DOI:
10.1039/C0MD00043D
(Concise Article)
Med. Chem. Commun., 2010,
1, 45-49
Received
12th April 2010
, Accepted 18th May 2010
First published on
9th June 2010
Abstract
The X-ray crystallographic structures of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundmesylate salts of a novel voltage-gated sodium channel-binding ligand R-(−)-BW202W92 and its much less active S-(+)-enantiomer (BW203W92) have been determined to establish their absolute configurations. Each enantiomer exists as two distinct atropisomeric forms in the solid state and the crystal structures for each enantiomer are stabilized by quite distinct patterns of intermolecular hydrogen bonding. Such structural differences will influence the pharmacological properties of the enantiomers and hence dictate their contrasting receptor binding properties.
Introduction
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundR-(−)-2,4-Diamino-6-fluoromethyl-5-(2,3,5-trichlorophenyl)pyrimidine (BW202W92, (I) in Fig. 1) is derived from chemical classes of compounds (triazines and pyrimidines) typified by agents such as COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundlamotrigine (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundLamictal™; a clinical antiepileptic agent), and sipatrigine (619C89; a neuroprotective agent).1,2 Electrophysiological studies3,4 have been used to demonstrate that BW202W92 is a potent/selective COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundsodium-ion channel blocker with no effect on N, P/Q and T type COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcalcium channels and a weak effect on R type COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcalcium channels. These different kinds of structurally homologous channels are distinguished by their physiological roles, inhibition by specific toxins, high- or low-voltage activation as well as activation/inactivation kinetics.5 BW202W92 is also, in an in vivo model of cerebral ischaemic/stroke damage, a potent neuroprotective agent.3 Recent binding and ligand binding displacement studies, using a [3H]-labelled form of BW202W92 as a ligand, have revealed a novel stereoselective drug binding site for COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundsodium channels in rat forebrain synaptosomes.6 Although the level of specific [3H]-BW202W92 binding in a standard incubation medium is relatively poor, the addition of low concentrations of tetrodotoxin (EC50 = 2–3 nM) greatly enhances the binding, apparently by increasing the affinity of the implicated binding sites. Tetrodotoxin-dependent binding is markedly stereoselective such that under these experimental conditions the S-(+)-enantiomer (BW203W92, (II) in Fig. 1) is up to 30-fold less potent, and extremely sensitive to inhibition by elevated K+ ion concentrations (IC50 = 5.9 mM). The latter effect has been ascribed to changes in membrane potential.6 This discovery has extremely important and wide-ranging implications: (i) the identification of a novel sodium channel binding site, and (ii) more importantly, that this site may represent a new target for drug intervention.6 A detailed structural study for BW202W92 is now warranted and will be of paramount importance to a broad spectrum of scientific disciplines, including medicinal chemistry, molecular physiology, pharmacology, and biophysics. To this end, we have determined the X-ray crystal structures for the mesylate salts of BW202W92 (R-form) and BW203W92 (S-form) to establish their absolute chiral configurations and critical spatial properties. The mesylate acid addition salts of the two enantiomers crystallized in different space groups, each comprising two crystallographically independent A and B molecules per asymmetric unit. Further, atropisomerism is exhibited by the molecules present in each structural arrangement. Herein we report the crystal structures of both the biochemically active R-form and the relatively inactive S-form and contrast their molecular presentations.
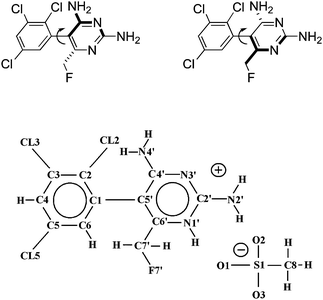 |
| | Fig. 1 (Top) Schematic chemical structure for the sterically hindered rotamers of I and II, respectively. (Bottom) General numbering scheme used for the enantiomeric compounds (R-form = BW202W92, S-form = BW203W92). Note that the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundpyrimidine ring is N1′-protonated in each of the mesylate acid addition salts. | |
Results and discussion
Experimental details are given in the ESI.† Crystal and structure refinement data for I and II are summarized in Table 1. Atom coordinates and equivalent isotropic thermal parameters are given in Tables S1A (I) and S1B (II), ESI,† respectively. Molecular geometry data are shown in Tables S2A (I) and S2B (II), ESI† (bond lengths and angles), and Tables S3A (I) and S4B (II), ESI† (torsion angles). COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundHydrogen bond geometries are tabulated in Tables S4A (I) and S4B (II), ESI,† respectively.
Table 1 Crystal data and structure refinement for the R-form (BW202W92; I) and S-form (BW203W92; II)
| Identification code |
BW202W92 (R) (I) |
BW203W92 (S) (II) |
|
The CAD4 diffractometer was set to discard extremely weak reflections in order to save collection time and as a precaution against excessive deterioration of the crystal.
|
| Empirical formula |
C12H12Cl3FN4O3S |
C12H12Cl3FN4O3S |
| Chemical formula |
C11H9Cl3FN4CH3SO3 |
C11H9Cl3FN4CH3SO3 |
| Formula weight |
417.67 |
417.67 |
|
T/K |
293(2) |
123(2) |
| Wavelength/Å |
1.54180 |
0.71073 |
| Crystal system |
Monoclinic |
Triclinic |
| Space group |
P21 |
P1 |
|
a/Å |
8.384(2) |
7.716(2) |
|
b/Å |
16.984(3) |
8.120(2) |
|
c/Å |
12.480(3) |
13.719(3) |
|
α (°) |
90 |
74.91(3) |
|
β (°) |
104.14(2) |
87.69(3) |
|
γ (°) |
90 |
89.83(3) |
| Volume/Å3 |
1723.4(7) |
829.2(3) |
| Z |
4 |
2 |
|
d
calcd (mg m−3) |
1.609 |
1.673 |
|
μ/mm−1 |
6.238 |
0.709 |
|
F(000) |
844 |
422 |
| Crystal size/mm |
0.30 × 0.16 × 0.16 |
0.30 × 0.20 × 0.20 |
|
θ range for |
|
|
| data collection (°) |
4.49 to 74.42 |
2.60 to 27.47 |
| Index ranges |
−10 ≤ h ≤ 10 |
−9 ≤ h ≤ 9 |
| −8 ≤ k ≤ 21 |
−10 ≤ k ≤ 10 |
| −6 ≤ l ≤ 15 |
−17 ≤ l ≤ 15 |
| Reflections collected |
3990 |
6456 |
| Independent reflections |
3192 [R(int) = 0.0496] 81.9% |
5302 [R(int) = 0.0733] 93.9% |
| Completeness to θ = 74.42°a |
81.9% |
93.9% |
| Refinement method |
Full-matrix least-squares on F2 |
Full-matrix least-squares on F2 |
| Data/restraints/parameters |
3192/1/450 |
5302/3/452 |
| Goodness-of-fit on F2 |
1.042 |
0.938 |
| Final R indices [I > 2σ(I)] |
R
1 = 0.0490 |
R
1 = 0.0614 |
| wR2 = 0.1254 |
wR2 = 0.1204 |
|
R indices (all data) |
R
1 = 0.0644 |
R
1 = 0.1248 |
| wR2 = 0.1324 |
wR2 = 0.1468 |
| Absolute structure parameter |
0.05(3) |
−0.10(9) |
| Extinction coefficient |
0.00150(16) |
0.0071(12) |
| Largest diff. peak and hole e.Å−3 |
0.339 and −0.510 |
0.384 and −0.351 |
For the R-form the absolute structure parameter7 is 0.05(3), indicating that the absolute configuration has been correctly assigned. Similarly for the S-form the absolute structure parameter7 is −0.10(9), which again confirms that the correct absolute configuration has been assigned. Furthermore, independent crystallization of the two enantiomeric molecules in different, and, more importantly, non-centrosymmetric space groups is a strong indication, although not a definite proof, that the R and S forms of this molecule are not easily interconvertible simply through rotation about the central bond.
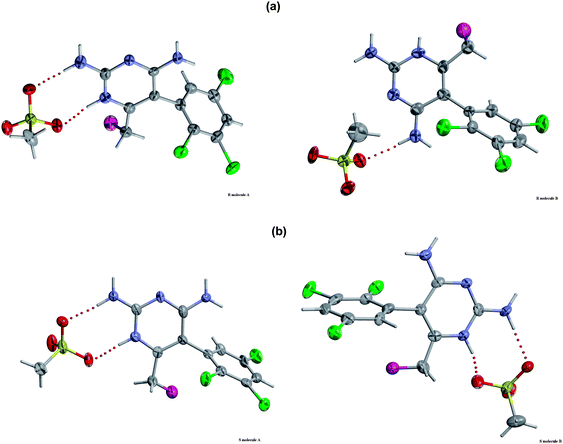
Fig. 1 shows the formula and atom numbering scheme used for the H[BW202W92]+ cation and associated counterion in the mesylate salt. ORTEP/RASTER-3D8,9 views of the molecules A and B in the R-form and S-form are shown in Fig. 2a and 2b, respectively. To aid comparison, the view direction is shown perpendicular to the pyrimidine ring.
 |
| | Fig. 2 ORTEP/RASTER-3D views of the paired A and B molecules in the crystal structures for the (a) R-enantiomer and (b) S-enantiomer, showing the 50% probability thermal motion ellipsoids. A common perpendicular orientation for the pyrimidine rings is used for comparison. For clarity, the mesylate counteranions are not shown. | |
Molecular geometry
Bond lengths in the R-form structure are determined to approximately within ±0.005 Å and bond angles to ±0.4°; corresponding values for the S-form were ±0.006 Å and to ±0.4°, respectively. Bond lengths and angles in each structure compare well with those found in organic compounds,10 exhibit no unusual values, and reveal no significant differences between corresponding regions of the different molecules. All ring atoms in each ring of both isomeric forms are coplanar (within 0.004 Å) and all bonded atoms for each ring are also positioned in the corresponding plane (i.e., within 0.004 Å).
Axial chirality and atropisomerism
The presence of two symmetry-independent molecules in the asymmetric unit of a crystal structure provides an ideal opportunity to assess variations in the molecular geometry, which here corresponds to differences in relative orientation for the rings in molecules A and B in both the R- and S-enantiomeric forms. The presence of three bulky flanking groups about the pivot bond C(1)-C(5′), Cl(2) on the benzene ring, and N(4′)H2 and C(7′)H2F(7′) on the pyrimidine ring, provides a very high barrier to free rotation such that two stable rotamers can exist without dynamic interchange at room temperature (Fig. 1). Interestingly this behaviour is observed for both the R- and S-enantiomers of this compound, with molecules A and B evident in each crystal structure. Fig. 3(a) shows views of molecules A and B in the R-enantiomer, respectively, viewed along the C(5′)–C(1) bond from the pyrimidine ring side, clearly showing that they are distinct atropisomers of this type. The distinction is characterized by the torsion angle τ = C(2)–C(1)–C(5′)–C(4′), which is −111.9(6)° in molecule A and −69.0(7)° in molecule B. Similarly, Fig. 3(b) shows molecules A and B in the S-enantiomer for which τ = 104.8(6)° in molecule A and 71.7(6)° in molecule B. Thus, both the R- and S-forms exhibit clear atropisomerism within their respective crystal forms. It is evident from Fig. 2 that the presence of three distinct chemical groups provides the basis for axial chirality in these compounds and, importantly, that the correct absolute configurations are assigned to each enantiomer (see crystallographic discussion, below). This is confirmed by the rotation direction of the priority sequence 1[N(4′)]–2[C(6′)]–3[Cl(2)], which is clockwise and anti-clockwise for the R- and S-forms, respectively. Formation of the two atropisomers in each compound presumably occurs during the synthesis procedure. Tetrodotoxin-dependent binding of this agent to rat forebrain synaptosomes has been shown to be stereoselective, as the less active S-(+)-enantiomer BW203W92 is up to 30-fold less potent on a dose basis compared to the active R-(−)-form.6 It is also likely that, in addition to this enantioselectivity, one atropisomeric form of the R-isomer (i.e., molecule A or B) may exhibit superior binding to the implicated bioreceptor.
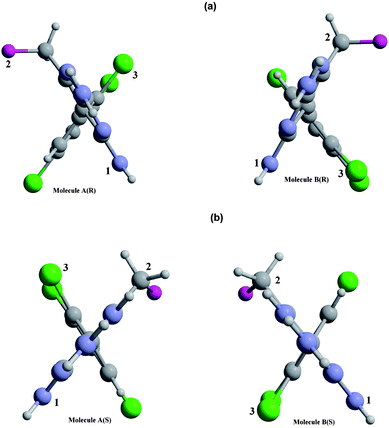 |
| | Fig. 3 Views for A and B molecules respectively in the (a) R-isomer and (b) S-isomer, viewed along the C(5′)–C(1) chiral axis and showing the absolute configurations. The ring planes are viewed edge-on and the interplane torsion is determined by C(2)–C(1)–C(5′)–C(4′). | |
Read more about this on ChemSpider
Download mol file of compoundH-bonding arrangement between molecule A or B and their associated COMPOUND LINKS
Read more about this on ChemSpider
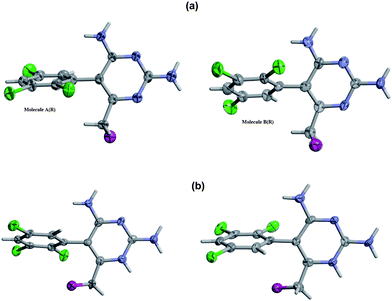
Download mol file of compoundmesylate counteranions. Fig. 5 reveals characteristically different extended networks of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundH-bonds for each of the enantiomeric forms. In the R-form, the BW202W92 molecules are COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundH-bonded to the O atoms of the mesylate through N(1′), N(2′) and N(4′). There is also a short contact between F(7′) and N(2′) of a neighbouring molecule. In contrast, for the S-enantiomer, the BW203W92 molecules form complementary hydrogen bonds between their N(4′)H2 groups in symmetry-related molecules, and also to the O atoms of the mesylate anion through N(1′), N(2′) and N(4′). Knowledge of these interactions will inform an understanding of why the R-form selectively binds to sodium channel receptors.
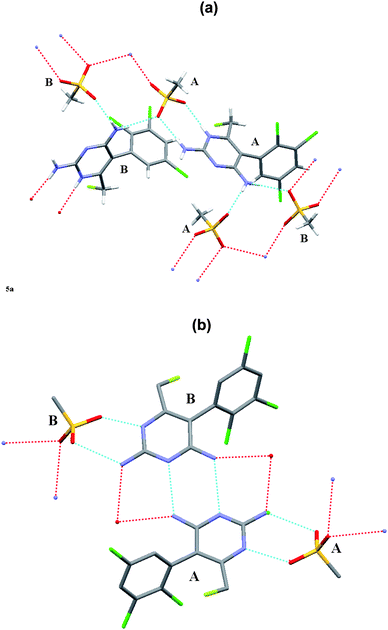 |
| | Fig. 5 Intermolecular hydrogen-bonded network patterns for the component molecules in the crystal structures for (a) the R-enantiomer and (b) the S-enantiomer. | |
Molecular modelling
Cronin et al.11 modelled the binding of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundlamotrigine using the known structure of the central helices on the potassium channel template and a model has also been determined for COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundlamotrigine binding to the sodium channel on the basis of mutation analysis.12 Currently, however, there is no suitable published structural template available exclusively for the sodium channel and particularly for brain sodium channel subtypes. In the absence of such a template this necessarily precludes any meaningful modelling studies for BW202W92 and BW203W92 receptor interactions.
It is anticipated that molecular modelling using each of the BW202W92 and BW203W92 ligand components would be more informative than previous attempted studies, as all other known members of the lamotrigine family crystallize in centrosymmetric space groups and thus contain equal contributions from their R and S enantiomers. On this basis, models derived from other structures, including COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundlamotrigine itself, will be unable to distinguish possible differences for the R and S modes of binding indicated from the marked contrast in drug behaviour evident for (I) and (II).
Conclusions
It is established from previous work that BW202W92 binds to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundsodium channels at a novel stereoselective site such that the R-form is biochemically potent whereas its S-enantiomer (BW203W92) is relatively inactive. In addition, other compounds of this COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundlamotrigine family of similar structure also display markedly different therapeutic preferences, e.g. for epilepsy, pain, or neurodegeneration. A major challenge for medicinal chemists is to account for such differences in drug activity at the molecular level. This structural study has highlighted that there are large and significant differences in both local structure and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundH-bonding for the R- and S-enantiomers of BW202W92. These structural differences are likely to reflect the different pharmacological behaviors of the two agents in vivo and their mechanism(s) for drug interaction at the receptor site. The detailed structural information obtained for the active/inactive forms of the two enantiomers of this drug provide a firm basis to discern subtle differences between agents of the important COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundlamotrigine class and to inform drug design for next-generation agents. In particular, the atropisomerism behaviour can now be exploited in dynamic molecular modelling studies of drug interaction with target receptors to gauge critical stereochemical factors that will be important for QSAR studies.
Acknowledgements
We thank Dr P. Barraclough (University of Greenwich) for the synthesis and provision of samples of BW202W92 and BW203W92 compounds. Low-temperature X-ray intensity data were collected using the EPSRC single-crystal X-ray data facility at the University of Southampton, UK.
References
-
M. J. Leach, A. D. Randall, A. Stefani and A. H. Hainsworth, in Antiepileptic Drugs; R. H. Levy, R. H. Mattson, B. S. Meldrum, E. Perucca, eds.; Lippincott, Williams & Wilkins: USA 2002, 363–369, 5th edition Search PubMed.
- A. H. Hainsworth, A. Stefani, P. Calabresi, T. W. Smith and M. J. Leach, CNS Drug Rev., 2000, 6, 111–134 CAS.
- L. Caputi, A. H. Hainsworth, F. Lavaroni, M. J. Leach, N. C. McNaughton, N. B. Mercuri, A. D. Randall, F. Spadoni, J. H. Swan and A. Stefani, Brain Res., 2001, 919, 259–268 CrossRef CAS.
- A. H. Hainsworth, N. C. McNaughton, A. Pereverzev, T. Schneider and A. D. Randall, Eur. J. Pharmacol., 2003, 467, 77–80 CrossRef CAS.
-
S. P. H. Alexander, A. Mathie and J. A. Peters, Brit. J. Pharm. (Guide to Receptors & Channels 4th Ed.) 2009, 158, Supplement 1, S127–S128; S146–S147 Search PubMed.
- D. R. Riddall, M. J. Leach and J. Garthwaite, Mol. Pharmacol., 2006, 69, 278–287 CAS.
- H. D. Flack, Acta Crystallogr., Sect. A: Found. Crystallogr., 1983, 39, 876–881 CrossRef.
-
C.L.Barnes ORTEP-3 for Windows-a version of ORTEP-III with a Graphical User Interface (GUI). J. Appl. Cryst., 30 ( 1997), 30, pp. 568–589 [Based on ORTEP-III (v 1.0.3) by C.K. Johnson and M.N. Burnett.] Search PubMed.
- E. A. Merrit and D. J. Bacon, Raster 3D Graphics version 2.7c., Methods Enzymol., 1997, 277, 505–524 CAS [Implemented in WinGX (qv) and generated by Ortep-3 for Windows.].
-
M. F. C. Ladd, R. A. Palmer, Structure Determination by X-ray Crystallography, Klewer-Plenum: N.Y., USA ( 2003) p503, 4th Edition Search PubMed.
- N. B. Cronin, A. O'Reilly, H. Duclohier and B. A. Wallace, J. Biol. Chem., 2003, 278, 10675–10682 CrossRef CAS.
- V. Yarov-Yarovoy, J. Brown, E. M. Sharp, J. J. Clare, T. Scheuer and W. A. Catterall, J. Biol. Chem., 2000, 276, 20–27 CrossRef.
|
| This journal is © The Royal Society of Chemistry 2010 |
Click here to see how this site uses Cookies. View our privacy policy here.