 7.46–7.89 (bs, 3H), 7.61 (m, 2H), 7.96 (s, 1H), 8.08 (s, 1H). 13C NMR (CDCl3, 100 MHz) δ (ppm): 68.44 (CH2), 113.1 (Ar), 117.9 (Ar), 121.9 (Ar), 130.4 (Ar), 147.0 (CH), 135.3 (C–C = N–NH–), 155.7 (C = NH), 158.7 (C-O-CH2). MS (EI), m/z (abundance, %): 326 (M+•, 5), 310 (7), 252 (10), 237(9), 221 (12), 177 (26), 161 (75), 133 (46), 119 (60), 107(30), 73 (100). (Found: C, 54.9; H, 4.0; N, 25.4. C15H14N6O3 requires C, 55.2; H, 4.3; N, 25.7%).
5-(2-Amidinohydrazonophenyloxymethyl)benzo[1,2-c]1,2,5-oxadiazole N-oxide (20). Yellow solid (77%), 212.2 °C (d). 1H NMR (CDCl3, 400 MHz) δ (ppm): 5.23 (s, 2H), 5.31 (s, 2H), 5.89 (s, 2H), 6.97 (t, 1H, J1 = 7.6 Hz, J2 = 7.2 Hz), 7.12 (t, 1H, J1 = 8.4 Hz, J2 = 9.2 Hz), 7.27 (t, 1H, J1 = 7.2 Hz, J2 = 8.0 Hz), 7.49–7.78 (bs, 3H), 8.00 (d, 1H, J = 7.6 Hz), 8.39 (s, 1H). 13C NMR (CDCl3, 100 MHz) δ (ppm): 69.2 (CH2), 113.7 (Ar), 122.0 (Ar), 126.3 (C–C = N–NH–), 126.5 (Ar), 129.8 (Ar), 139.0 (Ar), 156.0 (C-O-CH2), 161.3 (C = NH). MS (EI), m/z (abundance, %): 326 (M+•, 5), 310 (23), 252 (14), 237(9), 221 (11), 177 (34), 161 (82), 133 (41), 119 (64), 107(28), 73 (100). (Found: C, 55.3; H, 4.6; N, 25.3. C15H14N6O3 requires C, 55.2; H, 4.3; N, 25.7%).
5-(2-Methylamidinohydrazonophenyloxymethyl)benzo [1,2-c]1,2,5-oxadiazole N-oxide (25). Pale yellow solid (67%), 129.0 °C (d). 1H NMR (DMSO-d6, 400 MHz) δ (ppm): 2.56 (s, 3H), 5.32 (s, 2H), 7.08 (t, 3H, J1 = 7.2 Hz, J2 = 7.6 Hz), 7.27 (d, 1H, J = 8.4 Hz), 7.52–7.89 (bs, 3H), 7.57 (dt, 1H, J1 = 7.2 Hz, J2 = 8.4 Hz, J = 2.0 Hz), 7.65 (dd, 1H, J = 7.6 Hz, J = 1.6 Hz). 13C NMR (DMSO-d6, 100 MHz) δ (ppm): 40.2 (CH3), 69.2 (CH2), 114.1 (Ar), 121.5 (Ar), 128.6 (C–C = N–NH–), 130.2 (Ar), 134.3 (Ar), 155.8 (HNC=NH), 157.2 (C–C = N-NH-), 167.2 (CH3C=N). MS (EI), m/z (abundance, %): 267 (M+• - O - CH3N3, 54), 250 (7), 205 (9), 149 (61), 133 (100), 121 (50), 103 (21), 89 (48), 77 (26). IR, ν/cm−1: 3355, 3129, 3045, 2907, 1668, 1591, 1539, 1491, 1356, 1300, 1111, 774, 616. (Found: C, 56.3; H, 5.0; N, 24.6. C16H16N6O3 requires C, 56.5; H, 4.7; N, 24.7%).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). The precipitate was filtered and washed with ethanol.
4). The precipitate was filtered and washed with ethanol.
5-[4-(N4-Allylthiosemicarbazono)phenyloxymethyl]benzo[1,2-c]1,2,5-oxadiazole N-oxide, (15). Pale brown solid (73%), mp 176.7 °C (d). 1H NMR (DMSO-d6), 400 MHz) δ (ppm): 4.21 (t, 2H, J1 = 5.2 Hz, J2 = 5.6 Hz), 5.10 (dd, 1H, Jgem = 1.6 Hz, Jcis = 10.2 Hz), 5.15 (dd, 1H, Jgem = 1.6 Hz, Jtrans = 17.2 Hz), 5.25 (s, 2H), 5.91 (m, 1H), 7.11 (d, 1H, J = 8.8 Hz), 7.42–7.68 (bs, 3H), 7.79 (d, 1H, J = 8.8 Hz), 8.02 (s, 1H), 8.65 (t, 1H, J = 6.0 Hz), 11.40 (s, 1H). 13C NMR (DMSO-d6, 100 MHz) δ (ppm): 46.2 (CH2), 69.6 (CH2), 115.5 (Ar), 115.9 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 127.9 (C–C = N–NH–), 129.4 (Ar), 135.6 (C = C), 142.2, (C = NH), 159.6 (C-O-CH2), 177.4 (C = S). MS (ESI), m/z: 429 (M + 2Na+). IR, ν/cm−1: 3300, 3185, 3008, 2914, 1603, 1536, 1418, 1236, 1039, 927, 852, 743, 574. (Found: C, 56.6; H, 4.1; N, 18.0. C18H17N5O3S requires C, 56.4; H, 4.5; N, 18.3%).
C), 127.9 (C–C = N–NH–), 129.4 (Ar), 135.6 (C = C), 142.2, (C = NH), 159.6 (C-O-CH2), 177.4 (C = S). MS (ESI), m/z: 429 (M + 2Na+). IR, ν/cm−1: 3300, 3185, 3008, 2914, 1603, 1536, 1418, 1236, 1039, 927, 852, 743, 574. (Found: C, 56.6; H, 4.1; N, 18.0. C18H17N5O3S requires C, 56.4; H, 4.5; N, 18.3%).
5-[4-(N4-Phenylthiosemicarbazono)phenyloxymethyl] benzo[1,2-c]1,2,5-oxadiazole N-oxide (16). Yellow solid (51%), mp 180.0 °C (d). 1H NMR (DMSO-d6, 400 MHz) δ (ppm): 5.26 (s, 2H), 7.13 (d, 2H, J = 8.8 Hz), 7.21 (t, 1H, J1 = 7.6 Hz, J2 = 7.2 Hz), 7.37 (t, 2H, J1 = 7.6 Hz, J2 = 8.4 Hz), 7.56 (d, 2H, J = 7.6 Hz), 7.60–7.84 (bs, 3H), 7.89 (d, 2H, J = 8.8 Hz), 8.11 (s, 1H), 10.10 (s, 1H). 13C NMR (DMSO-d6, 100 MHz) δ (ppm): 68.5 (CH2), 115.5 (Ar), 125.7 (Ar), 126.3 (Ar), 127.7 (C–C = N–NH–), 128.5 (Ar), 129.8 (Ar), 139.6 (Ar), 143.1 (C = NH), 159.8 (C-O-CH2), 176.1 (C = S). MS (EI), m/z (abundance, %): 401 (M+• - H2O, 13), 268 (17), 252 (21), 237 (40), 151 (33), 133 (100), 119 (39), 93 (90). IR, ν/cm−1: 3246, 3141, 1689, 1600, 1539, 1369, 1247, 851, 743, 576. (Found: C, 59.9; H, 3.9; N, 16.5. C21H17N5O3S requires C, 60.1; H, 4.1; N, 16.7%).
5-(3-Thiosemicarbazonophenyloxymethyl)benzo[1,2-c]1,2,5-oxadiazole N-oxide (18). Pale brown solid (91%), mp 188.7 °C (d). 1H NMR (DMSO-d6, 400 MHz) δ (ppm): 5.24 (s, 2H), 7.10 (d, 1H, J = 7.4 Hz), 7.34 (m, 2H), 7.50–7.82 (bs, 3H), 7.62 (s, 1H), 7.95 (s, 1H), 8.10 (s, 1H), 8.28 (s, 1H), 11.50 (s, 1H). 13C NMR (DMSO-d6, 100 MHz) δ (ppm): 68.6 (CH2), 112.5 (Ar), 117.3 (Ar), 121.7 (Ar), 130.4 (Ar), 136.2 (C–C = N–NH–), 142.3 (C = NH), 158.6 (C-O-CH2), 162.8 (Bfx), 178.5 (C = S). MS (EI), m/z (abundance, %): 327 (M+• - 16, 8), 311 (3), 269 (10), 252 (21), 237 (25), 151 (30), 133 (100), 121 (39), 103 (20). IR ν: 3441, 3293, 3163, 3025, 1586, 1538, 1279, 1059, 941, 859, 790, 689. (Found: C, 52.5; H, 3.6; N, 20.2. C15H13N5O3S requires C, 52.5; H, 3.8; N, 20.4%).
5-[3-(N4-Allylthiosemicarbazono)phenyloxymethyl]benzo[1,2-c]1,2,5-oxadiazole N-oxide (19). Pale brown solid (80%), mp 149.6 °C (d). 1H NMR (DMSO-d6), 400 MHz) δ (ppm): 4.23 (t, 2H, J1 = 4.8 Hz, J2 = 5.2 Hz), 5.10 (dd, 1H, Jgem = 1.2 Hz, Jcis = 12 Hz), 5.16 (dd, 1H, Jgem = 1.2 Hz, Jtrans = 18 Hz), 5.24 (s, 2H), 5.93 (m, 1H), 7.12 (d, 1H, J = 6.8 Hz), 7.38 (d, 2H, J = 6.8 Hz), 7.56 (s, 1H), 7.64–7.88 (bs, 3H), 8.04 (s, 1H), 8.74 (t, 1H, J = 5.2 Hz), 11.60 (s, 1H). 13C NMR (DMSO-d6, 100 MHz) δ (ppm): 46.2 (CH2), 68.6 (CH2), 113.5 (Ar), 116.0 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 116.6 (Ar), 121.3 (Ar), 130.4 (Ar), 135.5 (C
C), 116.6 (Ar), 121.3 (Ar), 130.4 (Ar), 135.5 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 136.2 (C–C = N–NH–), 142.2, (C = NH), 158.6 (C-O-CH2), 177.7 (C = S). MS (ESI), m/z: 429 (M + 2Na+). (Found: C, 56.4; H, 4.9; N, 18.1. C18H17N5O3S requires C, 56.4; H, 4.5; N, 18.3%).
C), 136.2 (C–C = N–NH–), 142.2, (C = NH), 158.6 (C-O-CH2), 177.7 (C = S). MS (ESI), m/z: 429 (M + 2Na+). (Found: C, 56.4; H, 4.9; N, 18.1. C18H17N5O3S requires C, 56.4; H, 4.5; N, 18.3%).
5-(2-Thiosemicarbazonophenyloxymethyl)benzo[1,2-c]1,2,5-oxadiazole N-oxide (21). Yellow solid (79%), mp 215.0 °C (d). 1H NMR (DMSO-d6, 400 MHz) δ (ppm): 5.27 (s, 2H), 7.03 (dd, 1H, J1 = 7.6 Hz, J2 = 7.4 Hz), 7.20 (d, 1H, J1 = 8.4 Hz), 7.42 (dd, 1H, J1 = 7.4 Hz, J2 = 8.2 Hz), 7.49–7.79 (bs, 3H), 7.96 (s, 1H), 8.15 (s, 1H), 8.16 (d, 1H, 3J = 7.6 Hz), 8.62 (s, 1H), 11.50 (s, 1H). 13C NMR (DMSO-d6, 100 MHz) δ (ppm): 69.3 (CH2), 114.0 (Ar), 122.2 (Ar), 123.6 (C–C = N-NH-), 127.1 (Ar), 132.2 (Ar), 138.9 (CH), 157.1 (C-O-CH2), 163.2 (Bfx), 178.7 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). MS (EI), m/z (abundance, %): 327 (M+• - 16, 12), 311 (2), 269 (12), 252 (21), 237 (26), 151 (36), 133 (100), 121 (39), 103 (26). (Found: C, 52.2; H, 3.5; N, 20.1. C15H13N5O3S requires C, 52.5; H, 3.8; N, 20.4%).
S). MS (EI), m/z (abundance, %): 327 (M+• - 16, 12), 311 (2), 269 (12), 252 (21), 237 (26), 151 (36), 133 (100), 121 (39), 103 (26). (Found: C, 52.2; H, 3.5; N, 20.1. C15H13N5O3S requires C, 52.5; H, 3.8; N, 20.4%).
5-[2-(N4-Allylthiosemicarbazono)phenyloxymethyl]benzo[1,2-c]1,2,5-oxadiazole N-oxide (22). Pale brown solid (73%), mp 191.4 °C (d). 1H NMR (MeOH-d4, 400 MHz) δ (ppm): 4.36 (m, 2H), 5.11 (dd, 1H, Jgem = 1.6 Hz, Jcis = 10.0 Hz), 5.23 (dd, 1H, Jgem = 1.6 Hz, Jtrans = 17.2 Hz), 5.36 (s, 2H), 6.00 (m, 1H), 7.05 (t, 1H, J = 7.6 Hz), 7.24 (d, 1H, J = 8.0 Hz), 7.43 (dt, 1H, J1 = 7.2 Hz, J2 = 8.4 Hz, J = 1.6 Hz), 7.56–7.78 (bs, 3H), 8.06 (dd, 1H, J = 7.6 Hz, J = 1.6 Hz), 8.38 (s, 1H), 8.77 (s, 1H), 10.50 (s, 1H). 13C NMR (MeOH-d4, 100 MHz) δ (ppm): 45.9 (CH2), 69.3 (CH2), 113.5 (Ar), 115.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 121.9 (Ar), 123.6 (C–C = N–NH–), 126.5 (Ar), 131.7 (Ar), 135.2 (C
C), 121.9 (Ar), 123.6 (C–C = N–NH–), 126.5 (Ar), 131.7 (Ar), 135.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 138.0, (CH), 157.2 (C-O-CH2), 176.7 (C
C), 138.0, (CH), 157.2 (C-O-CH2), 176.7 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). MS (EI), m/z (abundance, %): 383 (M+•, 0.3), 366 (5), 268 (19), 254 (21), 237 (10), 133 (36), 115 (100). (Found: C, 56.1; H, 4.3; N, 17.9. C18H17N5O3S requires C, 56.4; H, 4.5; N, 18.3%).
S). MS (EI), m/z (abundance, %): 383 (M+•, 0.3), 366 (5), 268 (19), 254 (21), 237 (10), 133 (36), 115 (100). (Found: C, 56.1; H, 4.3; N, 17.9. C18H17N5O3S requires C, 56.4; H, 4.5; N, 18.3%).
5-[2-(N4-Phenylthiosemicarbazono)phenyloxymethyl]benzo[1,2-c]1,2,5-oxadiazole N-oxide (23). Pale brown solid (84%), mp 164.3 °C (d). 1H NMR (MeOH-d4, 400 MHz) δ (ppm): 5.38 (s, 2H), 7.08 (t, 1H, J = 7.6 Hz), 7.20 (t, 1H, J1 = 7.6 Hz, J2 = 7.2 Hz), 7.27 (d, 1H, J = 8.4 Hz), 7.37 (t, 2H, J = 8.0 Hz), 7.46 (dt, 1H, J = 7.6 Hz, J = 1.6 Hz), 7.52–7.72 (bs, 3H), 7.77 (t, 1H, J = 7.6 Hz), 8.20 (dd, 1H, J = 7.6 Hz, J = 1.6 Hz), 8.86 (s, 1H), 9.88 (s, 1H), 10.70 (s, 1H). 13C NMR (MeOH-d4, 100 MHz) δ (ppm): 69.4 (CH2), 113.5 (Ar), 122.0 (Ar), 123.4 (C–C = N–NH–), 125.1 (Ar), 125.5 (Ar), 127.0 (Ar), 128.5 (Ar), 131.8 (Ar), 138.7 (CH), 139.4 (Ar), 157.4 (C-O-CH2), 176.90 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). MS (EI), m/z (abundance, %): 419 (M+•, 0.2), 401 (10), 268 (19), 252 (17), 237 (44), 151 (33), 133 (94), 119 (29), 93 (100). (Found: C, 60.3; H, 4.4; N, 16.4. C21H17N5O3S requires C, 60.1; H, 4.1; N, 16.7%).
S). MS (EI), m/z (abundance, %): 419 (M+•, 0.2), 401 (10), 268 (19), 252 (17), 237 (44), 151 (33), 133 (94), 119 (29), 93 (100). (Found: C, 60.3; H, 4.4; N, 16.4. C21H17N5O3S requires C, 60.1; H, 4.1; N, 16.7%).
5-(2-Semicarbazonophenyloxymethyl)benzo[1,2-c]1,2,5-oxadiazole N-oxide (24). Pale brown solid (56%), mp 237.6 °C (d). 1H NMR (DMSO-d6, 400 MHz) δ (ppm): 5.26 (s, 2H), 6.44 (s, 2H), 7.02 (t, 1H, J1 = 7.2 Hz, J2 = 7.6 Hz), 7.17 (d, 1H, J = 8.8 Hz), 7.36 (t, 1H, J1 = 8.6 Hz, J2 = 7.6 Hz), 7.48–7.83 (bs, 3H), 7.49 (s, 1H), 8.04 (d, 1H, J1 = 7.2 Hz), 8.36 (s, 1H), 10.32 (s, 1H). 13C NMR (DMSO-d6, 100 MHz) δ (ppm): 69.3 (CH2), 114.0 (Ar), 122.2 (Ar), 124.3 (C–C = N–NH–), 126.6 (Ar), 131.2 (Ar), 135.6 (C = NH), 156.5 (C-O-CH2), 157.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). MS (EI), m/z (abundance, %): 311 (5), 252 (13), 237 (69), 221 (10), 133 (100), 121 (26), 107 (11). (Found: C, 54.8; H, 3.8; N, 21.1. C15H13N5O4 requires C, 55.0; H, 4.0; N, 21.4%).
O). MS (EI), m/z (abundance, %): 311 (5), 252 (13), 237 (69), 221 (10), 133 (100), 121 (26), 107 (11). (Found: C, 54.8; H, 3.8; N, 21.1. C15H13N5O4 requires C, 55.0; H, 4.0; N, 21.4%).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4).
4).
5-(4-Formylphenyloxymethyl)-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (12). Bordeaux solid (15%), mp 139.4–139.9 °C. 1H NMR (acetone-d6, 400 MHz) δ (ppm): 1.62 (s, 6H), 5.19 (s, 2H), 7.10 (d, 1H, J = 9.6 Hz), 7.28 (m, 4H), 7.93 (d, 2H, J = 8.8 Hz), 9.94 (s, 1H). 13C NMR (acetone-d6, 100 MHz) δ (ppm): 23.5 (CH3), 68.6 (CH2), 97.0 (C(CH3)2), 112.9 (Ar), 115.2 (Ar), 115.8 (Ar), 130.2 (Ar), 130.8 (Ar), 131.7 (Ar), 135.8 (C–C = N-NH-), 139.9 (Bfx), 163.1 (C-O-CH2), 190.4 (CHO). MS (EI), m/z (abundance, %): 312 (M+•, 37), 296 (40), 191 (32), 175 (61), 144 (52), 133 (100), 121 (7). (Found: C, 65.5; H, 5.1; N, 8.9. C17H16N2O4 requires C, 65.4; H, 5.2; N, 9.0%).
5-(3-Formylphenyloxymethyl)-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (13). Bordeaux solid (7%), mp 129.5–130.7 °C. MS (EI), m/z (abundance, %): 312 (M+•, 27), 296 (53), 265 (5), 191 (30), 175 (85), 144 (76), 133 (100), 121 (12). (Found: C, 65.2; H, 4.8; N, 8.7. C17H16N2O4 requires C, 65.4; H, 5.2; N, 9.0%).
5-(2-Acetylphenyloxymethyl)-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (14). Bordeaux solid (8%), mp 112.5–113.4 °C. 1H NMR (acetone-d6, 400 MHz) δ (ppm): 1.62 (s, 6H), 1.57 (s, 3H), 5.64 (s, 2H), 6.73 (d, 1H, J = 9.6 Hz), 7.30 (d, 1H, J = 9.6 Hz), 7.43 (t, 1H, J1 = 6.2, J2 = 7.4), 7.55 (dd, 2H, J = 8.4 Hz, J1 = 6.0 Hz, J2 = 4.4 Hz), 7.87 (s, 1H), 7.93 (d, 1H, J = 9.2 Hz). 13C NMR (acetone-d6, 100 MHz) δ (ppm): 23.6 (CH3), 68.5 (CH2), 96.7 (C(CH3)2), 112.2 (Ar), 113.2 (Ar), 116.1 (Ar), 121.3 (Ar), 123.9 (Ar), 128.7 (C–C = N-NH-), 130.7 (Ar), 136.2 (Ar), 140.6 (Bfx), 157.4 (C-O-CH2), 198.0 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). (Found: C, 65.9; H, 5.4; N, 8.4. C18H18N2O4 requires C, 66.2; H, 5.6; N, 8.6%).
O). (Found: C, 65.9; H, 5.4; N, 8.4. C18H18N2O4 requires C, 66.2; H, 5.6; N, 8.6%).
5-(4-Thiosemicarbazonophenyloxymethyl)-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (27). Bordeaux solid (34%), mp 122.2–123.0 °C. 1H NMR (CDCl3, 400 MHz) δ (ppm): 1.57 (s, 6H), 5.06 (s, 2H), 7.07 (m, 3H), 7.27 (d, 2H, J = 8.8 Hz), 7.77 (d, 2H, J = 8.8 Hz), 7.91 (s, 1H), 8.00 (s, 1H), 8.09 (s, 1H), 11.31 (s, 1H). 13C NMR (DMSO-d6, 100 MHz) δ (ppm): 24.5 (CH3), 68.8 (CH2), 97.6 (C(CH3)2), 113.5 (Ar), 115.9 (Ar), 116.6 (Ar), 128.3 (Ar), 129.8 (Ar), 131.7 (Ar), 136.4 (C–C = N-NH-), 141.5 (Bfx), 142.9 (CH), 160.0 (C-O-CH2), 178.61(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). MS (EI), m/z (abundance, %): 354 (M+• - 16 – 15, 3), 333 (10), 322 (16), 195 (40), 149 (73), 131(20), 121 (75), 57 (100). (Found: C, 55.8; H, 4.9; N, 18.0. C18H19N5O3S requires C, 56.1; H, 5.0; N, 18.2%).
S). MS (EI), m/z (abundance, %): 354 (M+• - 16 – 15, 3), 333 (10), 322 (16), 195 (40), 149 (73), 131(20), 121 (75), 57 (100). (Found: C, 55.8; H, 4.9; N, 18.0. C18H19N5O3S requires C, 56.1; H, 5.0; N, 18.2%).
5-[4-(N4-Allylthiosemicarbazono)phenyloxymethyl]-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (28). Bordeaux solid (31%), mp 121.2–122.1 °C. 1H NMR (acetone-d6, 400 MHz) δ (ppm): 1.62 (s, 6H), 4.35 (t, 2H, J1 = 4.6 Hz, J2 = 5.0 Hz), 5.10 (s, 2H), 5.11 (dd, 1H, Jgem = 1.6 Hz, Jcis = 10.0 Hz), 5.22 (dd, 1H, Jgem = 1.6 Hz, Jtrans = 17.2 Hz), 5.98 (m, 1H), 6.90 (d, 1H, J = 8.8 Hz), 7.11 (dd, 2H, J = 6.8 Hz, J = 2.0 Hz), 7.23 (d, 1H, J = 9.6 Hz), 7.63 (d, 1H, J = 8.8 Hz), 7.76 (d, 2H, J = 8.8 Hz), 8.15 (s, 1H). 13C NMR (acetone-d6, 100 MHz) δ (ppm): 23.5 (CH3), 46.0 (CH2), 68.3 (CH2), 96.9 (C(CH3)2), 112.7 (Ar), 115.1 (Ar), 115.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 115.7 (Ar), 125.9 (Ar), 127.7 (C–C = N-NH-), 128.8 (Ar), 128.9 (Ar), 130.2 (Ar), 134.9 (C
C), 115.7 (Ar), 125.9 (Ar), 127.7 (C–C = N-NH-), 128.8 (Ar), 128.9 (Ar), 130.2 (Ar), 134.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 140.3 (Bfx), 142.4 (CH), 159.8 (C-O-CH2), 176.7 (C
C), 140.3 (Bfx), 142.4 (CH), 159.8 (C-O-CH2), 176.7 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). MS (EI), m/z (abundance, %): 396 (M+• - C2H5, 4), 333 (10), 278 (17), 249 (3), 219 (15), 193 (12), 149 (62), 131(36), 57 (100). (Found: C, 59.1; H, 5.1; N, 16.2. C21H23N5O3S requires C, 59.3; H, 5.4; N, 16.5%).
S). MS (EI), m/z (abundance, %): 396 (M+• - C2H5, 4), 333 (10), 278 (17), 249 (3), 219 (15), 193 (12), 149 (62), 131(36), 57 (100). (Found: C, 59.1; H, 5.1; N, 16.2. C21H23N5O3S requires C, 59.3; H, 5.4; N, 16.5%).
5-[4-(N4-Phenylthiosemicarbazono)phenyloxymethyl]-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (29). Bordeaux solid (13%), mp 118.3–119.2 °C. 1H NMR (acetone-d6, 400 MHz) δ (ppm): 1.62 (s, 6H), 5.12 (s, 2H), 7.08 (d, 1H, J = 8.0 Hz), 7.14 (dd, 2H, J = 8.4 Hz, J1 = 2.0 Hz, J2 = 1.6 Hz), 7.22 (t, 2H, J1 = 9.0 Hz, J2 = 11 Hz), 7.29 (s, 1H), 7.37 (t, 2H, J1 = 7.6 Hz, J2 = 8.0 Hz), 7.76 (d, 2H, J = 7.6 Hz), 7.86 (d, 2H, J = 8.8 Hz), 8.23 (s, 1H). 13C NMR (acetone-d6, 100 MHz) δ (ppm): 23.5 (CH3), 68.3 (CH2), 96.9 (C(CH3)2), 112.7 (Ar), 115.1 (Ar), 115.7 (Ar), 124.6 (Ar), 125.0 (Ar), 128.1 (Ar), 129.2 (Ar), 130.2 (Ar), 135.8 (C–C = N-NH-), 140.4 (Bfx), 142.4 (CH), 159.9 (C-O-CH2), 176.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). MS (EI), m/z (abundance, %): 412 (M+• - 2 × 17 – 15, 5), 283 (3), 271(20), 207 (10), 177 (5), 145 (100), 135 (27), 93 (52). (Found: C, 62.2; H, 5.2; N, 14.9. C24H23N5O3S requires C, 62.5; H, 5.0; N, 15.2%).
S). MS (EI), m/z (abundance, %): 412 (M+• - 2 × 17 – 15, 5), 283 (3), 271(20), 207 (10), 177 (5), 145 (100), 135 (27), 93 (52). (Found: C, 62.2; H, 5.2; N, 14.9. C24H23N5O3S requires C, 62.5; H, 5.0; N, 15.2%).
5-(4-Semicarbazonophenyloxymethyl)-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (30). Bordeaux solid (25%), mp 123.3-124.1 °C. 1H NMR (DMSO-d6, 400 MHz) δ (ppm): 1.57 (s, 6H), 5.05 (s, 2H), 6.39 (s, 2H), 7.04 (m, 3H), 7.26 (d, 2H, J = 8.8 Hz), 7.68 (d, 2H, J = 8.8 Hz), 7.80 (s, 1H), 10.08 (s, 1H). (Found: C, 58.2; H, 4.9; N, 18.9. C18H19N5O4 requires C, 58.5; H, 5.2; N, 19.0%).
5-(3-Thiosemicarbazonophenyloxymethyl)-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (32). Bordeaux solid (20%), mp 116.4–117.2 °C. 1H NMR (acetone-d6, 400 MHz) δ (ppm): 1.62 (s, 6H), 5.09 (s, 2H), 7.08 (d, 1H, J = 9.6 Hz), 7.13 (m, 1H), 7.24 (2, 1H, J = 9.2), 7.55 (t, 1H, J = 1.2 Hz), 7.39 (m, 2H), 7.59 (d, 1H, J = 2.0 Hz), 8.17 (s, 1H). 13C NMR (acetone-d6, 100 MHz) δ (ppm): 23.5 (CH3), 68.3 (CH2), 96.9 (C(CH3)2), 112.5 (Ar), 112.6 (Ar), 115.7 (Ar), 116.7 (Ar), 121.1 (Ar), 129.9 (Ar), 130.3 (Ar), 136.0 (C–C = N-NH-), 140.5 (Bfx), 142.1 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH), 158.7 (C-O-CH2), 178.7 (C
NH), 158.7 (C-O-CH2), 178.7 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). MS (EI), m/z (abundance, %): 354 (M+• - 16 – 15, 6), 333 (13), 195 (29), 177 (8), 149 (100), 131 (10), 121 (32), 57 (11). (Found: C, 56.0; H, 5.0; N, 17.8. C18H19N5O3S requires C, 56.1; H, 5.0; N, 18.2%).
S). MS (EI), m/z (abundance, %): 354 (M+• - 16 – 15, 6), 333 (13), 195 (29), 177 (8), 149 (100), 131 (10), 121 (32), 57 (11). (Found: C, 56.0; H, 5.0; N, 17.8. C18H19N5O3S requires C, 56.1; H, 5.0; N, 18.2%).
5-(2-Amidinohydrazonophenyloxymethyl)-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (33). Bordeaux solid (22%), mp 125.7–126.6 °C. 1H NMR (acetone-d6, 400 MHz) δ (ppm): 1.62 (s, 6H), 5.24 (s, 2H), 7.16 (m, 2H), 7.25 (d, 1H, J = 9.6 Hz), 7.34 (m, 2H), 7.67 (dt, 1H, J1 = 7.6 Hz, J2 = 8.2 Hz, J1 = 1.6 Hz, J2 = 2.0 Hz), 7.81(dd, 1H, J = 7.6 Hz, J = 1.6 Hz), 7.97 (bs, 4H), 10.6 (s, 1H). MS (EI), m/z (abundance, %): 368 (M+•, 3), 354 (M+• −14, 7), 296 (18), 278 (24), 175 (77), 145 (83), 133 (84), 121 (100), 69 (91). (Found: C, 58.6; H, 5.2; N, 22.7. C18H20N6O3 requires C, 58.7; H, 5.5; N, 22.8%).
5-(2-Thiosemicarbazonophenyloxymethyl)-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (34). Bordeaux solid (30%), 124.5–125.8 °C. 1H NMR (CDCl3, 400 MHz) δ (ppm): 1.63 (s, 6H), 5.15 (s, 2H), 7.04 (s, 2H), 7.12 (d, 1H, J = 9.6 Hz), 7.22 (m, 2H), 7.40 (m, 3H), 7.98 (s, 1H), 8.76 (s, 1H). MS (EI), m/z (abundance, %): 354 (M+• - 16 – 15, 5), 333 (25), 195 (7), 177 (7), 149 (85), 131(20), 121 (100), 57 (69). (Found: C, 55.7; H, 4.7; N, 18.3. C18H19N5O3S requires C, 56.1; H, 5.0; N, 18.2%).
5-[2-(N4-Allylthiosemicarbazono)phenyloxymethyl]-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (35). Bordeaux solid (20%), mp 114.7–115.0 °C. 1H NMR (acetone-d6, 400 MHz) δ (ppm): 1.62 (s, 6H), 4.37 (t, 2H, J = 5.6 Hz), 5.11 (dd, 1H, Jgem = 1.6 Hz, Jcis = 10.0 Hz), 5.14 (s, 2H), 5.24 (dd, 1H, Jgem = 1.6 Hz, Jtrans = 17.2 Hz), 6.00 (m, 1H), 7.03 (t, 1H, J1 = 7.2 Hz, J2 = 8.0 Hz), 7.12 (d, 1H, J = 9.6 Hz), 7.21(dd, 2H, J = 11.8 Hz, J = 3.6 Hz), 7.31(s, 1H), 7.41(dt, 1H, J1 = 7.6 Hz, J2 = 8.0 Hz, J = 1.6 Hz), 8.04 (dd, 1H, J = 8.0 Hz, J = 1.6 Hz), 8.37 (s, 1H), 8.72 (s, 1H), 10.60 (s, 1H). 13C NMR (acetone-d6, 100 MHz) δ (ppm): 23.9 (CH3), 45.1 (CH2), 69.4 (CH2), 97.3 (C(CH3)2), 113.1 (Ar), 113.5 (Ar), 115.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 116.1 (Ar), 121.8 (Ar), 123.7 (Ar), 126.5 (Ar), 130.7 (Ar), 131.6 (Ar), 135.2 (C
C), 116.1 (Ar), 121.8 (Ar), 123.7 (Ar), 126.5 (Ar), 130.7 (Ar), 131.6 (Ar), 135.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 138.0 (CH), 140.7 (Bfx), 157.30 (C-O-CH2), 176.6 (C
C), 138.0 (CH), 140.7 (Bfx), 157.30 (C-O-CH2), 176.6 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). MS (EI), m/z (abundance, %): 396 (M+• - C2H5, 3), 333 (8), 278 (15), 249 (5), 219 (10), 193 (10), 149 (57), 131(35), 57 (100). (Found: C, 58.9; H, 5.2; N, 16.1. C21H23N5O3S requires C, 59.3; H, 5.4; N, 16.5%).
S). MS (EI), m/z (abundance, %): 396 (M+• - C2H5, 3), 333 (8), 278 (15), 249 (5), 219 (10), 193 (10), 149 (57), 131(35), 57 (100). (Found: C, 58.9; H, 5.2; N, 16.1. C21H23N5O3S requires C, 59.3; H, 5.4; N, 16.5%).
5-[2-(N4-Phenylthiosemicarbazono)formylphenyloxy methyl]-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (36). Bordeaux solid (14%), mp 122.8–123.3 °C. 1H NMR (acetone-d6, 400 MHz) δ (ppm): 1.62 (s, 6H), 5.17 (s, 2H), 7.19 (m, 1H), 7.24 (m, 5H), 7.36 (t, 2H, J1 = 7.6 Hz, J2 = 8.0 Hz), 7.44 (m, 1H), 7.78 (d, 2H, J1 = 8.0 Hz), 8.20 (dt, 1H, J1 = 7.2 Hz, J2 = 8.8 Hz, J = 1.6 Hz), 8.88 (s, 1H), 9.89 (s, 1H), 10.90 (s, 1H). 13C NMR (acetone-d6, 100 MHz) δ (ppm): 23.6 (CH3), 68.9 (CH2), 97.6 (C(CH3)2), 112.6 (Ar), 113.0 (Ar), 115.7 (Ar), 121.5 (Ar), 122.9 (C–C = N-NH-), 124.6 (Ar), 125.1 (Ar), 126.5 (Ar), 128.1 (Ar), 128.7 (Ar), 130.3 (Ar), 131.1 (Ar), 131.6 (Ar), 138.3, (C = NH), 139.4 (Ar), 140.2 (Bfx), 157.2 (C-O-CH2), 176.8 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). MS (EI), m/z (abundance, %): 412 (M+• - 2 × 17 – 15, 13), 283 (5), 271(16), 207 (2), 177 (12), 145 (100), 135 (21), 93 (49). (Found: C, 62.3; H, 4.8; N, 15.0. C24H23N5O3S requires C, 62.5; H, 5.0; N, 15.2%).
S). MS (EI), m/z (abundance, %): 412 (M+• - 2 × 17 – 15, 13), 283 (5), 271(16), 207 (2), 177 (12), 145 (100), 135 (21), 93 (49). (Found: C, 62.3; H, 4.8; N, 15.0. C24H23N5O3S requires C, 62.5; H, 5.0; N, 15.2%).
5-(2-Semicarbazonophenyloxymethyl)-2,2-dimethyl-2H-benzimidazole 1,3-di-N-oxide (37). Bordeaux solid (10%), mp 125.5–126.3 °C. 1H NMR (acetone-d6, 400 MHz) δ (ppm): 1.63 (s, 6H), 5.13 (s, 2H), 6.91 (d, 2H, J = 8.4 Hz), 7.24 (d, 2H, J1 = 8.0 Hz), 7.32 (s, 1H), 7.45 (d, 1H, J = 8.0 Hz), 8.04 (dd, 1H, J = 7.6 Hz, J = 1.6 Hz), 8.50 (s, 1H). 13C NMR (acetone-d6, 100 MHz) δ (ppm): 23.0 (CH3), 68.9 (CH2), 96.6 (C(CH3)2), 112.3 (Ar), 116.0 (Ar), 121.1 (Ar), 124.1 (C–C = N–NH–), 125.3 (Ar), 129.3 (Ar), 130.0 (Ar), 136.0 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH), 140.6 (C-O-CH2), 141.8 (Ar), 156.2 (Bfx), 158.0 (C
NH), 140.6 (C-O-CH2), 141.8 (Ar), 156.2 (Bfx), 158.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). MS (EI), m/z (abundance, %): 369 (M+•, 3), 336 (11), 279 (18), 237 (44), 175 (47), 145 (100), 133 (59). (Found: C, 58.4; H, 5.0; N, 18.8. C18H19N5O4 requires C, 58.5; H, 5.2; N, 19.0%).
O). MS (EI), m/z (abundance, %): 369 (M+•, 3), 336 (11), 279 (18), 237 (44), 175 (47), 145 (100), 133 (59). (Found: C, 58.4; H, 5.0; N, 18.8. C18H19N5O4 requires C, 58.5; H, 5.2; N, 19.0%).
Biology
Non-specific inhibition was evaluated in the presence of Triton X-100. A stock solution 0.02% (v/v) of the detergent was freshly prepared in 100 mM PBS or acetate buffer (pH 7.3 or 5.3, respectively) with EDTA and DTT to achieve the same final concentrations as in the standard assay. 50 μL of this solution was then added to 50 μL reaction mix to obtain 0.01% of Triton X-100 in the final reaction mix. This concentration of Triton X-100 was found not to interfere with CP activity.
Docking studies
The structures of the ligands, as protonated form for the amidinohydrazones and as neutral form for the thio- and semicarbazones, were built with standard bond lengths and angles using the molecular modeling package SYBYL 8.131 and their energies were minimized using the Conjugate Gradient algorithm with a conjugated gradient of <0.001 kcal mol−1 convergent criteria provided by the MMFF94 force field32 and MMFF94 electrostatic charges. The ligands considered were superimposed onto the inhibitor present in the reference structure (pdb code 1f29) but without forming any covalent bond with the enzyme. The ligand–receptor complex was subjected to energy minimization using the MMFF94 force field and MMFF94 electrostatic charges and their energies were minimized using the protocol previously indicated with a conjugate gradient of <0.1 kcal mol−1 convergent criteria. These complexes were the input structure for docking using the FlexiDock command.33 During the flexible docking analysis, the ligands and a sphere of 6 Å around the corresponding ligand were considered flexible. For each complex three flexible docking analyses were run. The default SYBYL FlexiDock parameters were utilized in all cases, with maximum and minimum iterations (MI) set for each complex according to MI = [N° of rotable bonds in the protein + N° of rotable bonds in the ligand + 6 × 1000 − 500, obtaining a series of model complexes. All conformations obtained with FlexiDock were clustered using a hierarchical cluster analysis taking into account the score and the distances of ligand moieties to key amino acids present in the active site of the enzyme. We chose the conformation with highest FlexiDock score (better interactions) and refined the minimization energy step using a conjugate gradient of <0.01 kcal mol−1 convergent criteria. Analysis of the refined receptor–ligand complex models was based on hydrogen bond, aromatic and hydrophobic interactions predicted with the LPC (Ligand Protein Contact) program34 and the values of ΔG binding and dissociation constants obtained from the difference accessible surface area method using the STC (Structural Thermodynamics Calculations) program.35Discussion and conclusions
We identified new benzofuroxan and benzimidazole 1,3-dioxide derivatives with interesting trypanosomicidal activities. Compounds 23, 35 and 37 exhibited relevant anti-T. cruzi activity, suggesting that a 2-substituted structural motif played a role that resulted in altering activities relative to those of the corresponding 4-substituted analogues, i.e., 16, 28 and 30, respectively (Table 1 and Fig. 3a). Furthermore, the erythrocyte cytotoxicity of derivative 23 was lower than that of either Nfx or AmpB (Table 1). | ||
| Fig. 3 a) In vitro anti-T. cruzi active moieties described here. b) A previous CP inhibitor and the best ones described here. | ||
The anti-T. cruzi profile of these derivatives did not appear to be highly correlated with CP inhibition, i.e. derivative 23 was unable to inhibit CP in the assay conditions (Table 3), and was the most active and selective anti-T. cruzi agent. However, some derivatives displayed interesting CP-inhibitory activities that allow us to corroborate some structure–activity features. On the one hand, no clear relationship between core scaffold, benzofuroxan or benzimidazole 1,3-dioxide, and CP inhibitory activity was observed, i.e., comparing inhibition of the pair 15 and 28 with that of the pair 21 and 34 (Table 3). For the first pair the benzimidazole 1,3-oxide derivative was the most active, whereas the opposite was the case for the second pair. On the other hand, there appeared to be a consistent relationship between both substituents’ type and position and CP inhibitory activity. In general, the thiosemicarbazones were the best CP inhibitors such that the order thiosemicarbazonyl > amidinohydrazonyl ∼ carbonyl > semicarbazonyl summarized the relative inhibitory activities. In reference to the position of these moieties, the 3-substituted derivatives exhibited better inhibitory activities, i.e., when comparing activities of 3-substituted derivatives (18, 19 and 32) the 4-substituted (15, 27 and 28) and 2-substituted derivatives (21, 22, 34 and 35) (Table 3). These results were consistent with previous reports describing other thiosemicarbazones (Fig. 3b).20,36 Theoretical results clearly indicated that 3-substituted derivatives were able to interact via hydrogen bonding and hydrophobic interactions with CP consistent with the more negative ΔG for the most stable conformers (Table 5) and our experimental data.
The most lipophilic derivatives were the most active against the whole parasite, i.e. hybrid compounds 16, 23, 35 and 36, with values of miLogP37 of 4.86, 4.82, 3.93 and 4.35 (Table 7), respectively, while the most hydrophilic derivatives were inactive, i.e., hybrid compounds 17, 24, 30 and 33, with values of miLogP of 2.67, 2.84, 2.42 and 2.17, respectively. When the molecular lipophilicity potential (MLP), virtual LogP,38 was calculated for the most active benzimidazole 1,3-dioxide derivative, 35, and the least active, 33, the difference in the lipophilicities was correlated with the relative inhibitory activities. The MLP analysis showed that the main differences were localized to the 2-phenyl substituents (Fig. 4). In addition, no relationship between miLogPs and CP inhibitory activity was found.
| Compd | miLogP | nON | nOHNH | MW | nviolations | TPSA |
|---|---|---|---|---|---|---|
| a miLogP, logarithm of compound partition coefficient between n-octanol and water; nON, number of hydrogen bond acceptors; nOHNH, number of hydrogen bond donors; MW, molecular weight; TPSA, topological polar surface area (Å2). | ||||||
| 8 | 3.28 | 6 | 0 | 270.2 | 0 | 77.8 |
| 10 | 3.23 | 6 | 0 | 270.2 | 0 | 77.8 |
| 11 | 3.33 | 6 | 0 | 284.3 | 0 | 77.8 |
| 16 | 4.86 | 8 | 2 | 419.5 | 0 | 97.1 |
| 18 | 3.40 | 8 | 3 | 343.4 | 0 | 111.1 |
| 19 | 4.43 | 8 | 2 | 383.4 | 0 | 97.1 |
| 23 | 4.82 | 8 | 2 | 419.5 | 0 | 97.1 |
| 32 | 2.94 | 8 | 3 | 385.4 | 0 | 117.1 |
| 35 | 3.93 | 8 | 2 | 425.5 | 0 | 103.2 |
| 36 | 4.35 | 8 | 2 | 461.5 | 0 | 103.2 |
| 37 | 2.37 | 9 | 3 | 369.4 | 0 | 134.2 |
| Nfx | 0.71 | 8 | 0 | 287.30 | 0 | 108.71 |
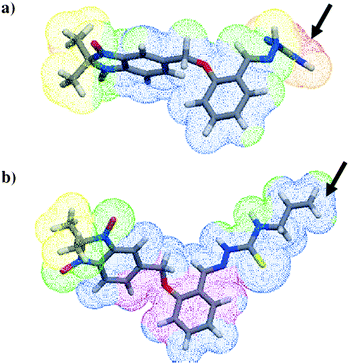 | ||
| Fig. 4 MLP maps, calculated projections of Broto-Moreau lipophilicity atomic constants on the molecular surface.38 Molecules 33 and 35 are represented as tubes with the traditional atom colors. MLP colors: red/yellow for hydrophilic regions; violet/blue for lipophilic regions; green for intermediate regions. The arrows show the polar substituent for 33 (a) and the lipophilic substituent for 35 (b). | ||
In terms of the hybrid derivatives drug-likeness properties, their conformity with the criteria of Lipinski's rule was analyzed.39 In this sense, we determined some properties, i.e., miLogP, number of donor and acceptor hydrogen bonds and molecular weight (Table 7),37 which determine whether these derivatives are similar to known drugs. All the most active derivatives, against whole parasite or CP, had properties in compliance with Lipinski's rule and some had better polar topological surface areas (TPSA,40Table 7) than the value for Nfx, distinguishing them as promising candidates for further drug development.
Further biological studies, QSAR studies, and in vivo activities are currently underway.
Acknowledgements
Financial support from RIDIMEDCHAG network (CYTED), from Collaborative Project UdelaR (Uruguay) – CSIC (Spain) (#2007UY0004) and from REDCLARA-AECID is acknowledged. We thank PEDECIBA-ANII for scholarships to AM, DB, and PH and PEDECIBA for a fellowship to AM. We thank Dr Graciela Mahler for the gift of the mbth reference compound.Notes and references
- http://www.who.int/tdr .
- J. Rodrigues Coura and S. L. Castro, Mem. Inst. Oswaldo Cruz, 2002, 97, 3–24 Search PubMed.
- C. J. Schofield, J. Jannin and R. Salvatella, Trends Parasitol., 2006, 22, 583–588 CrossRef.
- H. Cerecetto and M. González, Pharmaceuticals, 2010, 3, 810–838 Search PubMed.
- J. A. Castro, M. M. de Meca and L. C. Bartel, Hum. Exp. Toxicol., 2006, 25, 471–479 CrossRef CAS.
- Chagas Disease; Special Programme for Research and Training in Tropical Diseases (TDR), World Health Organization, Geneva, http://www.who.int/tdr/diseases/chagas/direction.htm Search PubMed.
- S. Nwaka and R. G. Ridley, Nat. Rev. Drug Discovery, 2003, 2, 919–928 CrossRef CAS.
- A. Cavalli and M. L. Bolognesi, J. Med. Chem., 2009, 52, 7339–7359 CrossRef CAS.
- L. Otero, M. Vieites, L. Boiani, A. Denicola, C. Rigol, L. Opazo, C. Olea-Azar, J. D. Maya, A. Morello, R. L. Krauth-Siegel, O. E. Piro, E. Castellano, M. González, D. Gambino and H. Cerecetto, J. Med. Chem., 2006, 49, 3322–3331 CrossRef CAS; M. Vieites, L. Otero, D. Santos, J. Toloza, R. Figueroa, E. Norambuena, C. Olea-Azar, G. Aguirre, H. Cerecetto, M. González, A. Morello, J. D. Maya, B. Garat and D. Gambino, J. Inorg. Biochem., 2008, 102, 1033–1043 CrossRef CAS; M. Vieites, L. Otero, D. Santos, C. Olea-Azar, E. Norambuena, G. Aguirre, H. Cerecetto, M. González, U. Kemmerling, A. Morello, J. D. Maya and D. Gambino, J. Inorg. Biochem., 2009, 103, 411–418 CrossRef CAS; A. Gerpe, I. Odreman-Nunez, P. Draper, L. Boiani, J. A. Urbina, M. González and H. Cerecetto, Bioorg. Med. Chem., 2008, 16, 569–577 CrossRef CAS; A. Gerpe, G. Álvarez, D. Benítez, L. Boiani, M. Quiroga, P. Hernández, M. Sortino, S. Zacchino, M. González and H. Cerecetto, Bioorg. Med. Chem., 2009, 17, 7500–7509 CrossRef CAS; A. Gerpe, L. Boiani, P. Hernández, M. Sortino, S. Zacchino, M. González and H. Cerecetto, Eur. J. Med. Chem., 2010, 45, 2154–2164 CrossRef CAS; W. Porcal, P. Hernández, M. Boiani, G. Aguirre, L. Boiani, A. Chidichimo, J. J. Cazzulo, N. E. Campillo, J. A. Paez, A. Castro, R. L. Krauth-Siegel, C. Davies, M. A. Basombrío, M. González and H. Cerecetto, J. Med. Chem., 2007, 50, 6004–6015 CrossRef CAS.
- W. Porcal, P. Hernandez, L. Boiani, M. Boiani, A. Ferreira, A. Chidichimo, J. J. Cazzulo, C. Olea-Azar, M. González and H. Cerecetto, Bioorg. Med. Chem., 2008, 16, 6995–7004 CrossRef CAS.
- M. Boiani, L. Piacenza, P. Hernández, L. Boiani, H. Cerecetto, M. González and A. Denicola, Biochem. Pharmacol., 2010, 79, 1736–1745 CrossRef CAS.
- G. B. Henderson, P. Ulrich, A. H. Fairlamb, I. Rosenmerg, M. Pereira, M. Sela and A. Cerami, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 5374–5378 CAS; N. Fujii, J. P. Mallari, E. J. Hansell, Z. Mackey, P. Doyle, Y. M. Zhou, J. Gut, P. J. Rosenthal, J. H. McKerrow and R. Kiplin Guy, Bioorg. Med. Chem. Lett., 2005, 15, 121–123 CrossRef CAS; R. Siles, S.-E. Chen, M. Zhou, K. G. Pinney and M. Lynn Trawick, Bioorg. Med. Chem. Lett., 2006, 16, 4405–4409 CrossRef CAS.
- C. Olea-Azar, C. Rigol, F. Mendizábal, H. Cerecetto, R. Di Maio, M. González, W. Porcal, A. Morello, Y. Repetto and J. D. Maya, Lett. Drug Des. Discovery, 2005, 2, 294–301 Search PubMed.
- J. F. Faucher, T. Baltz and K. G. Petry, Parasitol. Res., 1995, 81, 441–443 CrossRef CAS; M. Almeida-de-Faria, E. Freymuller, W. Colli and M. J. Alves, Exp. Parasitol., 1999, 92, 263–274 CrossRef CAS.
- M. Boiani, L. Boiani, A. Denicola, S. Torres de Ortiz, E. Serna, N. Vera de Bilbao, L. Sanabria, G. Yaluff, H. Nakayama, A. Rojas de Arias, C. Vega, M. Rolón, A. Gómez-Barrio, H. Cerecetto and M. Gonzalez, J. Med. Chem., 2006, 49, 3215–3224 CrossRef CAS; L. Boiani, C. Davies, C. Arredondo, W. Porcal, A. Merlino, A. Gerpe, M. Boiani, J. P. Pacheco, M. A. Basombrío, H. Cerecetto and M. González, Eur. J. Med. Chem., 2008, 43, 2229–2237 CrossRef CAS; L. Boiani, A. Gerpe, V. J. Arán, S. Torres de Ortiz, E. Serna, N. Vera de Bilbao, L. Sanabria, G. Yaluff, H. Nakayama, A. Rojas de Arias, J. D. Maya, A. Morello, H. Cerecetto and M. González, Eur. J. Med. Chem., 2009, 44, 1034–1040 CrossRef CAS.
- S. Choi, A. Isaacs, D. Clements, D. Liu, H. Kim, R. W. Scott, J. D. Winkler and W. F. DeGrado, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 6968–6973 CrossRef CAS.
- V. Yardley and S. L. Croft, Am. J. Trop. Med. Hyg., 1999, 61, 193–197 Search PubMed.
- B. Sahu, V. Chenna, K. L. Lathrop, S. M. Thomas, G. Zon, K. J. Livak and D. H. Ly, J. Org. Chem., 2009, 74, 1509–1516 CrossRef CAS.
- J. M. Pereira, R. P. Severino, P. C. Vieira, J. B. Fernandes, M. F. G. F. da Silva, A. Zottis, A. D. Andricopulo, G. Oliva and A. G. Corrêa, Bioorg. Med. Chem., 2008, 16, 8889–8895 CrossRef CAS; R. F. Freitas, I. M. Prokopczyk, A. Zottis, G. Oliva, A. D. Andricopulo, M. T. S. Trevisan, W. Vilegas, M. G. V. Silva and C. A. Montanari, Bioorg. Med. Chem., 2009, 17, 2476–2482 CrossRef CAS.
- N. Fujii, J. P. Mallari, E. J. Hansell, Z. Mackey, P. Doyle, Y. M. Zhou, J. Gut, P. J. Rosenthal, J. H. McKerrow and R. Kiplin Guy, Bioorg. Med. Chem. Lett., 2005, 15, 121–123 CrossRef CAS; A. C. Lima Leite, R. Souza de Lima, D. R.de M. Moreira, M. V.de O. Cardoso, A. C. Gouveia de Brito, L. M. Farias dos Santos, M. Zaldini Hernandes, A. Costa Kiperstok, R. Santana de Lima and M. B. P. Soares, Bioorg. Med. Chem., 2006, 14, 3749–3757 CrossRef.
- G. Veglia, M. Delfini, M. R. del Giudice, E. Gaggelli and G. Valensin, J. Magn. Reson., 1998, 130, 281–286 CrossRef CAS.
- K. Brak, P. S. Doyle, J. H. McKerrow and J. Ellman, J. Am. Chem. Soc., 2008, 130, 6404–6410 CrossRef CAS.
- L. S. Brinen, E. Hansell, J. Cheng, W. R. Roush, J. H. McKerrow and R. J. Fletterick, Structure, 2000, 8, 831–840 CrossRef CAS.
- L. Huang, L. S. Brinen and J. A. Ellman, Bioorg. Med. Chem., 2003, 11, 21–29 CrossRef CAS.
- M. E. McGrath, A. E. Eakin, J. C. Engel, J. H. McKerrow, C. S. Craik and R. J. Fletterick, J. Mol. Biol., 1995, 247, 251–259 CrossRef CAS.
- S. A. Gillmor, C. S. Craik and R. J. Fletterick, Protein Sci., 1997, 6, 1603–1611 CrossRef CAS.
- D. Bogdal, J. Pielichowsky and A. Boron, Synth. Commun., 1998, 28, 3029–3039 CAS; R. Kamakshi and B. S. R. Reddy, Aust. J. Chem., 2005, 58, 603–606 CrossRef CAS.
- N. Aggarwal and P. Mishra, J. Zhejiang Univ., Sci., 2005, 6b, 617–621 CrossRef CAS.
- L. Fernandez, M. Calderón, M. Martinelli, M. Strumia, H. Cerecetto, M. González, J. J. Silber and M. Santo, J. Phys. Org. Chem., 2008, 21, 1079–1085 CrossRef CAS; R. Hinojosa Valdez, L. T. Düsman Tonin, T. Ueda-Nakamura, B. P. Dias Filho, J. A. Morgado-Diaz, M. Sarragiotto and C. Vataru Nakamura, Acta Trop., 2009, 110, 7–14 CrossRef.
- R. S. Ferreira, C. Bryant, K. K. H. Ang, J. H. McKerrow, B. K. Shoichet and A. R. Renslo, J. Med. Chem., 2009, 52, 5005–5008 CrossRef CAS.
- SYBYL 8.1, Tripos Inc., 1699 South Hanley Rd., St. Louis, MO 63144, USA, 2008.
- M. Clark, R. D. Cramer III and N. Van Opdenbosch, J. Comput. Chem., 1989, 10, 982–1012 CrossRef CAS.
- R. Judson, Genetic algorithms and their use in chemistry, in Reviews in Computational Chemistry, ed. K. B. Lipkowitz and D. B. Boyd, vol. 10, VCH Publishers, New York, 1997, pp. 1–73 Search PubMed.
- V. Sobolev, A. Sorokine, J. E. Prilusky, E. Abola and M. Edelman, Bioinformatics, 1999, 15, 327–332 CrossRef CAS.
- P. Lavigne, J. R. Bagu, R. Boyko, L. Willard, C. F. B. Holmes and B. D. Sykes, Protein Sci., 2000, 9, 252–264 CAS.
- D. C. Greenbaum, Z. Mackey, E. Hansell, P. Doyle, J. Gut, C. R. Caffrey, J. Lehrman, P. J. Rosenthal, J. H. McKerrow and K. Chibale, J. Med. Chem., 2004, 47, 3212–3219 CrossRef CAS.
- Compounds properties calculation. Polar surface area (TPSA), miLog P, and violations of Lipinski's rule of five, were calculated using Molinspiration online property calculation toolkit: http://www.molinspiration.com/cgi-bin/properties Search PubMed.
- P. Gaillard, P. A. Carrupt, B. Testa and A. Boudon, J. Comput.-Aided Mol. Des., 1994, 8, 83–96 CAS; A. J. M. Carpy and N. Marchand-Geneste, SAR QSAR Environ. Res., 2003, 14, 329–337 Search PubMed.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 1997, 23, 3–25 CrossRef.
- P. Ertl, B. Rohde and P. Selzer, J. Med. Chem., 2000, 43, 3714–3717 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2010 |