SCOP/PHLPP and its functional role in the brain
Kimiko
Shimizu
a,
Scott M.
Mackenzie
bc and
Daniel R.
Storm
*b
aDepartment of Biophysics and Biochemistry, Graduate School of Science, University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-0033, Japan
bDepartment of Pharmacology, University of Washington, Seattle, WA 98195, USA. E-mail: dstorm@uw.edu
cGraduate Program in Neurobiology & Behavior, University of Washington, Seattle, WA 98195, USA
First published on 30th September 2009
Abstract
SCOP (suprachiasmatic nucleus (SCN) circadian oscillatory protein) was originally identified in 1999 in a differential display screen of the rat SCN for genes whose expression were regulated in a circadian manner (K. Shimizu, M. Okada, A. Takano and K. Nagai, FEBS Lett., 1999, 458, 363–369). The SCN is the principle pacemaker of the circadian clock, and expression of SCOP protein in the SCN was found to oscillate, increasing during the subjective night, even when animals were housed in constant darkness. SCOP interacts with and inhibits multiple proteins important for intracellular signaling, either by directly binding to K-Ras or by dephosphorylating p-Akt and p-PKC. Since the functions of K-Ras, Akt, and PKC are considerably divergent, SCOP may have several roles. We recently discovered that SCOP participates in the formation of long-term hippocampus-dependent memories, and other investigators have examined its role in cell proliferation and survival. In this review, we introduce SCOP from its molecular structure to its physiological functions, focusing mainly on its role in ERK1/2activation and memory consolidation.
 Kimiko Shimizu | Dr Kimiko Shimizu is an assistant professor in the Department of Biophysics and Biochemistry at the University of Tokyo. Her main interest is molecular mechanisms underlying brain functions with an emphasis on the physiological function of SCOP. She received a PhD in Science at Osaka University, Japan, after which she did postdoctoral studies at the University of Tokyo, Japan, and at the University of Washington, Seattle, USA. In 2007, she took her current position in Tokyo. |
 Scott M. Mackenzie | Scott M. Mackenzie has been a PhD student in Neurobiology and Behavior at the University of Washington, Seattle, since 2007 under the supervision of Professor Daniel R. Storm. His principle research interest is using behavioral pharmacology to investigate the role of signal transduction in the consolidation of hippocampus-dependent memories. He graduated with honors with a BS in Neurobiology and a BA in Psychology from the University of California, Irvine, in 2007. |
 Daniel R. Storm | Dr Daniel R. Storm has been a full professor of Pharmacology at the University of Washington Medical School since 1982. His main research interest is the molecular and cellular basis of neuroplasticity with a focus on memory formation and chemosensory mechanisms using transgenic mice as a model system. He received BS and MS degrees in Chemistry from the University of Washington and a PhD in Biochemistry from the University of California, Berkeley. After postdoctoral research at Harvard, he went to the Department of Biochemistry at the University of Illinois in 1973 and came to the University of Washington as an associate professor of Pharmacology in 1978. |
Structure and expression of SCOP
The SCOP gene is located on chromosome 1 in mice and encodes a transcript composed of 17 exons. SCOP is a large polypeptide (1687 amino acids in mice) containing a pleckstrin homology (PH) domain, a leucine-rich repeat (LRR), a protein phosphatase 2C (PP2C)-like domain, a glutamine (Q)-rich region, and a PDZ-binding domain (Fig. 1), suggesting a variety of roles in intracellular signaling. This domain composition has led to the alternate name “PHLPP” (PH-domain LRR protein phosphatase),1 and it was later determined that PHLPP encompasses a family of three proteins. PHLPP1α and PHLPP1β are products of the same gene but have different sizes due to separate start codons. PHLPP2 is a different gene product but has the same domain composition of PHLPP1.2 We detected only PHLPP1β (approximately 183 kDa) in mouse and rat brain lysates via immunoblot analysis, although we were able to detect the shorter PHLPP1α in HEK 293 cells. SCOP refers to PHLPP1β,2 and its amino acid and cDNA sequences are very highly conserved among mammals, including humans, mice, and rats.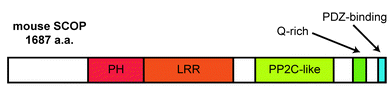 | ||
| Fig. 1 The domain composition of SCOP. Positions of the PH, LRR, PP2C-like, Q-rich, and PDZ-binding domains in the mouse SCOP protein are indicated. | ||
SCOP protein was readily detected only in the tissues of the central nervous system (Fig. 2),3 although microarray analysis and the distribution of PHLPP ESTs indicate that it is expressed in the majority of human tissues (http://genome-www.stanford.edu/). Immunoblot analysis revealed that SCOP is particularly enriched in the hippocampus and the piriform cortex. A marked immunoreactivity migrating slightly faster than the predicted size of 183 kDa was also detected in the rat testes, though its identity remains unclear. The intracellular localization of SCOP was examined using subcellular fractionation followed by immunoblot analysis.3 Comparable immunoreactivities for SCOP were detected in the nuclear, mitochondrial, and cytosolic fractions, potentially representing heterogeneous distribution of SCOP within the cell. Immunoblot analysis of primary cultured neurons and astrocytes from the rat cerebrum demonstrated that SCOP expression was highly concentrated in neurons, suggesting neuron-specific functions within the brain. Interestingly, we discovered that SCOP is present in membrane rafts,4 which are small, specialized domains that compartmentalize cellular processes and are enriched in certain lipids, proteins, and cholesterol. Membrane rafts are known to be important for effective signal transduction, and the PH domain probably participates in this membrane localization. Though SCOP exists in both raft and non-raft fractions, it may have different functions in these subcellular regions.
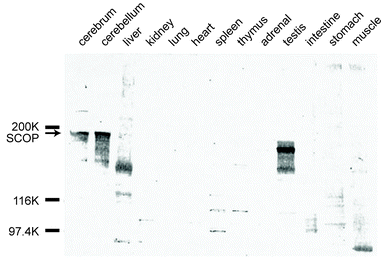 | ||
| Fig. 2 Immunoblot analysis of SCOP protein distribution. Protein extracts were prepared from various tissues of adult male rats. Marked immunoreactivity was observed only in the brain (cerebrum and cerebellum) and testes. Immunoreactivity of the testes migrated slightly faster than the predicted 183 kDa SCOP protein. | ||
Several regulatory mechanisms appear to tightly control SCOP expression. The level of SCOP protein in cultured primary hippocampal neurons decreased by approximately 50% within 5 minutes after brain-derived neurotrophic factor (BDNF)-induced degradation and recovered within 1 hour, suggesting rapid turnover.5 This degradation was mediated by calpain 1 and 2, which are Ca2+-activated cysteine proteases. In neurons, calpain 1 and 2 predominate among 15 identified isoforms and are found in distinct subcellular regions with different Ca2+ affinities, suggesting that they serve separate physiological roles. Ca2+ influx is a common feature of neuronal activity, and rapid regulation of SCOP protein levels by calpain 1/2 may be important for several activity-dependent phenomena. Some reports indicate that, like SCOP, calpain 2 localizes in the membrane rafts.6,7 The presence of other important Ca2+ signaling proteins in these rafts, including Ca2+ channels, receptor tyrosine kinases, and effectors of Ca2+ signaling (e.g., Ca2+-sensitive adenylyl cyclases and CaMKII), emphasizes the potential importance of SCOP in regulating signal transduction.
Molecular and physiological functions of SCOP
Inhibition of K-Ras
The LRR within SCOP is composed of 18 repeats of a short stretch with the relatively conserved but variable sequence LXXLXLXXNXLXXLPXXAXXL, where L, N, P, and A denote leucine, asparagine, proline, and aliphatic amino acids, respectively, and where X denotes any amino acid (Fig. 3).3 This LRR consensus sequence is known to be conserved in the yeast adenylyl cyclase, which contains 26 LRRs8 that are required for binding to and activation by Ras during vegetative growth.9,10 SOC-211 and SUR-812 have LRR domains homologous to the yeast adenylyl cyclase LRR and also bind to Ras, enabling them to regulate Ras-mediated signaling. Thus, we hypothesized that SCOP was involved in Ras-mediated signaling through an interaction between the SCOP LRR and Ras.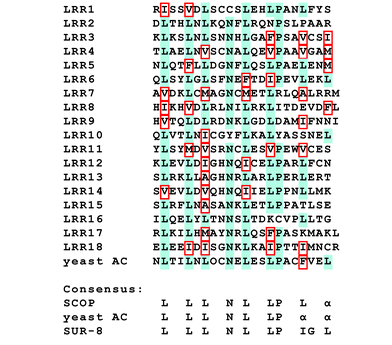 | ||
| Fig. 3 LRR sequence homology of SCOP, adenylyl cyclase, and SUR-8. Alignment of 18 SCOP LRRs and the consensus across SCOP, yeast adenylyl cyclase (AC), and human SUR-8 are shown. Residues conserved in the majority of LRRs are shaded in blue. Allowable substitutions for leucine residues are outlined in red. | ||
Acting as a molecular switch, Ras cycles between a GTP-bound active state and a GDP-bound inactive state viaGTP hydrolysis and GDP–GTP exchange. The SCOP LRR has no or very low affinity for both GDP- and GTPγS-bound forms of K-Ras,4 so it is most probable that the LRR domain binds to the nucleotide-free form of K-Ras present during GDP–GTP exchange (Fig. 4). Interestingly, we detected SCOP binding only with K-Ras and not with N-Ras or H-Ras despite very high homology among these Ras isoforms. One possible explanation is that only K-Ras and SCOP colocalize in the membrane rafts, and this is the only region in which we have found an interaction. However, an adaptor protein is not required as we previously confirmed that recombinant SCOP and recombinant K-Ras expressed in Escherichia coli can bind directly to each other in vitro.4 We predicted that the binding of SCOP to the nucleotide-free form of K-Ras inhibited its conversion to the GTP-bound active state, and, in fact, addition of a SCOP LRR peptide fragment into the raft fraction inhibited K-Ras GTP-binding activity in vitro.4 SCOP expression also suppresses downstream activation of ERK1 and ERK2, which are well-known downstream effectors of Ras.4 Ras–ERK signaling plays critical roles in cell proliferation, differentiation, and migration in response to extracellular signals. Although it is still unknown how many of the several ERK isoforms are regulated in this manner, SCOP may be one key to the compartmentalization and discrimination of their multiple functions.
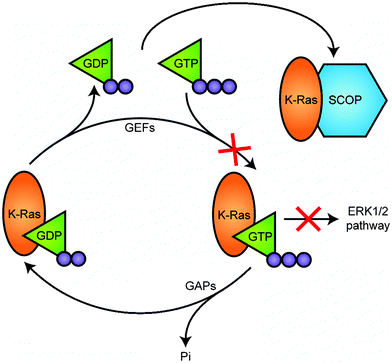 | ||
| Fig. 4 SCOP negatively regulates activation of K-Ras. Ras cycles between a GTP-bound active state and a GDP-bound inactive state viaGTP hydrolysis and GDP–GTP exchange steps. Guanine nucleotide exchange factors (GEFs) interact, reduce the affinity of Ras for GDP, facilitating formation of the nucleotide-free form, to which GTP binds for its activation. GTPase activating proteins (GAPs) increase the intrinsic GTPase activity of Ras and thereby promote its inactivation. SCOP is unique in that it binds to the nucleotide-free form to inhibit formation of the GTP-bound active form. | ||
Potential interactions with circadian clockwork
In mammals, peripheral clocks are synchronized by the master circadian clock located in the SCN of the ventral hypothalamus. SCOP was originally identified in a differential display screen for genes whose expression was regulated in a circadian manner within the rat SCN.3 SCOP protein expression in the SCN increased during subjective night (the active phase for rats and mice) from circadian time (CT) 12 to CT 21 during constant darkness (CT 0 refers to the start of the subjective day).3 Furthermore, the level of p-ERK in the SCN during subjective night is lower than during subjective day.13 This pattern of p-ERK expression is consistent with the oscillation of SCOP given that SCOP negatively regulates the K-Ras–ERK1/2 pathway. In the hypothalamic region outside the SCN, the level of SCOP protein was unchanged and consistent with the pattern of its mRNA expression. These findings suggest that the expression of SCOP protein is related to the generation or transmission of endogenous circadian oscillation in the SCN.The circadian rhythm is driven by transcription/translation-based feedback loops,14,15 and a critical feature of circadian biology is the ability of this clockwork to be reset by external stimuli, allowing animals to adjust their rhythm to changes in environmental conditions. Light is the strongest phase-adjusting stimulus that affects the circadian rhythm, and this signal is transduced by the secretion of glutamate from retinohypothalamic tract nerve terminals, followed by activation of NMDA receptors on SCN cells and subsequent Ca2+ influx.16,17 The intracellular signaling pathways that mediate light entrainment are not yet well understood, but recent studies have demonstrated that the Ras–ERK1/2signaling cascade makes an important contribution to the circadian clock of the rodent SCN. For example, light pulses during the subjective night produce an increase in Ras activity18 and a rapid up-regulation of ERK1/2phosphorylation13,19,20 in the SCN. This suggests that Ras and ERK1/2activation are involved in the signaling mechanisms that couple photic information to the circadian clock. Since SCOP is a negative regulator of the K-Ras–ERK1/2 pathway and degradation of SCOP is controlled in a Ca2+-dependent manner viacalpainproteases (Fig. 5), it is plausible that SCOP contributes to photic entrainment of the circadian clock in the SCN.
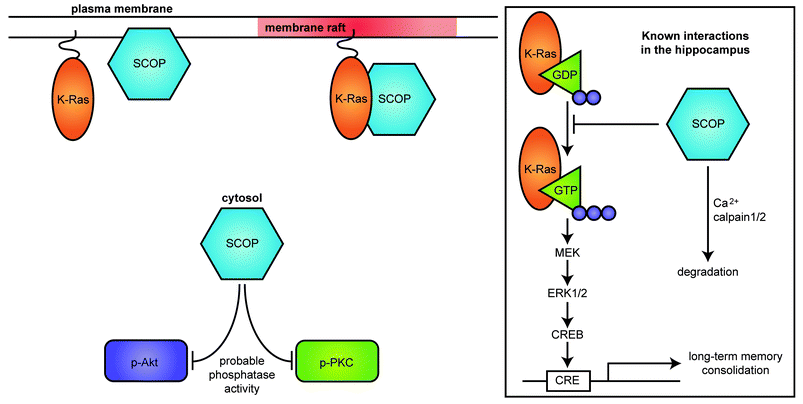 | ||
| Fig. 5 SCOP localization within the neuron. SCOP is expressed in the cytosol, on membranes, and in membrane rafts. SCOP colocalizes with and binds to K-Ras in membrane rafts. SCOP is also reported to have phosphatase activity in non-neuronal cells, which may have other functions in neurons. Inset: In the hippocampus, SCOP attenuates ERK1/2 activity by binding to the nucleotide-free form of K-Ras, thereby inhibiting the ERK signaling pathway. Calpain-mediated degradation of SCOP may contribute to activation of ERK1/2. | ||
Regulation of learning and memory
Negative regulation of ERK1/2 by SCOP is important for mechanistic studies of memory since ERK1/2 signaling is known to play a critical role in several forms of neuroplasticity, including long-term memory consolidation. ERK1/2activation mediates Ca2+ stimulation of cAMP response element (CRE)-mediated transcription21–23 and dendritic translation,24 and we have confirmed that SCOP inhibits CRE-mediated transcription in cultured primary hippocampal neurons.5 To determine if hippocampus-dependent memory is sensitive to changes in hippocampal SCOP expression in vivo, we made a tetracycline-inducible, SCOP overexpressing mouse strain using the CaMKIIα promoter to limit overexpression to the forebrain.5 SCOP overexpression completely blocked memory for novel objects when measured 24 hours after training (long-term memory) but had no effect when measured 8 minutes after training (short-term memory).5 Several studies have demonstrated that the hippocampus contributes to performance during object recognition tasks.25–27 However, overexpression of SCOP did not block memory for contextual fear conditioning, another hippocampus-dependent task, even though training for both learning paradigms reduced SCOP expression in wild-type mice.5 There are several possible reasons why novel object memory was more sensitive than fear conditioning to changes in SCOP expression in the hippocampus. For example, the training stimuli used in these two learning paradigms are very different: one allows the animal to observe of an object at will but the other surprises the animal with an electric shock, resulting in a stronger memory. Though both learning paradigms rely on hippocampal activity, other brain regions are also involved in the consolidation process for each (e.g., novel object memory is generally independent of the amygdala),28 and it remains unclear how much similarity exists between them.Infusing calpaininhibitors into area CA1 of the hippocampus also blocked long-term memory for novel objects with no effect on short-term memory, similar to the effects of overexpressing SCOP in transgenic animals.5 This was predicted from earlier experiments demonstrating that inhibition of calpain increases expression of SCOP in cultured primary hippocampal neurons. Since calpain regulates the expression of SCOP protein, transgenic mice with mutations in calpain could exhibit altered memory formation. A study of calpain 1 knockout mice showed no impairment for fear memory or LTP,29 but there are no data on memory for novel objects and there have been no reports of calpain 2 knockout mice. Though calpain 2 knockout or knockdown mice might show a deficit in memory for novel objects, calpain has many substrates, and potential interaction effects are an ongoing concern.
Many investigators have linked K-Ras and ERK1/2 to learning and memory. Although studies using knockout mice are difficult because loss of K-Ras produces an embryonic lethal mutation, K-Ras heterozygous mice are viable, and administering a MEK inhibitor to these animals produced a memory deficit.30ERK1/2activation is also required for the expression of long-term memory.31ERK1 knockout mice did not show significant impairment in learning,32ERK2 knockout mice are embryonic lethal, and ERK2 heterozygous mice are anatomically impaired. However, ERK2 knockdown mice, in which expression was partially (20–40%) reduced while the animals remained viable and fertile with normal appearance, exhibited a deficit in long-term memory for fear conditioning whereas short-term memory was intact.33 Overexpression of SCOP may induce memory impairment through its effects on ERK2 and related downstream signaling.
Regulation of cell proliferation and survival
In 2005, SCOP was found to have serine/threonine phosphatase activity in its PP2C-like domain.1 As described in the introduction, these investigators originated the alternative name PHLPP and have identified three isoforms to-date, of which SCOP is PHLPP1β. PHLPP specifically targeted the phosphorylation site at Ser473 of Akt, with negative consequences for the survival and proliferation of cultured cancer cells.1,2 These data suggested a possible role as a tumor suppressor, and it was later confirmed that overexpression of either PHLPP1 or 2 reduced tumor volume in vivo.34 However, PHLPP1 and 2 differentially regulate both ERK1/2 and Akt signaling. Unlike PHLPP1, overexpression of PHLPP2 did not affect phosphorylation of either MEK1/2 or ERK1/2, and while PHLPP1 specifically dephosphorylated Akt2 and 3, PHLPP2 targeted Akt1 and 3.2 The interaction between PHLPP and PKC has received less attention, but it is known that dephosphorylation of the hydrophobic motif of PKCβII by either PHLPP1 or 2 destabilizes the protein, leading to its degradation.35Questions for future research
SCOP is localized predominantly in the cytosol, though some attaches to the membrane and the membrane rafts. According to our theory, the presence of SCOP in membrane rafts is important for memory consolidation and possibly other neuronal phenomena because SCOP binds to and inhibits K-Ras activity in this region. It is possible that a reserve supply of SCOP exists to replenish any that degrades in membrane rafts and thus restore its inhibition of K-Ras as soon as possible. SCOP might be mobile within the cell, enabling it to stabilize protein levels across several subcellular regions. These and other possibilities mentioned above suggest that SCOP plays an important role in regulating learning and circadian processes.Other exciting avenues of research concern the phosphatase activity of SCOP. All research to-date examining this role has been performed in non-neuronal cells, though interactions with Akt and PKC may occur in neurons, as well. In the brain, the PI3K–Akt pathway seems to contribute to mechanisms of synaptic plasticity and consolidation of novel object memory,36 and there are some reports that PKC is also involved in synaptic plasticity and memory consolidation.37–39 Although we have published evidence that SCOP regulates consolidation of certain memories via K-Ras–ERK1/2 signaling, potential interactions with Akt and PKC in neurons might serve ancillary roles. In circadian biology, PKC is reported to be involved in photo-mediated entrainment of circadian rhythms.40 If SCOP helps mediate the stability of PKC in the SCN, then this may explain the oscillation of SCOP protein and mRNA expression of that region. Finally, SCOP may also have important roles during development. SCOP expression does not change during development and growth (embryonic day 14 to adult) even though many other neuron-specific molecules vary in their expression. Further investigation is necessary to determine what role, if any, SCOP has during development. Overall, research on SCOP is still at an early stage, and future studies will provide a fuller understanding of how SCOP mediates these multiple signal transduction pathways in the brains of developing and adult organisms.
References
- T. Gao, F. Furnari and A. C. Newton, Mol. Cell, 2005, 18, 13–24 CrossRef CAS.
- J. Brognard, E. Sierecki, T. Gao and A. C. Newton, Mol. Cell, 2007, 25, 917–931 CrossRef CAS.
- K. Shimizu, M. Okada, A. Takano and K. Nagai, FEBS Lett., 1999, 458, 363–369 CrossRef CAS.
- K. Shimizu, M. Okada, K. Nagai and Y. Fukada, J. Biol. Chem., 2003, 278, 14920–14925 CrossRef CAS.
- K. Shimizu, T. Phan, I. M. Mansuy and D. R. Storm, Cell (Cambridge, Mass.), 2007, 128, 1219–1229 CrossRef CAS.
- J. L. Hood, B. B. Logan, A. P. Sinai, W. H. Brooks and T. L. Roszman, Biochem. Biophys. Res. Commun., 2003, 310, 1200–1212 CrossRef CAS.
- L. A. Morford, K. Forrest, B. Logan, L. K. Overstreet, J. Goebel, W. H. Brooks and T. L. Roszman, Biochem. Biophys. Res. Commun., 2002, 295, 540–546 CrossRef CAS.
- T. Toda, I. Uno, T. Ishikawa, S. Powers, T. Kataoka, D. Broek, S. Cameron, J. Broach, K. Matsumoto and M. Wigler, Cell (Cambridge, Mass.), 1985, 40, 27–36 CrossRef CAS.
- J. Field, H. P. Xu, T. Michaeli, R. Ballester, P. Sass, M. Wigler and J. Colicelli, Science, 1990, 247, 464–467 CrossRef CAS.
- N. Suzuki, H. R. Choe, Y. Nishida, Y. Yamawaki-Kataoka, S. Ohnishi, T. Tamaoki and T. Kataoka, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 8711–8715 CrossRef CAS.
- L. M. Selfors, J. L. Schutzman, C. Z. Borland and M. J. Stern, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6903–6908 CrossRef CAS.
- D. S. Sieburth, Q. Sun and M. Han, Cell (Cambridge, Mass.), 1998, 94, 119–130 CrossRef CAS.
- K. Obrietan, S. Impey and D. R. Storm, Nat. Neurosci., 1998, 1, 693–700 CrossRef CAS.
- M. H. Hastings, A. B. Reddy and E. S. Maywood, Nat. Rev. Neurosci., 2003, 4, 649–661 CrossRef CAS.
- S. M. Reppert and D. R. Weaver, Annu. Rev. Physiol., 2001, 63, 647–676 CrossRef CAS.
- S. Michel, J. Itri, J. H. Han, K. Gniotczynski and C. S. Colwell, BMC Neurosci., 2006, 7, 15 CrossRef.
- J. M. Ding, D. Chen, E. T. Weber, L. E. Faiman, M. A. Rea and M. U. Gillette, Science, 1994, 266, 1713–1717 CrossRef CAS.
- G. Q. Butcher, B. Lee and K. Obrietan, J. Neurophysiol., 2003, 90, 3854–3863 CrossRef CAS.
- T. Serchov and R. Heumann, Chronobiol. Int., 2006, 23, 191–200 CrossRef.
- G. A. Pizzio, E. C. Hainich, G. A. Ferreyra, O. A. Coso and D. A. Golombek, NeuroReport, 2003, 14, 1417–1419 CrossRef CAS.
- J. Xing, D. D. Ginty and M. E. Greenberg, Science, 1996, 273, 959–963 CrossRef CAS.
- S. Impey, D. M. Smith, K. Obrietan, R. Donahue, C. Wade and D. R. Storm, Nat. Neurosci., 1998, 1, 595–601 CrossRef CAS.
- J. Athos, S. Impey, V. V. Pineda, X. Chen and D. R. Storm, Nat. Neurosci., 2002, 5, 1119–1120 CrossRef CAS.
- R. J. Kelleher, 3rd,, A. Govindarajan, H. Y. Jung, H. Kang and S. Tonegawa, Cell (Cambridge, Mass.), 2004, 116, 467–479 CrossRef.
- N. J. Broadbent, L. R. Squire and R. E. Clark, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14515–14520 CrossRef CAS.
- R. S. Hammond, L. E. Tull and R. W. Stackman, Neurobiol. Learn. Mem., 2004, 82, 26–34 CrossRef.
- M. N. de Lima, T. Luft, R. Roesler and N. Schroder, Neurosci. Lett., 2006, 405, 142–146 CrossRef.
- S. N. Moses, R. J. Sutherland and R. J. McDonald, Brain Res. Bull., 2002, 58, 517–527 CrossRef.
- M. Grammer, S. Kuchay, A. Chishti and M. Baudry, Neurobiol. Learn. Mem., 2005, 84, 222–227 CrossRef CAS.
- M. Ohno, P. W. Frankland, A. P. Chen, R. M. Costa and A. J. Silva, Nat. Neurosci., 2001, 4, 1238–1243 CrossRef CAS.
- C. M. Atkins, J. C. Selcher, J. J. Petraitis, J. M. Trzaskos and J. D. Sweatt, Nat. Neurosci., 1998, 1, 602–609 CrossRef CAS.
- J. C. Selcher, T. Nekrasova, R. Paylor, G. E. Landreth and J. D. Sweatt, Learn. Mem., 2001, 8, 11–19 CrossRef CAS.
- Y. Satoh, S. Endo, T. Ikeda, K. Yamada, M. Ito, M. Kuroki, T. Hiramoto, O. Imamura, Y. Kobayashi, Y. Watanabe, S. Itohara and K. Takishima, J. Neurosci., 2007, 27, 10765–10776 CrossRef CAS.
- J. Liu, H. L. Weiss, P. Rychahou, L. N. Jackson, B. M. Evers and T. Gao, Oncogene, 2009, 28, 994–1004 CrossRef CAS.
- T. Gao, J. Brognard and A. C. Newton, J. Biol. Chem., 2008, 283, 6300–6311 CrossRef CAS.
- J. M. Horwood, F. Dufour, S. Laroche and S. Davis, Eur. J. Neurosci., 2006, 23, 3375–3384 CrossRef.
- E. A. Van der Zee, P. G. Luiten and J. F. Disterhoft, Prog. Neuropsychopharmacol. Biol. Psychiatry, 1997, 21, 531–572 CrossRef CAS.
- E. J. Weeber, C. M. Atkins, J. C. Selcher, A. W. Varga, B. Mirnikjoo, R. Paylor, M. Leitges and J. D. Sweatt, J. Neurosci., 2000, 20, 5906–5914 CAS.
- S. C. Shalin, C. M. Hernandez, M. K. Dougherty, D. K. Morrison and J. D. Sweatt, Neuron, 2006, 50, 765–779 CrossRef CAS.
- B. Lee, A. Almad, G. Q. Butcher and K. Obrietan, Eur. J. Neurosci., 2007, 26, 451–462 CrossRef.
| This journal is © The Royal Society of Chemistry 2010 |
