Conversion of arginine to ornithine for improving the fragmentation pattern of peptides labeled with the N-terminal tris(2,4,6-trimethoxyphenyl)phosphonium group in tandem mass spectrometry†
Hiroki
Kuyama
*a,
Chihiro
Nakajima
ab,
Takashi
Nakazawa
c and
Osamu
Nishimura
a
aInstitute for Protein Research, Osaka University, Suita, 565-0871, Japan. E-mail: kuyama@protein.osaka-u.ac.jp; Fax: +81-6-6879-4320; Tel: +81-6-6879-4320
bLife Science Research Center, Shimadzu Corporation, Kyoto, 619-0237, Japan
cDepartment of Chemistry, Nara Women's University, Nara, 630-8506, Japan
First published on 24th September 2010
Abstract
We propose a method of converting arginine to ornithine residues by controlled hydrazinolysis, in order to facilitate the sequencing of peptides by tandem mass spectrometry (MS/MS). Whereas the presence of C-terminal arginine residue occurring in a tryptic peptide is generally desirable in MS and MS/MS analyses, that of arginine in the middle of a peptide often adversely affects the appearance of fragment peaks. This problem still arises in peptides, which are acylated with a reagent containing the tris(2,4,6-trimethoxyphenyl)phosphonium (TMPP) group at the N-termini. Using four model peptides, angiotensin III (RVYIHPF), a mucin-related peptide (APDTRPAPG), α-bag cell peptide (1–9) (APRLRFYSL), and [des-Pro2] bradykinin (RPGFSPFR), we optimized the protocol of hydrazinolysis to remove the guanidino group of arginine. Owing to this derivatization, we obtained much simpler MS/MS spectra composing mainly a-type ions characteristic of peptides tagged with the TMPP group. Formylation of the δ-amino group of ornithine further enhanced the efficacy of the derivatization, which would be applicable to the sequencing of non-tryptic peptides.
Introduction
Tandem mass spectrometry (MS/MS), in conjunction with peptide fragment fingerprinting (PFF) and peptide mass fingerprinting (PMF), is an established method for identifying proteins of interest.1 The fidelity of identification relies heavily on the quality and quantity of fragmentation data employed for searching protein and/or genomic databases. This approach is therefore of limited use for proteins compiled in databases. Understandably, further difficulties are imposed on de novo sequencing, due to the lack of a database.Even in the database search, it is difficult to identify proteins without determining the chemical structure, which mass spectrometry accesses less efficiently. The amino acid sequences of mature proteins (e.g., those proteolytically processed during/after translation, translated from alternatively spliced RNA, or enzymatically modified) do not match those coded from their DNAs. For conducting database search, it is indispensable to allow for the possible post-translational modifications. If the covalent structure of a protein appears to be altered extensively, there is no choice but to carry out de novo sequencing, as is the case of a protein not registered in any database.
The mass spectrometric de novo sequencing of peptides employs MS/MS spectra, without referring to any database of nucleotide or amino acid sequence. However, the pattern of fragmentation to be analyzed in MS/MS spectra is often so complex that unambiguous sequencing is very difficult to achieve.2 Because the complexity of MS/MS spectra arises due to the conglomeration of peaks resulting from a variety of fragmentation processes, the task of obtaining sequence information by searching for a set of fragment peaks would become less laborious if we could limit the observable fragment ions to those representing a well-defined ion series. The simplification of MS/MS spectra is desired particularly for de novo sequencing, in which the analysis relies solely on finding a consecutive match of mass difference between a pair of peaks to a known mass value of amino acid.
N-Succinimidyloxycarbonylmethyl tris(2,4,6-trimethoxyphenyl)phosphonium bromide (TMPP-Ac-OSu)3–6 is an acylating reagent to be used for selectively derivatizing the N-terminal α-amino group of peptides. The TMPP-acetylated peptides undergo fragmentation strongly biased towards creating a-type ions, making it easier to obtain sequence information even without employing an automated computational approach. Unfortunate exceptions include peptides containing arginine residue(s) in the middle of a peptide chain, as often exemplified by the poor appearance of fragment peaks, from which the amino acid sequence is to be derived.7
Peptides incorporating arginine residue exhibit a low degree of structurally informative fragmentation by MS/MS analysis. Arginine residue is the most basic of all genetically coded amino acids and easily undergoes protonation. Such a strong attachment of proton causes charge localization, which leads to a wide exploitation of this residue as a C-terminal charge tag naturally occurring in tryptic peptides. However, the presence of a non-terminal arginine residue often interferes with the measurement of sufficiently informative MS/MS spectra because it sequesters a proton that could otherwise promote the desired random cleavage of backbone bonds.7 To extend the applicability of mass spectrometric sequencing of peptides, several strategies to attenuate the strongly basic character of arginine have been suggested.8–12
In this work, we applied deguanidination of arginine toward TMPP-acetylated peptides incorporating arginine residue(s) in order to reduce the high basicity of arginine, and compared their fragmentation with those of non-treated counterparts to study its effects on the performance of sequencing by MS/MS analysis.
Experimental
Materials
N-Succinimidyloxycarbonylmethyl tris(2,4,6-trimethoxyphenyl)phosphonium bromide (TMPP-Ac-OSu) was obtained from Fluka (Switzerland). Sodium hydrogen carbonate (NaHCO3), acetonitrile, and trifluoroacetic acid (TFA) were purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). α-Cyano-4-hydroxycinnamic acid (CHCA, high-purity mass-spectrometric grade) was obtained from Shimadzu GLC (Tokyo, Japan). Three peptides used in this study (RVYIHPF, APRLRFYSL, and RPGFSPFR) were obtained from American Peptide Company, Inc. (Sunnyvale, CA). Another peptide (APDTRPAPG) was purchased from AnaSpec (San Jose, CA). Methanediphosphonic acid (MDPNA) was obtained from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan).Conversion of arginine to ornithine residue13
The peptide sample (100 pmol) was dissolved in 20 µL of 50 mM NaHCO3 containing 10% acetonitrile, and then 2 µL of 10 mM TMPP-Ac-OSu solution (in water–acetonitrile = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were added to the solution. The mixture was sonicated in a water bath for 5 min and incubated at 60 °C for 3 h, and was then dried using a vacuum centrifuge. Next, 10 µL of aqueous hydrazine (water
1) were added to the solution. The mixture was sonicated in a water bath for 5 min and incubated at 60 °C for 3 h, and was then dried using a vacuum centrifuge. Next, 10 µL of aqueous hydrazine (water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hydrazine hydrate = 1
hydrazine hydrate = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were added to the dried material and kept at 70 °C for 3 h. An aliquot (1 µL) of the solution was diluted with 10 µL of water and acidified with 10% TFA solution. The dilution was desalted using ZipTip (18C) and analyzed by MALDI-TOF MS.
1) were added to the dried material and kept at 70 °C for 3 h. An aliquot (1 µL) of the solution was diluted with 10 µL of water and acidified with 10% TFA solution. The dilution was desalted using ZipTip (18C) and analyzed by MALDI-TOF MS.
Formylation of δ-amino group in ornithine residue14
The hydrazinolysis solution was diluted with 100 µL of water and lyophilized, and then 20 µL of HCOOH–Ac2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were added to the dried residue and kept at 60 °C for 20 min. The reaction solution was dried over a vacuum concentrator. Next, 10 µL of aqueous NH2OH (1 M) were added to the residue, and the solution was gently shaken for 1 h at RT. An aliquot (1 µL) of the solution was diluted with 10 µL of water and acidified with 10% TFA solution. The dilution was desalted using ZipTip (18C) and used for MALDI-TOF MS analysis.
1) were added to the dried residue and kept at 60 °C for 20 min. The reaction solution was dried over a vacuum concentrator. Next, 10 µL of aqueous NH2OH (1 M) were added to the residue, and the solution was gently shaken for 1 h at RT. An aliquot (1 µL) of the solution was diluted with 10 µL of water and acidified with 10% TFA solution. The dilution was desalted using ZipTip (18C) and used for MALDI-TOF MS analysis.
MALDI-MS
MALDI mass spectra were recorded on an AXIMA CFR+ (Shimadzu/Kratos, Manchester, UK) reflectron time-of-flight mass spectrometer equipped with a nitrogen laser (337 nm, 3 ns pulse width). All measurements were performed in the positive-ion reflectron mode. The ion acceleration voltage was set to 20 kV, and the reflectron detector was operated at 24 kV. The flight path in the reflectron mode was 240 cm. MS/MS analysis was performed in the PSD mode.α-Cyano-4-hydroxycinnamic acid (CHCA) was used as a matrix, which was dissolved to saturation in 50% aqueous acetonitrile containing 0.05% TFA. We used MDPNA,15 which has been proven to be useful for MALDI analysis of non-purified, salt-containing samples as a matrix additive. An aliquot (0.4 µL) of the sample solution was mixed with an equivalent volume of matrix solution and co-matrix solution on the MALDI target plate, and analyzed after drying.
Result and discussion
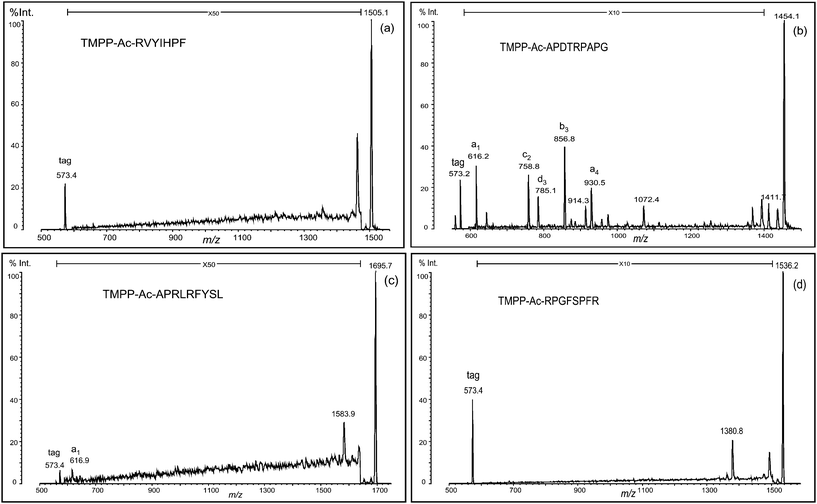
1. Conversion of arginine into ornithine and its side reactions
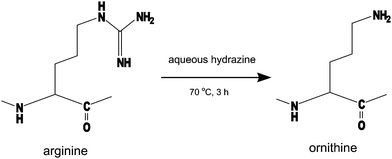
According to the mobile proton hypothesis of peptide fragmentation, the existence of arginine residue is undesirable for the sequence analysis by MS/MS, which should produce structurally informative fragment peaks covering the full length of a peptide chain uniformly.16 This is also the unfortunate case with TMPP-acetylated peptides as Fig. 1 shows that only a few fragment peaks scarcely observed in MS/MS spectra are far less sufficient to obtain sequence information. Although Dikler and co-workers successfully improved the fragmentation pattern of MS/MS spectra by modifying arginine with acetylacetone to N5-(4,6-dimethyl-2-pyrimidinyl)ornithine (Pyo),8 we found some difficulty in adapting the reaction to TMPP-acetylated peptides, which were derivatized very slowly8,17 (3 days at room temperature). In contrast, we converted arginine to ornithine by the reaction of TMPP-Ac-peptides with aqueous hydrazine at 70° for 3 h (Fig. 2), without causing serious side reactions. Anhydrous hydrazine is a reagent used in the Akabori reaction18,19 to identify the C-terminal amino acid residue, on the ground that it cleaves peptide bonds to convert all the amino acid residues (and a few short peptides) except for the C-terminal one into the corresponding hydrazides. In the present protocol, peptides were subjected to the reaction with aqueous hydrazine at much milder conditions, in which the removal of the guanidino group occurred in preference to other reactions such as the cleavage of peptide bonds (Fig. 3). While the hydrolysis or hydrazinolysis of amide bonds could be suppressed to a minimal level under the present reaction conditions, there appeared a few peptide linkages20 that must be regarded with some caution as the possible sites of cleavage to occur.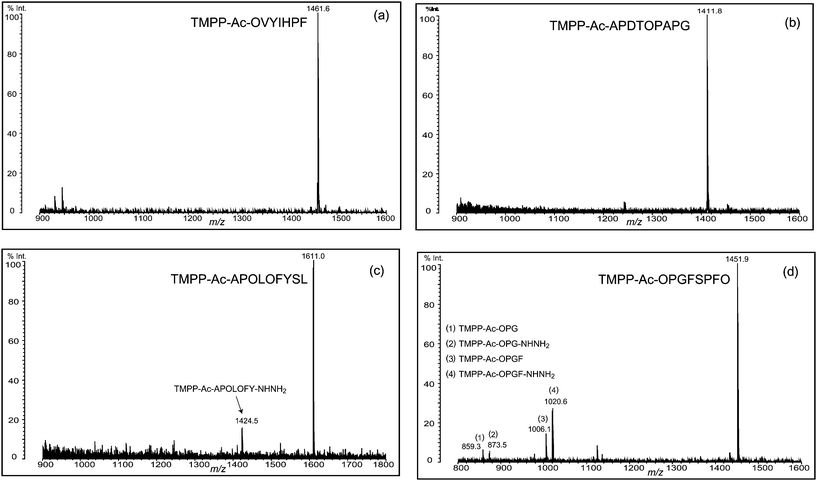 | ||
| Fig. 3 Deguanidination of four TMPP-acetylated peptides. Backbone cleavage was observed at the N-terminal of Ser (c and d) and the C-terminal of Gly (d), but non-cleaved desired peptide was the main product in the deguanidination reaction. Ornithine residue is indicated with O. | ||
That is, the following side products were detected in MALDI mass spectra and characterized by their MS/MS analysis (data not shown): (1) TMPP-Ac-OPGF (m/z 1006.1, Fig. 3d) from TMPP-Ac-RPGFSPFR, suggesting that the peptide bonds N-terminus to serine residue have been hydrolyzed; (2) TMPP-Ac-APOLOFY-NHNH2 (m/z 1424.5, Fig. 3c) and TMPP-Ac-OPGF-NHNH2 (m/z 1020.6, Fig. 3d) from TMPP-Ac-APRLRFYSL and TMPP-Ac-RPGFSPFR, respectively, as hydrozinolysis products created in a similar fashion as in (1); and (3) TMPP-Ac-OPG (m/z 859.3, Fig. 3d) and TMPP-Ac-OPG-NHNH2 (m/z 873.5, Fig. 3d) by the hydrolysis and hydrazinolysis, respectively, at the C-terminal side of glycine in TMPP-Ac-RPGFSPFR.
Note that these bonds are associated with glycine with the smallest side-chain structure and serine having the β-hydroxyl group, with which the cleavage reaction might be assisted in the manner of neighboring group participation. Owing to the use of aqueous hydrazine, the cleavage due to simultaneous hydrolysis and hydrazinolysis creates peptides derivatized with and without hydrazine at the C-terminal carboxyl group. Consequently, it is likely that a mass spectrum of deguanidination product displays a pair of small peaks corresponding to these peptides with the mass difference of 14 Da (see, for example, Fig. 3d), in addition to the main peak of a peptide, which contains ornithine converted from arginine. In favorable cases, the cleavage at these specific sites could be a useful alternative to a proteolytic digestion.
2. Fragmentation pattern of TMPP-Ac-peptides in MS/MS analysis
We first discuss the results obtained from a peptide incorporating an arginine residue at the N-terminus (RVYIHPF). The peptide was TMPP-acetylated at the α-amino group, then treated with aqueous hydrazine to convert arginine (R) to ornithine (O) residue. This conversion was smoothly processed to yield TMPP-Ac-OVYIHPF, and no side products were practically observed (Fig. 3a). The ornithinated peptide was analyzed by MALDI-PSD, and significant enhancement of fragmentation intensity was observed. The fragment peak at the C-terminal side of the ornithine residue was assigned to be a b-type ion. Hence, all fragment peaks assigned were b1, a2, a3, a4, a5, a6, and c6 (Fig. 4a). Comparison of the PSD spectrum with that of the non-treated peptide (TMPP-Ac-RVYIHPF) (Fig. 1a) indicated that this conversion effectively produced structurally informative fragmentation.
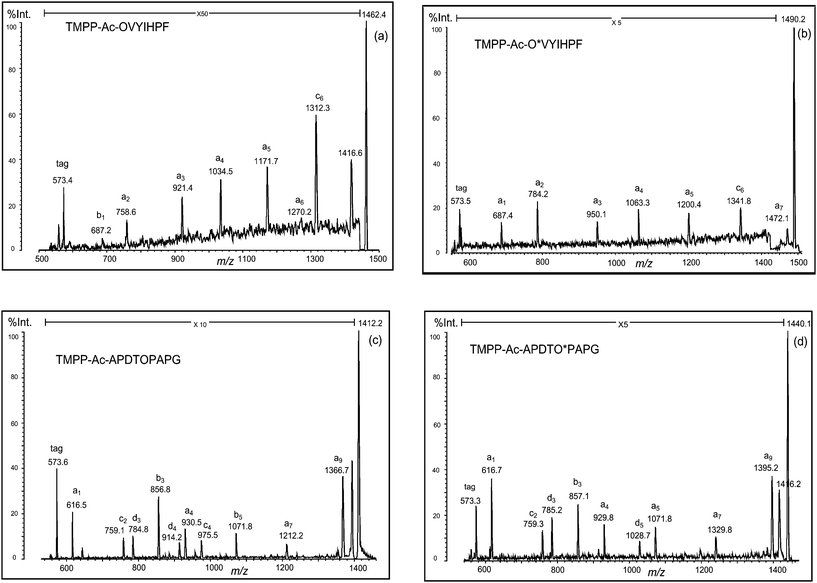 | ||
| Fig. 4 PSD spectra of deguanidinated peptides ((a) TMPP-Ac-OVYIHPF and (c) TMPP-Ac-APDTOPAPG) and their formylated forms ((b) TMPP-Ac-O*VYIHPF and (d) TMPP-Ac-APDTO*PAPG). O: ornithine residue and O*: formylated ornithine residue. | ||
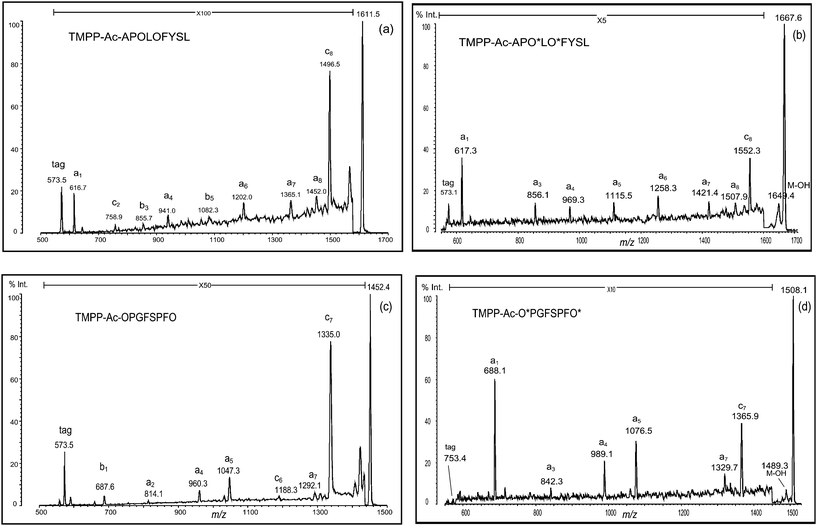 | ||
| Fig. 5 PSD spectra of deguanidinated peptides ((a) TMPP-Ac-APOLOFYSL and (c) TMPP-Ac-OPGFSPFO) and their formylated forms ((b) TMPP-Ac-APO*LO*FYS and (d) TMPP-Ac-O*PGFSPFO*). O: ornithine residue and O*: formylated ornithine residue. | ||
In a similar fashion, another peptide (RPGFSPFR) was derivatized using TMPP-Ac-OSu, and hydrazinolyzed (Fig. 3d). In this case, the desired deguanidinated peptide was the main product, and four side products were observed (Fig. 3d, peaks 1–4), though their relative intensities were weak. By MS/MS analysis of these products, they were characterized as hydrolyzed ((1) TMPP-Ac-OPG and (3) TMPP-Ac-OPGF) or hydrazinolyzed products ((2) TMPP-Ac-OPG-NHNH2 and (4) TMPP-Ac-OPGF-NHNH2) (Fig. 3d). MS/MS analysis of TMPP-Ac-OPGFSPFO also demonstrated a fragmentation-enhancing effect (Fig. 5c). While the non-treated peptide (TMPP-Ac-RPGFSPFR) produced no structurally informative fragmentation (Fig. 1d), the deguanidinated peptide produced mainly a-type ions and C-terminal cleavage at the ornithine or proline residue produced b- or c-type ion, respectively, as already observed in TMPP-Ac-OVYIHPF (Fig. 4a), TMPP-Ac-APDTOPAPG (Fig. 4c) and TMPP-Ac-APOLOFYSL (Fig. 5a).
Formylation followed by cleaving off over-formylation (i.e., cleavage of the formyl group attached to chemical groups other than the δ-amino group) was successfully performed to produce the desired products (Fig. Sa–d, ESI†). Although ions derived from the fragmentation at the C-terminal of the proline residue were masked in the noise, a set of a-type ions was detected, and the acylation of the δ-amino group completely eliminated different types of ions from a-type in fragmentation, except for the c-type ion that arose from the C-terminal cleavage of the penultimate residue from the C-terminus of the peptide. Thus, MS/MS spectra of these peptides indicated that the desired simplification in fragmentation was realized (Fig. 4b and d and Fig. 5b and d).
Conclusion
We applied the conversion reaction of arginine to ornithine to arginine-containing TMPP-acetylated peptides in order to improve fragmentation. Four arginine-containing peptides with TMPP-acetylation were subjected to MALDI-MS/MS analyses (PSD mode) after deguanidination of arginine and formylation of the resulting amino group of ornithine. A b-type ion arose in the fragmentation at the C-terminal side of the ornithine residue, and a c-type ion arose at the C-terminal side of the proline, with the exception of aspartic acid residue, which exhibited the characteristic triad peaks4 of cn−1-, dn-, and bn-type ions in PSD analysis. Therefore, these ornithine-containing peptides produced mainly a-type ions, and residue-dependent b-, c- and d-type ions, based on PSD analysis. These four types of ions arose in a regulated manner; hence, manual interpretation could be successfully performed. However, the occurrence of different incomplete ion series in fragmentation makes de novo interpretation of amino acid sequencing difficult. This problem can be solved by formylation of the δ-amino group of ornithine residue. With the acylation of ornithine residue, fragmentation was more simplified to produce a-type ions in PSD analysis. This technique can serve as an alternative for obtaining the structurally informative fragmentation of TMPP-acetylated, arginine-containing peptides.Acknowledgements
The authors are grateful to Prof. S. Tsunasawa for his valuable suggestions and constant encouragement. This work was supported in part by a Grant-in-Aid for Scientific Research (C) (Grant 22510230 to H.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.References
- R. Aebersold and M. Mann, Nature, 2003, 422, 198–207 CrossRef CAS.
- A. V. Laboda, A. N. Krutchinsky, M. Bromirski, W. Ens and K. G. Standing, Rapid Commun. Mass Spectrom., 2000, 14, 1047–1057 CrossRef CAS.
- Z. H. Huang, J. Wu, K. D. W. Roth, Y. Yang, D. A. Gage and J. T. Watson, Anal. Chem., 1997, 69, 137–144 CrossRef CAS.
- P. C. Liao, Z. H. Huang and J. Allison, J. Am. Soc. Mass Spectrom., 1997, 8, 501–509 CrossRef CAS.
- Z. H. Huang, T. Shen, J. Wu, D. A. Gage and J. T. Watson, Anal. Biochem., 1999, 268, 305–317 CrossRef CAS.
- T. L. Shen and J. Allison, J. Am. Soc. Mass Spectrom., 2000, 11, 145–152 CrossRef CAS.
- S. R. Carr and C. Cassady, J. Mass Spectrom., 1997, 32, 959–967 CrossRef CAS.
- S. Dikler, J. W. Kelly and D. H. Russell, J. Mass Spectrom., 1997, 32, 1337–1349 CrossRef CAS.
- B. Spengler, F. Luetzenkirchen, S. Metzger, P. Chaurand, R. Kaufmann, W. Jeffery, M. Bartlet-Jones and D. J. C. Pappin, Int. J. Mass Spectrom. Ion Processes, 1997, 169–170, 127–140 CrossRef.
- A. Leitner and W. Lindner, Anal. Chim. Acta, 2005, 528, 165–173 CrossRef CAS.
- A. Foettinger, A. Leitner and W. Lindner, J. Mass Spectrom., 2006, 41, 623–632 CrossRef CAS.
- H. Kuyama, K. Sonomura, K. Shima, O. Nishimura and S. Tsunasawa, Rapid Commun. Mass Spectrom., 2008, 22, 2063–2072 CrossRef CAS.
- M. M. Shemyakin, E. I. Vinogradova, Yu.A. Ovchinikov, A. A. Kiryushkin, M. Yu. Feigina, N. A. Aldanova, Yu.B. Alakhof, V. M. Lipkin, B. V. Rosinov and L. A. Fonina, Tetrahedron, 1969, 25, 5785–5806 CrossRef CAS.
- H. Kuyama, C. Nakajima, T. Nakazawa, O. Nishimura and S. Tsunasawa, Proteomics, 2009, 9, 4063–4070 CrossRef CAS.
- H. Kuyama, K. Sonomura and O. Nishimura, Rapid Commun. Mass Spectrom., 2008, 22, 1109–1116 CrossRef CAS.
- R. Boyd and A. Somogyi, J. Am. Soc. Mass Spectrom., 2010, 21, 1275–1278 CrossRef CAS , and references cited therein.
- H. F. Gilbert, III and M. H. O'Leary, Biochemistry, 1975, 14, 5179–5199.
- S. Akabori, K. Ohno and K. Narita, Bull. Chem. Soc. Jpn., 1952, 25, 214–218 CAS.
- A. K. Bose, Y. H. Ing, N. Lavlinskaia, C. Sareen, B. N. Pramanik, P. L. Bartner, Y. H. Liu and L. Heimark, J. Am. Soc. Mass Spectrom., 2002, 13, 839–850 CrossRef CAS.
- A. Honegger, G. J. Hughes and K. J. Wilson, Biochem. J., 1981, 199, 53–59 CAS.
- H. Kuyama, K. Shima, K. Sonomura, M. Yamaguchi, E. Ando, O. Nishimura and S. Tsunasawa, Proteomics, 2008, 8, 1539–1550 CrossRef CAS.
- H. Kuyama, K. Sonomura, O. Nishimura and S. Tsunasawa, Anal. Biochem., 2008, 380, 291–296 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Formylation of δ-amino group(s) in ornithine residue(s). See DOI: 10.1039/c0ay00439a |
| This journal is © The Royal Society of Chemistry 2010 |