Synthesis, modeling, and crystallographic study of 3,4-disubstituted-1,2,5-oxadiazoles and evaluation of their ability to decrease STAT3 activity †
Dae-Seop
Shin
a,
Daniela
Masciocchi
b,
Arianna
Gelain
b,
Stefania
Villa
b,
Daniela
Barlocco
*b,
Fiorella
Meneghetti
b,
Alessandro
Pedretti
b,
Young-Min
Han
a,
Dong Cho
Han
a,
Byoung-Mog
Kwon
*a,
Laura
Legnani
c and
Lucio
Toma
*c
aLaboratory of Chemical Biology and Genomics, Korea Research Institute of Bioscience and Biotechnology, and Department of Biomolecular Science, Korea University of Science and Technology, 52 Uen-Dong Yusung-Ku, Daejeon 305-600, South Korea. E-mail: kwonbm@kribb.re.kr; Fax: +82-42-861-2675; Tel: +82-42-860-4557
bDipartimento di Scienze Farmaceutiche “Pietro Pratesi”, Università degli Studi di Milano, via L. Mangiagalli 25, 20133 Milano, Italy. E-mail: daniela.barlocco@unimi.it; Fax: +39-02-503-19359; Tel: +39-02-503-19367
cDipartimento di Chimica Organica, Università degli Studi di Pavia, Via Taramelli 10, 27100 Pavia, Italy. E-mail: lucio.toma@unipv.it; Fax: +39-0382-98-7323; Tel: +39-0382-98-7843
First published on 28th May 2010
Abstract
STAT3 (Signal Transducer and Activator of Transcription 3) is a transcription factor constitutively activated by aberrant upstream tyrosine kinase activities in a broad spectrum of human solid and blood tumors, thus suggesting that its inhibition could represent an interesting molecular target for cancer therapy. With the aim to disclose novel scaffolds for compounds active on STAT3 the potential of the 1,2,5-oxadiazole ring was explored and several new compounds substituted at positions 3 and 4 of the heterocycle were synthesized. When tested in a dual-luciferase assay, using HCT-116 cells, some compounds showed a significant inhibition value towards STAT3. So, to give support to the biological results, modeling and crystallographic studies of representative terms of the new series were performed.
Introduction
Signal Transducer and Activator of Transcription 3 (STAT3) is a member of the STAT family, latent, cytosolic transcription factors that directly relate signals from the plasma membrane to the nucleus. At present, seven members have been described: STAT1 to STAT4, STAT5a, STAT5b and STAT6. These proteins transmit cytoplasmic signals from growth factors and polypeptide cytokines, that have receptors with intrinsic or associated Jak tyrosine kinase activity, to the nucleus.1 Non-receptor cytoplasmic tyrosine kinases that signal through STATs include Abl (Abelson leukemia protein) and Src (sarcoma)-related kinases.2 The STATs have SH2 domains, and they are recruited to phosphorylated receptors via this domain. STAT3 then becomes phosphorylated on Tyr705 by Jak kinases, Src, Abl or the kinase activity of the receptor. After phosphorylation, it dimerizes, forming homodimers and/or heterodimers with STAT1, by reciprocal phosphotyrosine SH2 interaction.3 The dimers then translocate into the nucleus, bind to response elements in gene promoters, and enhance the transcription of target genes.4Persistently activated (Tyr phosphorylated) STATs, especially STAT3 and STAT5, have been reported in a wide variety of human tumors, suggesting that their inhibitors might be widely effective as anticancer therapies. In particular STAT3 is persistently activated in almost all head and neck cancers, multiple myelomas, hepatocellular carcinoma, lymphomas and leukemia; thus this protein has the potential to be a leading target for cancer therapy.5
At present, some compounds, able to inhibit STAT3 signaling either by blocking the upstream tyrosine kinases responsible for its activation6–12 or by direct inhibition of the STAT protein,13–20 have been described.
Several screening systems are known for evaluation of STAT3 activity. Amongst others, cell-based STAT3 (Tyr705) ELISA system and luciferase system, including dual-luciferase assay. The latter is a rapid, convenient and sensitive method, easily applicable for high-throughput screening.21
On these bases, some of us installed a dual-luciferase assay system with cancer cell line harboring abnormally activated STAT3 to find inhibitors of this pathway and disclosed the oxadiazole ureido derivative AVS-0288 (I), as agent for prevention and treatment of cancer.22 In the search of other compounds, structurally related to I and able to lower STAT3 activity, we investigated the effects of a series of structural modifications, summarized in structure II (Chart 1): i) replacement of the chlorophenyl group at position 4 of the heterocycle with different moieties (R3 = CH3, COOCH3, CH2OH, p-HO-Ph, etc.); ii) elimination of the trifluoromethyl group on the N-phenyl (R1 = H); iii) methylation of the ureido moiety (R2 = H, CH3, Y = CONH, CONCH3); iv) substitution of urea with a carboxyamido or sulfonamido function (Y = CO, SO2); v) insertion of a methylene between the group of item iv and the phenyl linked to it (n = 1). Here we describe the synthetic, biological, and structural studies of these new compounds.
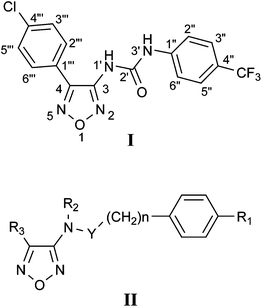 | ||
| Chart 1 | ||
Results and discussion
Synthesis
We synthesized three series of novel 3,4-disubstituted-1,2,5-oxadiazoles which differ for their main “Y” functionality: the ureido (1), carboxyamido (2), and sulfonamido (3) derivatives reported in Chart 2.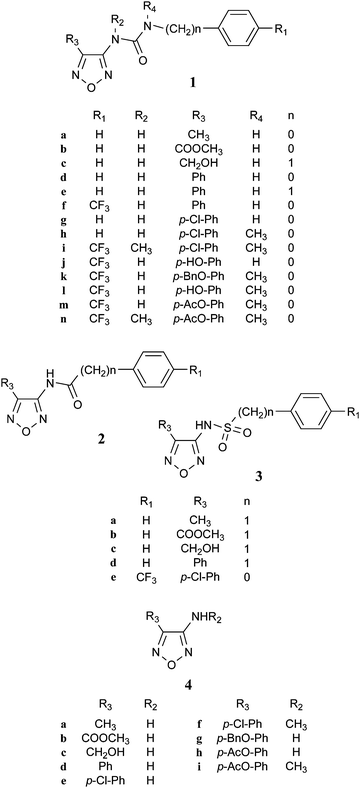 | ||
| Chart 2 | ||
The key intermediates for their synthesis were 3-amino-1,2,5-oxadiazoles differently substituted at position 4 (Chart 2). Most of them (4a,b,d,e,g,h) were prepared according to literature methods,23–25 whereas compound 4c was obtained from 4b by reduction with LiAlH4 and 4f and 4i were synthesized as 4e and 4h, respectively, using methyl hydroxylamine instead of hydroxylamine.
Two different approaches were used to obtain the target ureido compounds 1a–n. The first one, applied to the synthesis of 1a–f, is based on the reaction of the appropriate oxadiazole 4a–d with the required isocyanates in toluene or acetonitrile in the presence of triethylamine (TEA) under microwave irradiation (Scheme 1). In the case of 4c the alcoholic function was protected by methyl chloroformate prior to condensation with the isocyanate. Deprotection with K2CO3 in methanol gave the desired 1c. The second approach, applied to the synthesis of 1g–n, is based on the reaction of the appropriate oxadiazole 4e–i with a suitable amine in the presence of triphosgene and N,N-diisopropylethylamine (DIEA) in tetrahydrofuran (Scheme 1).26 The two p-hydroxyphenyl derivatives 1j and 1l were obtained by deprotection with 10% Pd–C and 1,4-cyclohexadiene in DMF.27,28
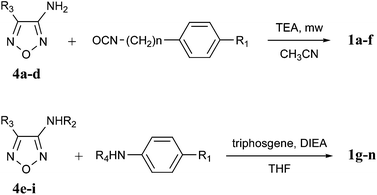 | ||
| Scheme 1 Synthesis of compounds 1. | ||
The carboxyamido compounds 2a–e were prepared from the required oxadiazole 4a–e by reaction with a suitable acyl chloride in tetrahydrofuran or toluene–diethyl ether using NaHCO3 or pyridine as base (Scheme 2). In the case of 2b the condensing agent 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydro-chloride (EDAC) in dichloromethane was used.
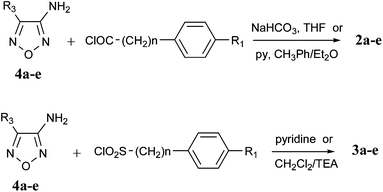 | ||
| Scheme 2 Synthesis of compounds 2 and 3. | ||
Finally, the sulfonamido compounds 3a–e were obtained by reaction of the appropriate oxadiazole 4a–e (protected when necessary) with the suitable sulfonyl chloride in pyridine or dichloromethane/triethylamine (Scheme 2).
Biological studies
All the synthesized compounds 1–3 were evaluated in a dual-luciferase assay to determine their ability to lower STAT3 activity. Colorectal carcinoma HCT-116 cells were co-transfected with pSTAT-TA-Luc carrying firefly luciferase gene, which is dependently expressed by STAT3 activity, and pRL-TK carrying renilla luciferase gene, which is independently expressed by STAT3 activity. Beetle luciferin and coelenterazine were used as substrates for the two enzymes, respectively. Cells were treated with the tested compounds at concentration of 2 μM and incubated for 24 h. The results, reported in Table 1 as percentage inhibition, clearly indicate that, though just one compound (1l) showed equivalent activity to I, in the ureido series a number of derivatives were found to be active and an attempted SAR could be done. In particular, the presence of a (substituted) phenyl at position 4 of the heterocycle proved to be essential; in fact, compounds 1a–c were completely inactive. The substitution pattern on both phenyl rings seems to be critical. The unsubstituted 1d has better properties of both 1f and 1g, which lack, respectively, the chloro and the trifluoromethyl group of I. Insertion of a methylene between the urea and the phenyl is detrimental (see 1evs.1d). By contrast, monomethylation of the ureido chain improves activity (see 1hvs.1g) and led to the most potent compound of this series (1l). The presence of an additional methyl group on the ureido moiety does not seem to exert a significant effect (see 1nvs.1m). Finally, substitution of the ureido moiety by either a carboxyamido (2a–e) or a sulfonamido group (3a–e) lowers activity making the resulting compounds poorly active or even inactive.| Compd | Inhibitory activity (%) | Compd | Inhibitory activity (%) |
|---|---|---|---|
| I | 45 | 2a | 6 |
| 1a | 2 | 2b | 9 |
| 1b | −13 | 2c | 14 |
| 1c | −14 | 2d | 11 |
| 1d | 36 | 2e | −7 |
| 1e | 11 | 3a | 6 |
| 1f | −16 | 3b | 7 |
| 1g | −25 | 3c | −2 |
| 1h | 25 | 3d | −38 |
| 1i | 8 | 3e | −3 |
| 1j | 24 | ||
| 1k | 21 | ||
| 1l | 51 | ||
| 1m | 32 | ||
| 1n | 26 |
For compound I, the luciferase assay was repeated at different concentrations, ranging from 0.5 to 100 μM, allowing the determination of its dose-dependent inhibition activity, reported in Fig. 1. After treatment with I, cells were incubated for 24 h. IC50 of luciferase activity was about 2.5 μM.
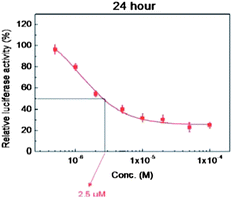 | ||
| Fig. 1 Dose-dependent effect of I on STAT3 activity after incubation for 24 h. | ||
In addition, growth inhibition by I was tested in breast and colon cancer cell lines. When cells were incubated with I for 24 h at different concentrations, ranging from 0.5 to 100 μM, cell proliferation of HCT-116, MDA-MB-468, MDA-MB-231, and SW620 lines was reduced to about 50% at the highest concentrations (Fig. 2A). Moreover, incubation for 48 h (Fig. 2B) allowed an almost complete inhibition of proliferation at the highest concentration. Fig. 2C shows a rounded and floated morphology of HCT-116 cells after treatment with I demonstrating that the compound induces apoptosis in HCT-116 cells. In order to confirm apoptosis by compound I, western blotting of apoptotic marker proteins was carried out. Cleaved caspase-3 and PARP were remarkably increased by 10 μM I (Fig. 2D).
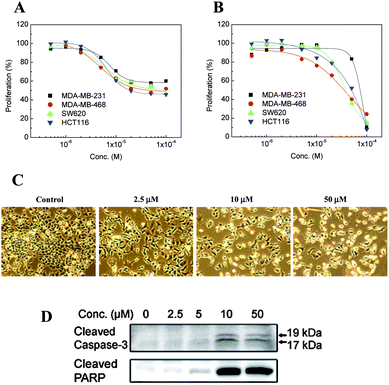 | ||
| Fig. 2 Inhibitory effect on proliferation of several cancer cell lines at different concentrations of I after incubation for 24 (A) or 48 h (B). Morphology of HCT-116 cells after treatment with I (C). Increase in cleaved caspase-3 and PARP after treatment with 10 μM I (D). | ||
Modeling studies
A modeling study of representative terms of each series of tested compounds was undertaken in order to give theoretical support to the above biological results. The study started with compound I. All its degrees of conformational freedom were considered with particular attention to the different arrangements of the ureido moiety (trans/trans, TT; trans/cis, TC; cis/trans, CT; and cis/cis, CC) described by the torsional angles τ1 (C3-N1′-C2′-N3′) and τ2 (N1′-C2′-N3′-C1′′). The geometries were optimized at the B3LYP level with the 6-311+G(d,p) basis set [6-311+G(2df,p) for the chlorine atom].29 The energy of the optimized conformations was recalculated in solvent using a polarizable continuum model (PCM) to take into account the effect of water.30Several minimum energy conformations were located. In Table 2 the gas-phase and the water-solvated energies of I are reported, together with the corresponding percentage contributions to the overall population and selected torsional angles. On the basis of the ureido moiety geometry, all the located conformers were grouped into four families, characterized by similar values of τ1 and τ2, while the members of each family differ for the other degrees of conformational freedom of the molecule: i) the orientation of the oxadiazole ring respect to the ureido moiety (τ3, N2-C3-N1′-C2′); ii) the single bond between the chlorophenyl group and C4 of the heterocycle (τ4, C3-C4-C1′′′-C2′′′); iii) the single bond between the trifluoromethylphenyl group and N3 of urea (τ5, C2′-N3′-C1′′-C2”). In Fig. 3 are pictured the three-dimensional plots of all the members of the TT family, differing in the orientation of the oxadiazole ring and/or the chlorophenyl group, and the most representative of the other three ones.
| E rel gas phase/kcal mol−1 | % | E rel water/kcal mol−1 | % | τ 1 (°) | τ 2 (°) | τ 3 (°) | τ 4 (°) | τ 5 (°) | |
|---|---|---|---|---|---|---|---|---|---|
| a τ 1: C3-N1′-C2′-N3′. b τ 2: N1′-C2′-N3′-C1′′. c τ 3: N2-C3-N1′-C2′. d τ 4: C3-C4-C1′′′-C2′′′. e τ 5: C2′-N3′-C1′′-C2”. | |||||||||
| TT family | |||||||||
| TT-1 | 8.79 | 0.0 | 0.84 | 16.7 | −174 | −174 | 43 | 42 | 3 |
| TT-2 | 6.28 | 0.0 | 0.00 | 68.6 | 175 | −177 | −124 | 33 | 0 |
| TT-3 | 9.50 | 0.0 | 2.22 | 1.6 | −178 | −176 | 32 | −49 | 4 |
| TT-4 | 6.72 | 0.0 | 1.51 | 5.3 | 174 | −177 | −110 | −46 | 6 |
| 0.0 | 92.2 | ||||||||
| TC family | |||||||||
| TC-1 | 10.01 | 0.0 | 5.35 | 0.0 | 175 | −11 | −33 | −41 | −40 |
| TC-2 | 7.76 | 0.0 | 3.52 | 0.2 | −172 | −1 | 124 | −33 | −42 |
| TC-3 | 10.57 | 0.0 | 5.80 | 0.0 | 180 | −15 | −28 | 46 | −39 |
| TC-4 | 11.08 | 0.0 | 5.79 | 0.0 | 172 | −7 | −28 | −44 | 55 |
| TC-5 | 7.74 | 0.0 | 4.71 | 0.0 | −170 | −4 | 109 | 45 | −43 |
| TC-6 | 8.46 | 0.0 | 5.29 | 0.0 | −168 | 8 | 122 | −32 | 49 |
| TC-7 | 8.46 | 0.0 | 6.38 | 0.0 | −166 | 9 | 105 | 42 | 49 |
| 0.0 | 0.2 | ||||||||
| CT family | |||||||||
| CT-1 | 0.00 | 100.0 | 1.31 | 7.5 | −2 | −179 | −1 | −43 | 0 |
| CT-2 | 8.80 | 0.0 | 4.14 | 0.1 | 2 | 174 | 115 | −27 | −7 |
| 100.0 | 7.6 | ||||||||
| CC family | |||||||||
| CC-1 | 9.92 | 0.0 | 5.43 | 0.0 | −45 | −23 | 2 | −35 | −31 |
| CC-2 | 10.94 | 0.0 | 6.33 | 0.0 | −27 | −34 | 141 | −34 | −22 |
| CC-3 | 10.58 | 0.0 | 7.17 | 0.0 | −43 | −30 | 15 | 48 | −25 |
| CC-4 | 13.57 | 0.0 | 9.80 | 0.0 | −20 | −39 | 117 | 26 | −28 |
| 0.0 | 0.0 | ||||||||
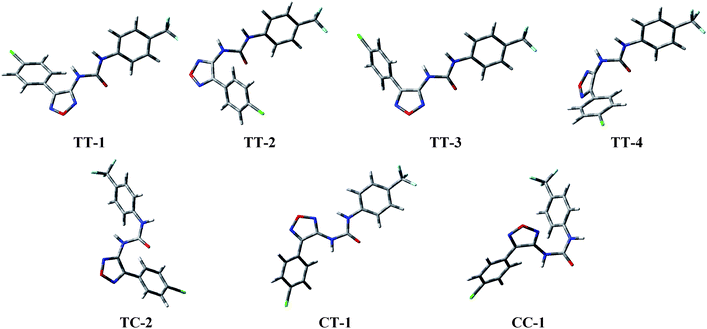 | ||
| Fig. 3 3D Plot of representative members of the conformational families of the compound I. | ||
In the gas phase only the most stable conformer CT-1, which presents the cis/trans arrangement around the two amide bonds (τ1 = −2°, τ2 = −179°), was populated, because of the presence of an intramolecular H-bond between the N atom of the amide trans orientated and one of the N atoms of the oxadiazole ring (τ3 = −1°).
The solvent deeply influenced the relative energy values and hence the percentage values. In fact in water the trans/trans arrangement was largely preferred and the overall population of each conformational family was 92.2 (TT), 0.2 (TC), 7.6 (CT), and 0.0% (CC). All the members of the most populated TT family showed a planar arrangement of the trifluoromethylphenyl group respect to the ureido moiety (τ5 = 0 ÷ 6°), analogously to the gas phase favored CT-1 conformation (τ5 = 0°).
It should be noted that the differences in the conformational preferences of I, which is predicted to adopt a cis/trans conformation in vacuo and a trans/trans conformation in a polar solvent, are confirmed by 1H-NMR spectroscopy. In fact, depending on the solvent used (CDCl3vs. DMSO), different resonance patterns are observed.31
Then, we extended the modeling study to the poorly active 2d and the inactive 3d, in which the ureido moiety is replaced, respectively, by an amido or a sulfonamido group. The presence of a methylene group guarantees the same bond distance as in I between the left and right ends of the molecules. Calculations were performed at the same level as above [6-311+G(2df,p) for the sulfur atom] and all the degrees of conformational freedom were considered. To put in evidence analogies and differences, we superimposed the preferred conformations of the three compounds in vacuo and solvent through rms fitting of the atoms of the oxadiazole ring of I with the corresponding atoms of 2d and 3d (Fig. 4). The superimposition of the active compound with the two analogues clearly shows for 2d and 3d a perpendicular arrangement (τ5 ≈ 90°) of the trifluoromethylphenyl group respect to the amido/sulfonamido moiety. Moreover, the energy cost to force τ1 of compound 3d to planar arrangements may be estimated through the profile of rotation around this angle that shows a relative energy > 10 kcal mol−1 in correspondence of values close to 0° and 180°. A similar behavior is observed for rotation around the bond defined by τ5 both for 2d and 3d. Conversely, deviation from planarity is very demanding in the case of I, as an orthogonal arrangement at τ1 and τ2 requires > 20 kcal mol−1. Thus, the lack of the planarity seems to be the main factor for the reduction of the inhibitory activity in the 2 and 3 series, though the chemical difference between the “Y” groups in the three series cannot be neglected.
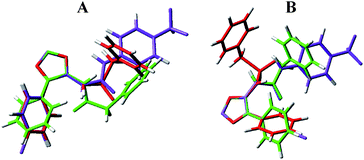 | ||
| Fig. 4 Overlay of the preferred conformations in vacuo (A) and solvent (B) of compounds I (violet), 2d (green) and 3d (red), obtained through rms fitting of the atoms of the oxadiazole ring. | ||
Crystallographic studies
To go into structural investigation thoroughly, the above biological and modeling studies were complemented by a crystallographic analysis. Compound I was characterized by an X-ray diffraction study and compared with the theoretical calculations. In the solid state (Fig. 5), I has the two amide bonds in trans conformation like in several N,N′-disubstituted ureas, in which the N–H bonds point in opposite direction to that of the carbonyl.32 This molecular feature corresponds to the trans/trans arrangement calculated in solution, as defined by the τ1 and τ2 values of −170(1)° and 177(1)° respectively, while the torsional angle τ3 of 26(1)° in the crystal structure slightly differs from the value of the modeled TT-1 conformer in water (43°). In the solid state, the chlorophenyl ring is less rotated with respect to the oxadiazole, as shown by the torsional angle τ4 of 25(1)° compared with the value determined for TT-1 (42°). The major conformational difference found between the crystal state structure and TT-1 is the orientation of the trifluorophenyl moiety with respect to the ureido group, as shown by the torsional angle τ5 of −36(1)° (3° in the theoretical conformation). Nevertheless, the dihedral angle of 13(1)° between the oxadiazole and the distal phenyl group indicates the overall planarity of the compound and the superimposition of the crystal structure on the TT-1 conformer (Fig. 6) clearly shows the similarity of the solid state and the structure calculated using simulated water. In the crystal packing, I is involved in a three-dimensional network of intermolecular interaction of type N–H⋯N, N–H⋯O, C–H⋯F, Cπ–H⋯N, and π–π stacking interaction between the trifluorophenyl rings at 4.07(1) Å.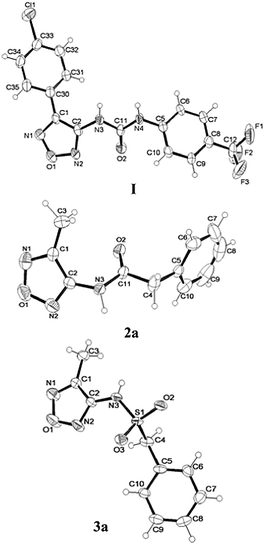 | ||
| Fig. 5 ORTEP33 view of I, 2a, 3a and the relative arbitrary atom-numbering scheme (thermal ellipsoids at 40% probability). | ||
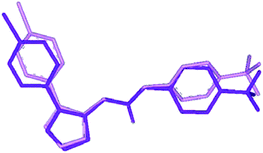 | ||
| Fig. 6 Overlay of the crystal structure of I (violet) on the TT-1 conformation (pink) obtained through rms fitting of the atoms of the ureido group. Hydrogen atoms are omitted for the sake of clarity. | ||
Besides I, representative terms of the other two synthesized series, 2a and 3a, were structurally characterized and compared with compound I. They correspond to the modeled compounds with the exception that they bear a methyl instead of an aryl group at position 4 of the oxadiazole ring. An ORTEP33 view of the analyzed compounds is presented in Fig. 5.
In compound 2a, the oxadiazole and the phenyl moieties are inclined toward each other as indicated by a value of 63(1)° of the dihedral angle between the mean plane of the rings. In the crystal packing the molecules are linked by hydrogen bonds type N–H⋯O and by Cπ–H⋯N intermolecular interactions, that are weaker than those in I. Weak π–π stacking between the oxadiazole and the phenyl ring is also present (centroid-to-centroid distance: at 4.31(1) Å).
Finally, compound 3a presents the oxadiazole oriented almost parallel to the distal phenyl moiety (dihedral angle of 13(1)°) as found in I. It has a less extended chain conformation with respect to 2a, as shown by values of the torsional angles corresponding to τ1 of −73(1)° and 178(1)°, respectively. The bond distance of 1.367(6) Å between the sulfonamido nitrogen atom and the carbon atom of the oxadiazole ring (N3 and C2 in Fig. 5) may indicate slight conjugation. The π–π interactions at 3.77(1) Å between the oxadiazole and the benzene stabilize the crystal packing together with hydrogen bonds linking N–H with a sulfonamido oxygen atom of the centrosymmetrically related molecule. The latter forms dimers further connected through intermolecular interactions type Cπ–H⋯O along the crystallographic a axis, giving rise to a three-dimensional network. The superimposition of the crystallographic structure of I, 2a, and 3a through the best fit of the oxadiazole ring presented in Fig. 7 clearly shows their main conformational differences.
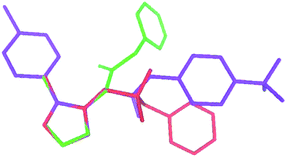 | ||
| Fig. 7 Superimposition of the crystal structures of I (violet), 2a (green) and 3a (red) achieved by overlaying the oxadiazole rings. Hydrogen atoms are omitted for the sake of clarity. | ||
Conclusions
Three series of compounds (1–3) which incorporate an 1,2,5-oxadiazole unit attached to either a substituted phenylurea or a carboxyamido or sulfonamido group were designed and synthesized. Several substituents at position 4 of the heterocyclic ring were introduced. Moreover, methylation of the ureido moiety was considered. All compounds were submitted to a dual-luciferase assay to evaluate their ability in lowering STAT3 activity. The results indicate the importance of a (p-substituted)-phenyl at position 4. Monomethylation of the phenylurea lead to the most potent derivative 1l, while the insertion of a second methyl did not improve the activity.Modeling studies on I showed a deep influence of the solvent on the conformational population, suggesting a large preference of the ureido moiety for a trans/trans or a cis/trans arrangement in polar and non polar solvents, respectively. In all the cases, however, an almost planar arrangement of the structural entities of the molecule was observed suggesting planarity as an essential requisite for activity.
Finally, crystallographic studies on representative terms highlighted the less extended conformation in 2a and 3a with respect to I, giving support to the molecular modeling calculations.
Experimental
General
Phenyl isocyanate, benzyl isocyanate, 4-(trifluoromethyl)-phenyl isocyanate, 4-(trifluoromethyl)benzoyl chloride, phenylacetyl chloride, phenylmethanesulfonyl chloride, 4-(trifluoromethyl)benzenesulfonyl chloride, 4-chloro-benzaldehyde, 4-(benzyloxy)-benzaldehyde, 4-(acetyloxy)-benzaldehyde, aniline, N-methylaniline, 4-(trifluoromethyl)-aniline, and 4-trifluoromethyl-N-methylaniline were purchased from Sigma-Aldrich and were used without any further purification. Compounds 4 were synthesized according to literature methods.23–25 All reactions involving air-sensitive reagents were performed under nitrogen atmosphere. The Biotage-Initiator microwave synthesizer was used. Melting points were determined in open capillary tubes on a Büchi Melting Point B-540. 1H NMR spectra were acquired at room temperature on a Varian 300 MHz Oxford instrument. Chemical shifts are expressed in ppm from tetramethylsilane resonance in the indicated solvent (TMS: 0.0 ppm). Reaction were monitored by thin layer chromatography (TLC) on aluminium-backed Silica Gel 60 plates (0.2 mm, Merck). Intermediates and final compounds were purified by flash chromatography using Merck Silica Gel 60 (70–230 mesh). The purity of final compounds were determined by HPLC analysis and were ≥95%. All new compounds gave satisfactory C, H, N elemental analyses (± 0.4%).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) to afford (4-amino-1,2,5-oxadiazol-3-yl)methyl methylcarbonate as a yellow/orange oil (5) (141 mg, 94%). A mixture of compound 5 (50 mg, 0.29 mmol) and benzyl isocyanate (0.40 mmol, 0.05 mL) in toluene (1 mL) in a sealed vial was irradiated in a microwave synthesizer at 300 Watts at a temperature of 180 °C for 40 min. After cooling, [4-(3-benzylureido)-1,2,5-oxadiazol-3-yl]methyl methyl carbonate (6) precipitated as a white jelly-like solid, was collected by filtration and washed with toluene (27 mg, 31%). Deprotection of 6 with K2CO3 in methanol gave 1c. The crude product was purified by flash chromatography.
3) to afford (4-amino-1,2,5-oxadiazol-3-yl)methyl methylcarbonate as a yellow/orange oil (5) (141 mg, 94%). A mixture of compound 5 (50 mg, 0.29 mmol) and benzyl isocyanate (0.40 mmol, 0.05 mL) in toluene (1 mL) in a sealed vial was irradiated in a microwave synthesizer at 300 Watts at a temperature of 180 °C for 40 min. After cooling, [4-(3-benzylureido)-1,2,5-oxadiazol-3-yl]methyl methyl carbonate (6) precipitated as a white jelly-like solid, was collected by filtration and washed with toluene (27 mg, 31%). Deprotection of 6 with K2CO3 in methanol gave 1c. The crude product was purified by flash chromatography.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5) to give (4-phenylmethylsulfonamido-1,2,5-oxadiazole-3-yl)methyl-methyl-carbonate (7) (55 mg, 50%). Deprotection of 7 with K2CO3 in methanol gave 3c. The crude product was purified by flash chromatography.
0.5) to give (4-phenylmethylsulfonamido-1,2,5-oxadiazole-3-yl)methyl-methyl-carbonate (7) (55 mg, 50%). Deprotection of 7 with K2CO3 in methanol gave 3c. The crude product was purified by flash chromatography.
Cell culture
The cancer cell lines were obtained from American Type Culture Collection. Human breast cancer cell lines (MDA-MB-468, MDA-MB-231), human colon cancer cell line (SW620) were maintained in RPMI 1640 (Gibco/BRL). Human colon cancer cell line (HCT-116) was maintained in McCoy's 5A (Gibco/BRL). All culture media were supplemented with 10% heat-inactivated fetal bovine serum (Gibco/BRL). Cell cultures were maintained at 37 °C under a humidified atmosphere of 5% CO2 in an incubator.Transient transfection and dual-luciferase assays
HCT-116 cells were seeded at a density of 10 × 105 cells in 100 mm2 culture plate. On the following day, the cells were co-transfected with pSTAT3-TA-Luc (27 μg/plate) and internal control plasmid pRL-TK (9 μg/plate) containing the Renilla luciferase gene. All plasmids used in this experiment were purchased from Promega. The transfection was carried out using TransFectin (Bio-Rad) according to the manufacturer's protocol. After 5 h of transfection, the cells were trypsinized and seeded onto sterilized black bottom 96-well plates at a density of 1 × 104 cells per well. On the following day, cells were treated with test compounds and incubated for 24 h. Firefly and Renilla luciferase activity was measured using dual-light reporter gene assay kit (Promega) on Wallac Victor2 (Perkin-Elmer, Inc., Wellesley, MA). Renilla luciferase activity was determined to calibrate transfection efficiency and cytotoxicity of chemicals. Relative STAT3 activity was calculated by dividing the firefly luciferase activity with Renilla luciferase activity in each transfection experiment.Cell proliferation assay
Cells were seeded at a density of 5,000 cells per well in 96-well plates in RPMI 1640 or McCoy's medium containing 10% FBS. Cells were replenished with fresh complete medium containing either test compound or 0.1% DMSO. After incubation for 24 or 48 h, the cell proliferation reagent WST-1 (Roche Applied Science) was added to each well. WST-1 formazan was quantitatively measured at 450 nm using an enzyme-linked immunosorbent assay reader (Bio-Rad).Western blotting
Cell lysates were prepared by RIPA lysis buffer (50 mM Tris, pH 7.0, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1% deoxycholic acid, 0.1% SDS, 30 mM Na2HPO4, 50 mM NaF, and 1 mM NaVO4) containing protease inhibitor cocktail (Roche Applied Science). Proteins (40 μg) were resolved by 7% or 15% SDS–PAGE and transferred to PVDF membrane (Millipore). The membrane were blocked with 5% nonfat dried milk in TBS-T (50 mM Tris–HCl, pH 7.6, 150 mM NaCl, and 0.1% Tween 20) and probed with cleaved caspase-3 (Asp175) and cleaved PARP (Asp214) primary antibody for 3 h. These antibodies were purchased from Cell Signaling Technology (Danvers, MA). The blot was washed, exposed to HRP-conjugated anti-rabbit IgG for 1 h, and examined by chemiluminescence POD reagents (Roche Applied Science).Computational methods
A systematic search of the conformational space of compounds I, 2d, and 3d was performed using the Gaussian03 program package through optimizations in the gas-phase at the B3LYP level with the 6-311+G(d,p) basis set [6-311+G(2df,p) for the chlorine and sulfur atoms].29 In the case of I a starting geometry, presenting the TT arrangement at the ureido moiety and the τ3, τ4, and τ5 torsional angles assuming values close to 0°, was optimized allowing to obtain conformer TT-1. Then, the energy profiles for rotation around the single bonds defined by τ3, τ4, and τ5 were determined with a step size of 30°. All the combinations of the observed minima were used to generate a number of starting geometries optimized as above. The same procedure was repeated for the TC, CT, and CC arrangements of I and for compounds 2d and 3d. The energy of the optimized conformations was recalculated in water using a polarizable continuum model (PCM).30X-Ray crystallographic analysis
The crystals of I and 3a were obtained by crystallization from methanol as colorless prisms, while 2a crystallized from water as transparent needles. They were mounted on an Enraf Nonius CAD-4 diffractometer using Mo-Kα (λ = 0.71073 Å) radiation at 293(2)K. The lattice parameters were determined by least-squares refinements of 25 high angle reflections. The structures were solved by direct methods34 and the refinements were carried out by full-matrix least-squares. All non-H-atoms were refined anisotropically. Hydrogen atoms of 3a were detected in a difference Fourier synthesis and refined with isotropic thermal factors, while all hydrogen atoms in 2a were introduced at calculated positions, in their described geometries and allowed to ride on the attached carbon atom with fixed isotropic thermal parameters (1.2Ueq of the parent carbon atom). Refinements were carried out with SHELX-97.35 Geometrical calculations were carried out with the program PARST.36CCDC-739433 (I), CCDC-739434 (2a) and CCDC-739435 (3a) numbers contain the supplementary crystallographic data for this paper (excluding structure factors). These data can be obtained free of charge viahttp://www.ccdc.cam.ac.uk/conts/retrieving.html (or from the Cambridge Crystallographic Data Centre, 12, Union Road, Cambridge CB21EZ, UK; fax: (+44)1223-336-033; or mailto:deposit@ccdc.cam.ac.uk).
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , a = 5.589(2) Å, b = 10.096(2) Å, c = 10.981(2) Å, α = 70.09(1)°, β = 86.17(1)°, γ = 74.03(1)°, V = 559.9(9) Å3, Z = 2, Dc = 1.502 Mg m−3, R = 0.0542 (2803 reflections), wR2 = 0.1300, T = 293(2) K, GOF = 0.944. The reflections were collected in the range 2.23° ≤ θ ≤ 26.99° employing a 0.15 × 0.16 × 0.14 mm crystal.
, a = 5.589(2) Å, b = 10.096(2) Å, c = 10.981(2) Å, α = 70.09(1)°, β = 86.17(1)°, γ = 74.03(1)°, V = 559.9(9) Å3, Z = 2, Dc = 1.502 Mg m−3, R = 0.0542 (2803 reflections), wR2 = 0.1300, T = 293(2) K, GOF = 0.944. The reflections were collected in the range 2.23° ≤ θ ≤ 26.99° employing a 0.15 × 0.16 × 0.14 mm crystal.
Acknowledgements
Authors thank the Universities of Milan and Pavia (PUR and FAR grants, respectively), the National Chemical Genomics Research Program and the Center for Biological Modulators of the 21st Century Frontier Research Program for DCH and BMK for the financial support.Notes and references
- J. E. Darnell, Science, 1997, 277, 1630–1635 CrossRef CAS.
- M. V. Karamouzis, P. A. Konstantinopoulos and A. G. Papavassiliou, J. Mol. Med., 2007, 85, 427–436 CrossRef CAS.
- K. Imada and W. J. Leonard, Mol. Immunol., 2000, 37, 1–11 CrossRef CAS.
- R. Buettner, L. B. Mora and R. Jove, Clin. Cancer Res., 2002, 8, 945–954 CAS.
- R. Catlett-Falcone, W. S. Dalton and R. Jove, Curr. Opin. Oncol., 1999, 11, 490–496 CrossRef CAS.
- S. Nam, R. Buettner, J. Turkson, D. Kim, J. Q. Cheng, S. Muehlbeyer, F. Hippe, S. Vatter, K.-H. Merz, G. Eisenbrand and R. Jove, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 5998–6003 CrossRef CAS.
- A. Kotha, M. Sekharam, L. Cilenti, K. Siddiquee, A. Khaled, A. S. Zervos, B. Carter, J. Turkson and R. Jove, Mol. Cancer Ther., 2006, 5, 621–629 CrossRef CAS.
- M. A. Blaskovich, J. Sun, A. Cantor, J. Turkson, R. Jove and S. M. Sebti, Cancer Res., 2003, 63, 1270–1279 CAS.
- B. B. Aggarwal, A. Kumar and A. C. Bharti, Anticancer Res., 2003, 23, 363–398 CAS.
- S.-C. Chen, Y.-L. Chang, D. I. Wang and J.-J. Cheng, Br. J. Pharmacol., 2006, 148, 226–232 CrossRef CAS.
- L. Costantino and D. Barlocco, Curr. Med. Chem., 2008, 15, 834–843 CrossRef CAS.
- J. Turkson, S. Zhang, J. Palmer, H. Kay, J. Stanko, L. B. Mora, S. Sebti, H. Yu and R. Jove, Mol. Cancer Ther., 2004, 3, 1533–1542 CAS.
- J. Turkson, D. Ryan, J. S. Kim, Y. Zhang, Z. Chen, E. Haura, A. Laudano, S. Sebti, A. D. Hamilton and R. Jove, J. Biol. Chem., 2001, 276, 45443–45455 CrossRef CAS.
- S. Fletcher, J. Turkson and P. T. Gunning, ChemMedChem, 2008, 3, 1159–1168 CrossRef CAS.
- J. Turkson, J. S. Kim, S. Zhang, J. Yuan, M. Huang, M. Glenn, E. Haura, S. Sebti, A. D. Hamilton and R. Jove, Mol. Cancer Ther., 2004, 3, 261–269 CAS.
- P. T. Gunning, W. P. Katt, M. Glenn, K. Siddique, J. S. Kim, R. Jove, S. M. Sebti, J. Turkson and A. D. Hamilton, Bioorg. Med. Chem. Lett., 2007, 17, 1875–1878 CrossRef CAS.
- J. Schust, B. Sperl, A. Hollis, T. U. Mayer and T. Berg, Chem. Biol., 2006, 13, 1235–1242 CrossRef CAS.
- K. Siddiquee, S. Zhang, W. C. Guida, M. A. Blaskovich, B. Greedy, H. R. Lawrence, M. L. R. Yip, R. Jove, M. M. McLaughlin, N. J. Lawrence, S. M. Sebti and J. Turkson, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 7391–7396 CrossRef CAS.
- D. Bhasin, K. Cisek, T. Pandharkar, N. Regan, C. Li, B. Pandit, J. Lin and P.-K. Li, Bioorg. Med. Chem. Lett., 2008, 18, 391–395 CrossRef CAS.
- W. Hao, Y. Hu, C. Niu, X. Huang, C.-P. B. Chang, J. Gibbons and J. Xu, Bioorg. Med. Chem. Lett., 2008, 18, 4988–4992 CrossRef CAS.
- B. A. Sherf, S. L. Navarro, R. R. Hannah and K. V. Wood, Promega Notes Magazine, 1996, 57, 2–8 Search PubMed.
- B. M. Kwon, D. C. Han, K. H. Son, D. S. Shin and J. Lee. US Pat., 2008/00514442 A1, 2008.
- A. B. Sheremetev, Yu. L. Shamshina and D. E. Dmitriev, Russ. Chem. Bull., 2005, 54, 1032–1037 Search PubMed.
- A. B. Sheremetev, N. S. Aleksandrova, D. E. Dmitriev, B. B. Averkiev and M. Yu, J. Heterocycl. Chem., 2005, 42, 519–525 CrossRef CAS.
- A. B. Sheremetev, Russ. Chem. Bull., 2005, 54, 1057–1059 Search PubMed.
- J. L. Castro, R. G. Ball, H. B. Broughton, M. G. N. Russell, D. Rathbone, A. P. Watt, R. Baker, K. L. Chapman, A. E. Fletcher, S. Patel, A. J. Smith, G. R. Marshall, W. Ryecroft and V. G. Matassa, J. Med. Chem., 1996, 39, 842–849 CrossRef CAS.
- D. E. Thurston, V. S. Murty, D. R. Langley and G. B. Jones, Synthesis, 1990, 81–84 CrossRef CAS.
- A. M. Felix, E. P. Heimer, T. J. Lambros, C. Tzougraky and J. Meienhofer, J. Org. Chem., 1978, 43, 4194–4196 CrossRef CAS.
- (a) C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter, 1988, 37, 785–789 CrossRef CAS; (b) A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- (a) E. Cancés, B. Mennucci and J. Tomasi, J. Chem. Phys., 1997, 107, 3032–3042 CrossRef; (b) M. Cossi, V. Barone, R. Cammi and J. Tomasi, Chem. Phys. Lett., 1996, 255, 327–335 CrossRef CAS; (c) V. Barone, M. Cossi and J. Tomasi, J. Comput. Chem., 1998, 19, 404 CrossRef CAS.
- The energy of the conformations optimized in vacuo was recalculated choosing also CHCl3 and DMSO as solvents. The overall population of each conformational family, recalculated in CHCl3, was 19.6, 0.2, 80.2, and 0.0% for the TT, TC, CT, CC families, respectively. The corresponding percentages in DMSO were 97.3, 0.2, 2.5, and 0.0%.
- X. Zhao, Y.-L. Chang, F. W. Fowler and J. W. Lauher, J. Am. Chem. Soc., 1990, 112, 6627–6634 CrossRef CAS.
- L. J. Farruja, J. Appl. Crystallogr., 1999, 32, 837–838 CrossRef.
- A. Altomare, M. C. Burla, M. Camalli, G. Cascarano, C. Giacovazzo, A. Gagliardi and G. Polidori, J. Appl. Cryst., 1994, 27, 435 CrossRef.
- G. M. Sheldrick, SHELX-97, Program for the Refinement of Crystal Structure, University of Göttingen, Germany, 1997 Search PubMed.
- M. Nardelli, J. Appl. Crystallogr., 1995, 28, 659 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Physico-chemical data and 1H NMR spectra of compounds I and 1–3. Summary data from the modeling study of compounds 2d and 3d. Crystallographic data of I, 2a and 3a. See DOI: 10.1039/c0md00057d |
| This journal is © The Royal Society of Chemistry 2010 |
