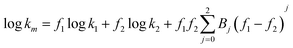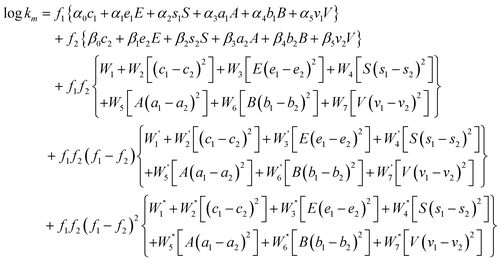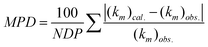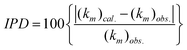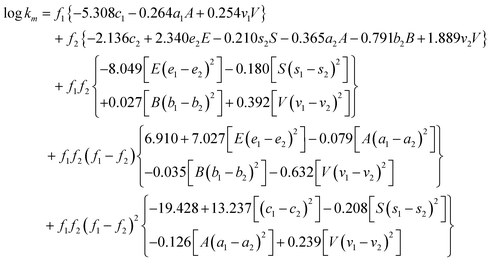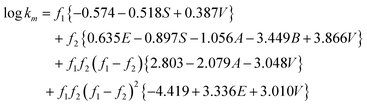DOI:
10.1039/C0AY00254B
(Paper)
Anal. Methods, 2010,
2, 1286-1297
Received
19th April 2010
, Accepted 15th June 2010
First published on
12th July 2010
Abstract
An extension to the Jouyban-Acree model was proposed to calculate the retention factor of analytes in RPLC with hydro-organic solvent mixtures as mobile phase by using the Abraham solvent coefficients and Abraham solute parameters. The accuracy of the proposed method was checked by computing the mean percentage deviation as a criterion. The proposed method provides an ab initio prediction (without employing any experimental retention data of an analyte) method with an acceptable prediction error for the retention data of various analytes based on their chemical structures. The accuracy of the proposed method was also compared with that of a previously reported model and provided comparable results with the advantage of modeling the effects of various organic modifiers using a single equation.
1. Introduction
Reverse phase liquid chromatography (RPLC) is the most widely used separation technique in pharmaceutical/chemical analysis. Despite of this wide range of applications, separations are still being developed using a non-systematic manner (trial and error) which is time consuming and leads to non-optimum conditions. In an attempt to overcome this problem many efforts have been made in order to predict the retention factor as the most important variables governing the separations, and some models were developed.1–11 Among these, the models which are based on linear free energy relationships (LFER) have been used over two decades to study solute retention in RPLC. Vitha and Carr12 reviewed the applications of these models and evaluated the different chemical interactions which affect the retention and selectivity in chromatographic separations. Torres-Lapasio and co-workers13 compared a number of models predicting the retention factor as a function of solvation parameters and mobile phase composition. They used a set of 146 organic compounds of diverse nature, eluted with methanol and acetonitrile as organic modifier, and concluded that the poor quality of the general solvation parameter models should be improved and tend to target the prediction quality of individual models. The main limitation of the Torres-Lapasio model is that it treats each solvent composition as a separate system and this may cause trouble in predicting the retention behavior by interpolation techniques.
In the previous studies,14–16 the Jouyban-Acree models were developed to represent the retention factor of analytes in binary,15 ternary16 and quaternary14 mobile phases as a function of the mobile phase compositions. Using this model, it is possible to optimize the concentration of organic modifier of the mobile phase for each analyte, however, the generated model is valid for only one analyte. The general form of the Jouyban-Acree model for representing the retention factor of analytes in a binary solvent mobile phase is:
| | 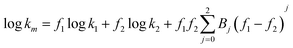 | (1) |
where
k is the
retention factor of the
analyte,
f denotes the volume fraction of the
solvent in the binary
solvent mobile phase, subscripts
m, 1 and 2 are the mixed
solvent mobile phase, components 1 and 2, respectively,
Bj is the model constant which represents various
solvent–
solvent and
analyte–
solvent interactions and is calculated by using a no intercept least square analysis for each
analyte separately.
15 The model produced reasonably accurate predictions after training by a minimum number of
experimental data points. The required
retention data in mixed
solvent mobile phases (even a minimum number of
experimental data) to train the Jouyban-Acree model is a limitation for the model and any attempt to overcome this limitation could improve its practical applicability. The aim of this work is to provide a model to simulate the
retention data of
analytes in hydro-organic
mobile phases using the Abraham solvation parameters of the
analytes and the
solvents. Using such models, one is able to predict the
retention data of an
analyte employing the computed chemical descriptors. The models could provide rational starting conditions considering the
solvent composition of the
mobile phase and save time and cost of method development.
2. Experimental
The details of the experimental data sets collected from the literature including names of analytes, organic modifiers, number of data points in each set, the references and the mean percentage deviations are listed in Table 1. All data were obtained using a 100 × 5 mm I.D., column packed with Spherisorb ODS 5-μm. The Abraham solvent coefficients of water, acetonitrile and methanol are listed in Table 2. The Abraham solvation parameters of the analytes are reported in Table 3. In addition to the experimentally derived solvation parameters, descriptors can be computed using Pharma-Algorithms web-based software,19 this makes predictive procedures presented in this study more feasible.
Table 1 Details of experimental data of analytes, the organic modifier, the references, the number of data points in each set (NDP) and the mean percentage deviation (MPD)
| Analyte |
Organic modifier |
NDP
|
Reference |
MPD
|
MPD
|
MPD
|
MPD
|
|
The MPDs calculated for back-calculated data sets using eqn (7).
The MPDs calculated for predicted data sets using the trained eqn (4) by experimental data of five references and one reference left out method.
The MPDs calculated for predicted data sets using eqn (8) or (9).
The MPDs calculated for predicted data sets using eqn (10) or (11).
The excluded data sets with the lowest MPD.
The excluded data sets with the highest MPD.
All data were obtained using a 100 × 5 mm I.D., column packed with Spherisorb ODS 5-μm.
|
| 1-Bromo-2-nitrobenzene |
Acetonitrile |
4 |
1
|
18.0 |
10.2 |
18.2 |
22.1 |
| 1-Bromo-2-nitrobenzene |
Methanol |
4 |
1
|
38.7 |
38.9 |
24.3 |
25.9 |
| 1-Phenyl-1-propanol |
Acetonitrile |
6 |
5
|
13.9 |
21.9 |
15.3 |
15.4 |
| 1-Phenyl-1-propanol |
Methanol |
5 |
5
|
10.1 |
12.7 |
13.7 |
5.4 |
| 1-Phenyl-1-propene |
Acetonitrile |
6 |
5
|
13.2 |
15.0 |
16.1 |
33.2 |
| 1-Phenyl-1-propene |
Methanol |
5 |
5
|
16.4 |
18.5 |
15.5 |
25.8 |
| 1-Phenyl-2-butanone |
Acetonitrile |
6 |
3
|
18.9 |
17.1 |
16.0 |
17.5 |
| 1-Phenyl-2-butanone |
Methanol |
5 |
3
|
8.5 |
8.5 |
7.2 |
5.7 |
| 1,2-Dihydroxybenzene |
Acetonitrile |
6 |
6
|
21.6 |
39.9 |
24.8 |
50.0 |
| 1,2-Dihydroxybenzene |
Methanol |
5 |
6
|
6.2 |
15.1 |
20.9 |
38.3 |
| 1,2-Dimethylbenzene |
Acetonitrile |
6 |
6
|
9.2 |
5.5 |
7.3 |
19.6 |
| 1,2-Dimethylbenzene |
Methanol |
5 |
6
|
6.0 |
13.3 |
10.8 |
19.4 |
| 1,3-Dihydroxybenzene |
Acetonitrile |
6 |
6
|
23.2 |
41.7 |
24.0 |
52.2 |
| 1,3-Dihydroxybenzene |
Methanol |
3 |
6
|
3.8 |
—e |
—e |
—e |
| 1,3-Dimethylbenzene |
Acetonitrile |
6 |
6
|
5.2 |
6.7 |
12.3 |
19.9 |
| 1,3-Dimethylbenzene |
Methanol |
5 |
6
|
13.0 |
19.6 |
16.8 |
23.5 |
| 1,4-Dihydroxybenzene |
Acetonitrile |
6 |
6
|
19.8 |
38.3 |
20.9 |
69.3 |
| 1,4-Dihydroxybenzene |
Methanol |
3 |
6
|
28.6 |
18.9 |
10.7 |
88.9 |
| 1,4-Dimethylbenzene |
Acetonitrile |
6 |
6
|
4.7 |
—e |
—e |
—e |
| 1,4-Dimethylbenzene |
Methanol |
5 |
6
|
15.8 |
22.2 |
19.4 |
25.0 |
| 2-Aminophenol |
Acetonitrile |
6 |
6
|
30.2 |
47.6 |
30.6 |
50.3 |
| 2-Aminophenol |
Methanol |
5 |
6
|
21.4 |
10.3 |
8.7 |
46.5 |
| 2-Bromoaniline |
Acetonitrile |
4 |
1
|
11.9 |
18.7 |
12.4 |
20.2 |
| 2-Bromoaniline |
Methanol |
3 |
1
|
14.7 |
10.3 |
5.9 |
6.4 |
| 2-Bromophenol |
Acetonitrile |
6 |
6
|
65.0 |
—f |
—f |
—f |
| 2-Bromophenol |
Methanol |
5 |
6
|
88.8 |
—f |
—f |
—f |
| 2-Bromotoluene |
Acetonitrile |
6 |
6
|
9.8 |
5.9 |
9.8 |
23.0 |
| 2-Bromotoluene |
Methanol |
5 |
6
|
8.9 |
14.5 |
12.9 |
26.3 |
| 2-Chlorophenol |
Acetonitrile |
6 |
6
|
55.3 |
30.9 |
40.2 |
40.2 |
| 2-Chlorophenol |
Methanol |
5 |
6
|
75.4 |
—f |
—f |
—f |
| 2-Chlorotoluene |
Acetonitrile |
6 |
6
|
13.3 |
7.2 |
7.5 |
22.3 |
| 2-Chlorotoluene |
Methanol |
5 |
6
|
5.3 |
13.9 |
11.2 |
24.6 |
| 2-Hydroxyacetophenone |
Acetonitrile |
6 |
6
|
34.5 |
27.2 |
29.6 |
22.7 |
| 2-Hydroxyacetophenone |
Methanol |
5 |
6
|
41.9 |
36.6 |
24.7 |
23.5 |
| 2-Hydroxybenzaldehyde |
Acetonitrile |
6 |
6
|
15.0 |
16.0 |
10.1 |
13.9 |
| 2-Hydroxybenzaldehyde |
Methanol |
5 |
6
|
80.5 |
—f |
—f |
—f |
| 2-Hydroxybenzamide |
Acetonitrile |
6 |
6
|
16.0 |
15.4 |
15.3 |
34.0 |
| 2-Hydroxybenzamide |
Methanol |
5 |
6
|
23.2 |
24.1 |
17.2 |
27.9 |
| 2-Hydroxybenzonitrile |
Acetonitrile |
6 |
6
|
27.4 |
16.2 |
22.2 |
36.2 |
| 2-Hydroxybenzonitrile |
Methanol |
3 |
6
|
58.0 |
54.6 |
63.9 |
92.5 |
| 2-Methoxyphenol |
Acetonitrile |
6 |
6
|
42.3 |
20.3 |
33.8 |
39.3 |
| 2-Methoxyphenol |
Methanol |
5 |
6
|
93.8 |
—f |
—f |
—f |
| 2-Methylacetophenone |
Acetonitrile |
6 |
6
|
7.3 |
6.8 |
4.1 |
2.5 |
| 2-Methylacetophenone |
Methanol |
5 |
6
|
15.7 |
18.8 |
14.3 |
10.2 |
| 2-Methylanisole |
Acetonitrile |
6 |
6
|
7.9 |
12.4 |
15.7 |
17.1 |
| 2-Methylanisole |
Methanol |
5 |
6
|
5.4 |
8.7 |
9.1 |
12.5 |
| 2-Methylphenol |
Acetonitrile |
6 |
6
|
34.9 |
16.7 |
24.3 |
24.2 |
| 2-Methylphenol |
Methanol |
5 |
6
|
38.8 |
29.2 |
30.6 |
29.2 |
| 2-Nitroaniline |
Acetonitrile |
4 |
1
|
4.4 |
—e |
—e |
—e |
| 2-Nitroaniline |
Methanol |
3 |
1
|
14.9 |
11.7 |
3.8 |
8.4 |
| 2-Nitrotoluene |
Acetonitrile |
6 |
6
|
16.3 |
13.9 |
11.4 |
14.7 |
| 2-Nitrotoluene |
Methanol |
5 |
6
|
17.4 |
10.2 |
5.8 |
4.5 |
| 2-Phenyl-2-propanol |
Acetonitrile |
6 |
5
|
34.2 |
45.7 |
39.2 |
33.2 |
| 2-Phenyl-2-propanol |
Methanol |
5 |
5
|
21.8 |
30.4 |
31.6 |
24.7 |
| 2-Phenylethanol |
Acetonitrile |
6 |
3
|
21.4 |
14.5 |
18.9 |
38.2 |
| 2-Phenylethanol |
Methanol |
5 |
3
|
6.2 |
5.0 |
8.2 |
17.0 |
| 2-Phenylethyl bromide |
Acetonitrile |
6 |
3
|
10.3 |
11.0 |
11.3 |
24.3 |
| 2-Phenylethyl bromide |
Methanol |
5 |
3
|
16.1 |
14.8 |
12.7 |
27.5 |
| 2-Phenylethyl chloride |
Acetonitrile |
6 |
3
|
7.7 |
5.0 |
3.6 |
24.1 |
| 2-Phenylethyl chloride |
Methanol |
5 |
3
|
4.4 |
—e |
—e |
—e |
| 2-Phenylphenol |
Acetonitrile |
6 |
6
|
25.9 |
15.4 |
9.6 |
24.4 |
| 2-Phenylphenol |
Methanol |
5 |
6
|
29.3 |
11.7 |
7.6 |
22.3 |
| 2-Phenyltoluene |
Acetonitrile |
5 |
6
|
8.4 |
37.2 |
19.6 |
63.8 |
| 2-Phenyltoluene |
Methanol |
4 |
6
|
16.4 |
20.2 |
23.2 |
64.6 |
| 2-Tolualdehyde |
Acetonitrile |
6 |
6
|
16.8 |
10.9 |
8.3 |
7.4 |
| 2-Tolualdehyde |
Methanol |
5 |
6
|
39.0 |
27.9 |
16.3 |
14.2 |
| 2-Toluamide |
Acetonitrile |
6 |
6
|
16.1 |
18.6 |
10.1 |
39.6 |
| 2-Toluamide |
Methanol |
5 |
6
|
23.7 |
19.5 |
24.7 |
31.5 |
| 2-Toluidine |
Acetonitrile |
6 |
6
|
13.5 |
27.6 |
13.8 |
19.2 |
| 2-Toluidine |
Methanol |
5 |
6
|
36.9 |
24.6 |
9.5 |
24.9 |
| 2-Tolunitrile |
Acetonitrile |
6 |
6
|
34.4 |
26.1 |
21.4 |
20.0 |
| 2-Tolunitrile |
Methanol |
5 |
6
|
46.4 |
35.5 |
29.8 |
27.6 |
| 2,4-Dimethylphenol |
Acetonitrile |
4 |
1
|
19.8 |
27.6 |
15.7 |
11.0 |
| 2,4-Dimethylphenol |
Methanol |
4 |
1
|
12.6 |
16.2 |
18.9 |
14.6 |
| 2,5-Dimethylphenol |
Acetonitrile |
4 |
1
|
13.6 |
21.8 |
9.3 |
8.9 |
| 2,5-Dimethylphenol |
Methanol |
3 |
1
|
17.6 |
22.0 |
25.4 |
20.8 |
| 2,6-Dimethyl-4-nitrophenol |
Acetonitrile |
4 |
1
|
29.7 |
65.4 |
31.3 |
33.5 |
| 2,6-Dimethyl-4-nitrophenol |
Methanol |
3 |
1
|
64.5 |
114.0 |
129.8 |
139.3 |
| 3-Aminophenol |
Acetonitrile |
6 |
6
|
25.4 |
44.7 |
23.5 |
49.2 |
| 3-Aminophenol |
Methanol |
3 |
6
|
55.3 |
37.2 |
6.5 |
86.6 |
| 3-Bromoaniline |
Acetonitrile |
4 |
1
|
5.6 |
12.8 |
6.3 |
13.6 |
| 3-Bromoaniline |
Methanol |
3 |
1
|
19.2 |
16.0 |
9.6 |
4.7 |
| 3-Bromophenol |
Acetonitrile |
6 |
6
|
11.4 |
7.5 |
7.9 |
5.4 |
| 3-Bromophenol |
Methanol |
5 |
6
|
13.1 |
15.3 |
10.1 |
11.5 |
| 3-Bromotoluene |
Acetonitrile |
6 |
6
|
5.9 |
4.1 |
8.7 |
23.9 |
| 3-Bromotoluene |
Methanol |
5 |
6
|
16.1 |
21.1 |
19.4 |
29.7 |
| 3-Chlorophenol |
Acetonitrile |
6 |
6
|
14.5 |
8.9 |
6.2 |
8.7 |
| 3-Chlorophenol |
Methanol |
5 |
6
|
7.1 |
12.0 |
6.5 |
6.8 |
| 3-Chlorotoluene |
Acetonitrile |
6 |
6
|
10.6 |
6.5 |
7.6 |
22.9 |
| 3-Chlorotoluene |
Methanol |
4 |
6
|
8.2 |
16.6 |
14.2 |
20.1 |
| 3-Hydroxyacetophenone |
Acetonitrile |
6 |
6
|
11.4 |
13.1 |
13.4 |
28.8 |
| 3-Hydroxyacetophenone |
Methanol |
5 |
6
|
11.3 |
8.2 |
5.9 |
25.9 |
| 3-Hydroxybenzaldehyde |
Acetonitrile |
5 |
6
|
5.1 |
—e |
—e |
—e |
| 3-Hydroxybenzaldehyde |
Methanol |
5 |
6
|
4.7 |
—e |
—e |
—e |
| 3-Hydroxybenzonitrile |
Acetonitrile |
6 |
6
|
12.1 |
15.3 |
16.0 |
22.6 |
| 3-Hydroxybenzonitrile |
Methanol |
5 |
6
|
20.1 |
20.6 |
13.7 |
18.2 |
| 3-Methoxyphenol |
Acetonitrile |
6 |
6
|
22.5 |
12.2 |
17.8 |
25.9 |
| 3-Methoxyphenol |
Methanol |
5 |
6
|
29.7 |
27.0 |
29.2 |
34.5 |
| 3-Methylacetophenone |
Acetonitrile |
6 |
6
|
9.7 |
8.8 |
7.5 |
4.4 |
| 3-Methylacetophenone |
Methanol |
5 |
6
|
16.5 |
19.3 |
14.3 |
10.8 |
| 3-Methylanisole |
Acetonitrile |
6 |
6
|
7.0 |
4.4 |
4.7 |
13.3 |
| 3-Methylanisole |
Methanol |
5 |
6
|
6.7 |
6.0 |
6.9 |
4.9 |
| 3-Methylphenol |
Acetonitrile |
6 |
6
|
33.1 |
18.9 |
23.5 |
21.6 |
| 3-Methylphenol |
Methanol |
5 |
6
|
19.6 |
12.0 |
13.2 |
16.4 |
| 3-Nitroaniline |
Acetonitrile |
4 |
1
|
20.3 |
25.3 |
19.5 |
31.5 |
| 3-Nitroaniline |
Methanol |
3 |
1
|
11.3 |
12.2 |
19.1 |
26.1 |
| 3-Nitrobenzyl alcohol |
Acetonitrile |
4 |
1
|
15.6 |
16.3 |
10.7 |
30.2 |
| 3-Nitrobenzyl alcohol |
Methanol |
3 |
1
|
14.0 |
13.8 |
15.6 |
32.4 |
| 3-Nitrophenol |
Acetonitrile |
6 |
6
|
16.0 |
18.1 |
18.0 |
20.0 |
| 3-Nitrophenol |
Methanol |
5 |
6
|
26.0 |
25.1 |
20.6 |
18.5 |
| 3-Nitrotoluene |
Acetonitrile |
6 |
6
|
14.2 |
12.7 |
9.2 |
15.7 |
| 3-Nitrotoluene |
Methanol |
4 |
6
|
7.6 |
4.9 |
4.3 |
3.8 |
| 3-Phenyl-1-propanol |
Acetonitrile |
6 |
5
|
22.5 |
31.8 |
26.1 |
20.9 |
| 3-Phenyl-1-propanol |
Methanol |
5 |
5
|
9.6 |
12.0 |
12.4 |
5.4 |
| 3-Phenyl-1-propene |
Acetonitrile |
6 |
5
|
10.9 |
12.1 |
16.8 |
31.2 |
| 3-Phenyl-1-propene |
Methanol |
5 |
5
|
10.3 |
14.0 |
7.2 |
23.7 |
| 3-Phenyl-1-propionamide |
Acetonitrile |
6 |
3
|
22.9 |
16.3 |
14.9 |
32.9 |
| 3-Phenyl-1-propionamide |
Methanol |
5 |
3
|
52.9 |
50.8 |
45.4 |
44.5 |
| 3-Phenyl-1-propionitrile |
Acetonitrile |
6 |
3
|
7.8 |
11.7 |
7.4 |
10.0 |
| 3-Phenyl-1-propionitrile |
Methanol |
5 |
3
|
21.6 |
12.5 |
19.4 |
18.1 |
| 3-Phenyl-1-propyl bromide |
Acetonitrile |
5 |
3
|
16.3 |
10.2 |
14.9 |
25.1 |
| 3-Phenyl-1-propyl bromide |
Methanol |
5 |
3
|
33.9 |
26.6 |
20.1 |
37.2 |
| 3-Phenyl-1-propyl chloride |
Acetonitrile |
6 |
3
|
7.4 |
4.8 |
9.4 |
33.7 |
| 3-Phenyl-1-propyl chloride |
Methanol |
5 |
3
|
26.6 |
23.2 |
9.6 |
34.3 |
| 3-Phenylphenol |
Acetonitrile |
6 |
6
|
13.1 |
14.7 |
10.8 |
24.3 |
| 3-Phenylphenol |
Methanol |
5 |
6
|
26.8 |
9.7 |
6.0 |
24.2 |
| 3-Phenyltoluene |
Acetonitrile |
5 |
6
|
4.3 |
—e |
—e |
—e |
| 3-Phenyltoluene |
Methanol |
4 |
6
|
25.5 |
15.1 |
6.5 |
50.1 |
| 3-Tolualdehyde |
Acetonitrile |
6 |
6
|
10.4 |
14.6 |
9.3 |
8.5 |
| 3-Tolualdehyde |
Methanol |
5 |
6
|
25.7 |
15.7 |
6.9 |
7.1 |
| 3-Toluamide |
Acetonitrile |
6 |
6
|
10.0 |
8.2 |
10.2 |
38.2 |
| 3-Toluamide |
Methanol |
5 |
6
|
21.8 |
17.7 |
21.3 |
21.7 |
| 3-Toluidine |
Acetonitrile |
6 |
6
|
7.3 |
20.8 |
7.9 |
18.2 |
| 3-Toluidine |
Methanol |
5 |
6
|
42.0 |
29.4 |
12.7 |
25.2 |
| 3-Tolunitrile |
Acetonitrile |
6 |
6
|
10.4 |
5.6 |
3.1 |
2.7 |
| 3-Tolunitrile |
Methanol |
5 |
6
|
23.9 |
14.5 |
9.1 |
7.6 |
| 4-Aminophenol |
Acetonitrile |
6 |
6
|
14.6 |
37.4 |
12.1 |
63.7 |
| 4-Aminophenol |
Methanol |
3 |
6
|
70.2 |
—f |
—f |
—f |
| 4-Bromophenol |
Acetonitrile |
6 |
6
|
5.1 |
5.3 |
2.9 |
4.2 |
| 4-Bromophenol |
Methanol |
5 |
6
|
17.6 |
20.2 |
16.1 |
18.7 |
| 4-Bromotoluene |
Acetonitrile |
6 |
6
|
6.3 |
4.3 |
8.5 |
24.1 |
| 4-Bromotoluene |
Methanol |
5 |
6
|
14.6 |
19.7 |
17.6 |
28.9 |
| 4-Chlorophenol |
Acetonitrile |
6 |
6
|
7.9 |
7.1 |
4.5 |
5.6 |
| 4-Chlorophenol |
Methanol |
5 |
6
|
14.4 |
19.7 |
15.5 |
16.5 |
| 4-Chlorotoluene |
Acetonitrile |
6 |
6
|
7.9 |
6.5 |
9.1 |
22.8 |
| 4-Chlorotoluene |
Methanol |
5 |
6
|
13.6 |
21.2 |
17.4 |
26.3 |
| 4-Hydroxyacetophenone |
Acetonitrile |
6 |
6
|
10.7 |
10.2 |
14.2 |
43.4 |
| 4-Hydroxyacetophenone |
Methanol |
5 |
6
|
6.4 |
5.0 |
4.6 |
23.4 |
| 4-Hydroxybenzaldehyde |
Acetonitrile |
5 |
6
|
6.5 |
6.6 |
4.9 |
37.9 |
| 4-Hydroxybenzaldehyde |
Methanol |
3 |
6
|
32.9 |
31.8 |
36.0 |
68.0 |
| 4-Hydroxybenzonitrile |
Acetonitrile |
6 |
6
|
7.0 |
10.1 |
10.2 |
27.7 |
| 4-Hydroxybenzonitrile |
Methanol |
4 |
6
|
9.2 |
7.9 |
6.2 |
14.2 |
| 4-Methoxyphenol |
Acetonitrile |
6 |
6
|
15.3 |
5.9 |
12.0 |
30.3 |
| 4-Methoxyphenol |
Methanol |
5 |
6
|
30.3 |
26.8 |
23.8 |
39.0 |
| 4-Methylacetophenone |
Acetonitrile |
6 |
6
|
11.0 |
9.2 |
9.5 |
5.7 |
| 4-Methylacetophenone |
Methanol |
5 |
6
|
17.8 |
20.6 |
14.4 |
10.5 |
| 4-Methylphenol |
Acetonitrile |
6 |
6
|
44.2 |
29.1 |
33.4 |
30.8 |
| 4-Methylphenol |
Methanol |
5 |
6
|
27.5 |
19.4 |
22.0 |
21.4 |
| 4-Nitrobenzyl alcohol |
Acetonitrile |
4 |
1
|
8.2 |
8.6 |
6.0 |
26.7 |
| 4-Nitrobenzyl alcohol |
Methanol |
3 |
1
|
9.0 |
8.3 |
10.5 |
27.0 |
| 4-Nitrophenol |
Acetonitrile |
6 |
6
|
16.8 |
14.7 |
12.1 |
19.8 |
| 4-Nitrophenol |
Methanol |
4 |
6
|
27.5 |
27.5 |
24.8 |
14.7 |
| 4-Nitrotoluene |
Acetonitrile |
6 |
6
|
10.6 |
8.5 |
6.1 |
10.9 |
| 4-Nitrotoluene |
Methanol |
5 |
6
|
5.5 |
6.6 |
5.5 |
7.3 |
| 4-Phenyl-1-butanol |
Acetonitrile |
4 |
1
|
8.3 |
27.9 |
13.6 |
11.8 |
| 4-Phenyl-1-butanol |
Methanol |
3 |
1
|
15.1 |
10.1 |
13.9 |
9.9 |
| 4-Phenyl-1-butyronitrile |
Acetonitrile |
6 |
3
|
11.0 |
8.8 |
8.4 |
11.5 |
| 4-Phenyl-1-butyronitrile |
Methanol |
5 |
3
|
8.6 |
9.1 |
8.7 |
11.0 |
| 4-Phenyl-2-butanone |
Acetonitrile |
6 |
3
|
15.5 |
14.1 |
8.7 |
9.3 |
| 4-Phenyl-2-butanone |
Methanol |
5 |
3
|
6.2 |
5.4 |
14.5 |
9.9 |
| 4-Phenylphenol |
Acetonitrile |
6 |
6
|
16.3 |
10.8 |
7.3 |
20.2 |
| 4-Phenylphenol |
Methanol |
5 |
6
|
30.3 |
10.6 |
6.0 |
20.7 |
| 4-Phenyltoluene |
Acetonitrile |
5 |
6
|
10.4 |
14.2 |
6.8 |
49.2 |
| 4-Phenyltoluene |
Methanol |
4 |
6
|
32.0 |
15.9 |
10.0 |
52.6 |
| 4-Tolualdehyde |
Acetonitrile |
4 |
6
|
12.4 |
15.0 |
6.6 |
6.5 |
| 4-Tolualdehyde |
Methanol |
3 |
6
|
19.2 |
26.0 |
12.7 |
12.1 |
| 4-Toluamide |
Acetonitrile |
6 |
6
|
10.5 |
12.5 |
7.6 |
32.2 |
| 4-Toluamide |
Methanol |
5 |
6
|
37.3 |
21.2 |
25.1 |
23.3 |
| 4-Toluidine |
Acetonitrile |
6 |
6
|
11.9 |
14.5 |
3.1 |
18.8 |
| 4-Toluidine |
Methanol |
5 |
6
|
25.0 |
33.4 |
18.1 |
25.3 |
| 4-Tolunitrile |
Acetonitrile |
6 |
6
|
5.9 |
6.7 |
4.0 |
3.2 |
| 4-Tolunitrile |
Methanol |
5 |
6
|
46.2 |
11.4 |
7.0 |
5.3 |
| 4-t-Butylphenol |
Acetonitrile |
6 |
1
|
12.9 |
48.5 |
11.9 |
25.2 |
| 4-t-Butylphenol |
Methanol |
5 |
1
|
20.4 |
10.1 |
11.2 |
34.3 |
| 5-Phenyl-1-pentanol |
Acetonitrile |
4 |
1
|
14.6 |
44.1 |
16.7 |
17.9 |
| 5-Phenyl-1-pentanol |
Methanol |
3 |
1
|
74.2 |
—f |
—f |
—f |
| Acetophenone |
Acetonitrile |
7 |
2
|
32.5 |
16.1 |
22.7 |
24.7 |
| Acetophenone |
Methanol |
6 |
2
|
38.5 |
37.0 |
17.4 |
19.5 |
| α-4-Dibromoacetophenone |
Acetonitrile |
4 |
1
|
12.2 |
8.2 |
10.7 |
9.8 |
| α-4-Dibromoacetophenone |
Methanol |
4 |
1
|
19.2 |
16.5 |
13.1 |
14.7 |
| Aniline |
Acetonitrile |
7 |
2
|
7.7 |
20.1 |
10.9 |
32.8 |
| Aniline |
Methanol |
6 |
2
|
62.0 |
47.0 |
17.9 |
45.0 |
| Anisole |
Acetonitrile |
6 |
2
|
20.1 |
8.7 |
3.5 |
4.3 |
| Anisole |
Methanol |
5 |
2
|
39.2 |
26.8 |
12.3 |
12.5 |
| Benzaldehyde |
Acetonitrile |
6 |
2
|
22.4 |
10.4 |
6.6 |
15.3 |
| Benzaldehyde |
Methanol |
5 |
2
|
59.2 |
41.4 |
13.8 |
25.5 |
| Benzamide |
Acetonitrile |
7 |
2
|
14.3 |
20.8 |
13.5 |
50.6 |
| Benzamide |
Methanol |
6 |
2
|
23.2 |
23.5 |
35.0 |
41.4 |
| Benzene |
Acetonitrile |
7 |
2
|
30.5 |
18.3 |
2.8 |
11.1 |
| Benzene |
Methanol |
6 |
2
|
44.0 |
24.0 |
5.9 |
11.7 |
| Benzonitrile |
Acetonitrile |
7 |
2
|
15.7 |
12.0 |
4.5 |
14.0 |
| Benzonitrile |
Methanol |
6 |
2
|
35.9 |
24.0 |
8.9 |
18.6 |
| Benzyl acetate |
Acetonitrile |
5 |
6
|
17.8 |
17.8 |
16.5 |
18.7 |
| Benzyl acetate |
Methanol |
5 |
6
|
12.8 |
12.7 |
13.2 |
15.5 |
| Benzyl alcohol |
Acetonitrile |
7 |
2
|
24.7 |
14.6 |
14.3 |
47.1 |
| Benzyl alcohol |
Methanol |
6 |
2
|
26.2 |
21.7 |
10.4 |
35.8 |
| Benzylbromide |
Acetonitrile |
7 |
2
|
15.0 |
9.4 |
10.9 |
22.5 |
| Benzylbromide |
Methanol |
6 |
2
|
22.6 |
30.4 |
14.5 |
25.7 |
| Benzylchloride |
Acetonitrile |
7 |
2
|
14.6 |
26.6 |
21.0 |
26.7 |
| Benzylchloride |
Methanol |
6 |
2
|
6.3 |
15.5 |
19.1 |
23.2 |
| Benzylcyanide |
Acetonitrile |
7 |
2
|
12.8 |
7.7 |
5.0 |
9.0 |
| Benzylcyanide |
Methanol |
6 |
2
|
60.0 |
58.6 |
36.1 |
32.9 |
| Biphenyl |
Acetonitrile |
6 |
2
|
6.3 |
6.9 |
7.3 |
42.4 |
| Biphenyl |
Methanol |
5 |
2
|
14.7 |
9.5 |
7.7 |
39.4 |
| Bromobenzene |
Acetonitrile |
7 |
2
|
18.5 |
14.5 |
7.5 |
21.3 |
| Bromobenzene |
Methanol |
6 |
2
|
15.3 |
8.3 |
11.0 |
18.3 |
| Butyrophenone |
Acetonitrile |
7 |
2
|
9.5 |
17.6 |
9.8 |
19.5 |
| Butyrophenone |
Methanol |
6 |
2
|
8.4 |
29.1 |
18.6 |
31.8 |
| Chlorobenzene |
Acetonitrile |
7 |
2
|
25.7 |
13.9 |
5.1 |
21.3 |
| Chlorobenzene |
Methanol |
6 |
2
|
21.7 |
10.9 |
6.7 |
14.0 |
| Dimethyl phthalate |
Acetonitrile |
4 |
1
|
7.3 |
7.0 |
1.9 |
13.5 |
| Dimethyl phthalate |
Methanol |
3 |
1
|
6.1 |
13.8 |
19.2 |
12.9 |
| Ethyl-3-phenylpropionate |
Acetonitrile |
5 |
1
|
16.0 |
11.1 |
16.4 |
18.1 |
| Ethyl-3-phenylpropionate |
Methanol |
3 |
1
|
35.1 |
20.9 |
13.8 |
9.7 |
| Ethyl benzoate |
Acetonitrile |
4 |
1
|
7.9 |
7.2 |
9.9 |
10.0 |
| Ethyl benzoate |
Methanol |
4 |
1
|
2.8 |
—e |
—e |
—e |
| Ethylphenylacetate |
Acetonitrile |
4 |
1
|
18.3 |
32.8 |
17.5 |
25.0 |
| Ethylphenylacetate |
Methanol |
4 |
1
|
18.6 |
30.8 |
41.6 |
44.9 |
| Ethylbenzene |
Acetonitrile |
6 |
3
|
3.2 |
—e |
—e |
—e |
| Ethylbenzene |
Methanol |
5 |
3
|
6.4 |
14.8 |
10.0 |
21.4 |
| Heptanophenone |
Acetonitrile |
7 |
2
|
20.9 |
29.5 |
12.1 |
62.5 |
| Heptanophenone |
Methanol |
6 |
2
|
43.4 |
67.5 |
6.8 |
102.5 |
| Hexanophenone |
Acetonitrile |
7 |
2
|
13.2 |
25.6 |
4.9 |
48.3 |
| Hexanophenone |
Methanol |
6 |
2
|
32.1 |
52.8 |
4.1 |
75.8 |
| Isobutylbenzene |
Acetonitrile |
6 |
5
|
18.4 |
14.2 |
23.7 |
37.6 |
| Isobutylbenzene |
Methanol |
4 |
5
|
33.7 |
30.5 |
15.1 |
44.6 |
| Isopropylbenzene |
Acetonitrile |
6 |
5
|
47.8 |
49.4 |
33.8 |
42.4 |
| Isopropylbenzene |
Methanol |
5 |
5
|
14.3 |
16.4 |
6.6 |
32.1 |
| Methyl-2-hydroxybenzoate |
Acetonitrile |
6 |
6
|
16.1 |
11.0 |
13.3 |
8.1 |
| Methyl-2-hydroxybenzoate |
Methanol |
5 |
6
|
5.2 |
7.8 |
5.4 |
5.9 |
| Methyl-2-Methylbenzoate |
Acetonitrile |
6 |
6
|
26.6 |
31.4 |
23.7 |
26.9 |
| Methyl-2-Methylbenzoate |
Methanol |
5 |
6
|
14.5 |
22.1 |
24.5 |
21.5 |
| Methyl-3-hydroxybenzoate |
Acetonitrile |
6 |
6
|
10.5 |
14.6 |
13.5 |
22.4 |
| Methyl-3-hydroxybenzoate |
Methanol |
5 |
6
|
16.1 |
7.7 |
6.0 |
10.5 |
| Methyl-3-Methylbenzoate |
Acetonitrile |
6 |
6
|
13.9 |
14.7 |
8.8 |
13.8 |
| Methyl-3-Methylbenzoate |
Methanol |
5 |
6
|
9.2 |
5.1 |
3.5 |
14.1 |
| Methyl-3-phenylpropionate |
Acetonitrile |
6 |
3
|
13.8 |
8.5 |
11.7 |
14.6 |
| Methyl-3-phenylpropionate |
Methanol |
5 |
3
|
30.5 |
29.2 |
14.2 |
16.8 |
| Methyl-4-hydroxybenzoate |
Acetonitrile |
6 |
6
|
19.5 |
24.5 |
23.5 |
31.5 |
| Methyl-4-hydroxybenzoate |
Methanol |
5 |
6
|
13.0 |
8.5 |
17.0 |
14.5 |
| Methyl-4-methylbenzoate |
Acetonitrile |
6 |
6
|
13.6 |
14.5 |
8.2 |
13.2 |
| Methyl-4-methylbenzoate |
Methanol |
5 |
6
|
10.3 |
6.2 |
3.5 |
14.0 |
| Methyl-4-phenylbutyrate |
Acetonitrile |
6 |
3
|
18.0 |
9.5 |
12.0 |
17.6 |
| Methyl-4-phenylbutyrate |
Methanol |
5 |
3
|
43.9 |
38.3 |
16.9 |
25.8 |
| Methylphenylacetate |
Acetonitrile |
7 |
3
|
18.3 |
5.8 |
3.9 |
6.5 |
| Methylphenylacetate |
Methanol |
6 |
3
|
16.0 |
8.7 |
14.0 |
4.9 |
| Methylbenzoate |
Acetonitrile |
6 |
2
|
5.6 |
19.8 |
7.8 |
14.1 |
| Methylbenzoate |
Methanol |
5 |
2
|
4.7 |
—e |
—e |
—e |
|
n-Butylbenzene |
Acetonitrile |
6 |
3
|
22.2 |
15.5 |
26.8 |
37.2 |
|
n-Butylbenzene |
Methanol |
5 |
3
|
43.5 |
40.1 |
23.7 |
48.0 |
|
N-Ethylaniline |
Acetonitrile |
4 |
1
|
15.6 |
19.3 |
15.0 |
20.9 |
|
N-Ethylaniline |
Methanol |
4 |
1
|
24.5 |
23.0 |
16.8 |
16.6 |
|
N-Methylbenzamide |
Acetonitrile |
5 |
1
|
12.8 |
12.8 |
8.2 |
33.7 |
|
N-Methylbenzamide |
Methanol |
5 |
1
|
8.7 |
8.6 |
12.3 |
32.6 |
|
n-Propylbenzene |
Acetonitrile |
3 |
5
|
20.4 |
10.4 |
19.9 |
29.0 |
|
n-Propylbenzene |
Methanol |
5 |
5
|
34.8 |
27.8 |
15.4 |
35.6 |
|
n-Propyl-4-hydroxybenzoate |
Acetonitrile |
6 |
1
|
11.1 |
73.7 |
28.7 |
23.4 |
|
n-Propyl-4-hydroxybenzoate |
Methanol |
5 |
1
|
26.1 |
10.5 |
11.1 |
25.5 |
|
N,N-Dimethylbenzamide |
Acetonitrile |
5 |
1
|
12.4 |
15.1 |
3.9 |
30.9 |
|
N,N-Dimethylbenzamide |
Methanol |
5 |
1
|
12.1 |
12.5 |
20.5 |
23.1 |
| Nitrobenzene |
Acetonitrile |
7 |
2
|
14.1 |
11.1 |
3.5 |
9.2 |
| Nitrobenzene |
Methanol |
6 |
2
|
22.1 |
10.7 |
9.3 |
15.2 |
| Phenacyl bromide |
Acetonitrile |
4 |
1
|
10.1 |
9.2 |
6.5 |
5.9 |
| Phenacyl bromide |
Methanol |
4 |
1
|
30.7 |
37.1 |
28.6 |
28.4 |
| Phenol |
Acetonitrile |
7 |
2
|
18.1 |
6.3 |
9.9 |
25.9 |
| Phenol |
Methanol |
6 |
2
|
33.3 |
21.4 |
12.2 |
34.3 |
| Phenylacetaldehyde |
Acetonitrile |
6 |
3
|
17.9 |
12.1 |
9.5 |
20.1 |
| Phenylacetaldehyde |
Methanol |
5 |
3
|
93.0 |
—e |
—e |
—e |
| Phenylacetamide |
Acetonitrile |
6 |
3
|
12.9 |
14.5 |
8.8 |
46.5 |
| Phenylacetamide |
Methanol |
5 |
3
|
22.7 |
25.7 |
29.6 |
33.2 |
| Propiophenone |
Acetonitrile |
7 |
2
|
16.4 |
10.8 |
12.9 |
10.8 |
| Propiophenone |
Methanol |
6 |
2
|
18.2 |
29.6 |
16.1 |
13.3 |
|
s-Butylbenzene |
Acetonitrile |
6 |
5
|
14.1 |
12.8 |
17.5 |
39.6 |
|
s-Butylbenzene |
Methanol |
4 |
5
|
24.4 |
20.8 |
6.5 |
44.0 |
|
t-Butylbenzene |
Acetonitrile |
6 |
5
|
93.1 |
—f |
—f |
—f |
|
t-Butylbenzene |
Methanol |
4 |
5
|
92.7 |
—f |
—f |
—f |
| Thymol |
Acetonitrile |
5 |
1
|
7.2 |
28.5 |
5.1 |
14.9 |
| Thymol |
Methanol |
5 |
1
|
26.8 |
17.5 |
18.2 |
32.0 |
| Toluene |
Acetonitrile |
7 |
2
|
14.1 |
18.5 |
8.8 |
16.6 |
| Toluene |
Methanol |
6 |
2
|
16.3 |
5.1 |
9.2 |
8.9 |
| Valerophenone |
Acetonitrile |
7 |
2
|
9.3 |
26.3 |
8.7 |
36.4 |
| Valerophenone |
Methanol |
6 |
2
|
15.7 |
33.7 |
16.9 |
56.5 |
Table 2 The Abraham solvent coefficients used in this work taken from a ref. 17
| Solvent |
c |
e |
s |
a |
b |
v |
| Acetonitrile |
0.413 |
0.077 |
0.326 |
−1.566 |
−4.391 |
3.364 |
| Methanol |
0.329 |
0.299 |
−0.671 |
0.08 |
−3.389 |
3.512 |
| Water |
−0.994 |
0.577 |
2.549 |
3.813 |
4.841 |
−0.869 |
Table 3 The Abraham solute parameters of the analytes investigated in this work taken from a ref. 18
| Analyte |
E |
S |
A |
B |
V |
| 1-Bromo-2-nitrobenzene |
1.18 |
1.32 |
0.00 |
0.26 |
1.07 |
| 1-Phenyl-1-propanol |
0.78 |
0.83 |
0.30 |
0.66 |
1.20 |
| 1-Phenyl-1-propene |
0.91 |
0.72 |
0.00 |
0.18 |
1.10 |
| 1-Phenyl-2-butanone |
0.75 |
1.14 |
0.00 |
0.66 |
1.30 |
| 1,2-Dihydroxybenze |
0.97 |
1.07 |
0.85 |
0.52 |
0.83 |
| 1,2-Dimethylbenzene |
0.66 |
0.56 |
0.00 |
0.16 |
1.00 |
| 1,3-Dihydroxybenze |
0.98 |
1.00 |
1.10 |
0.58 |
0.83 |
| 1,3-Dimethylbenzene |
0.62 |
0.52 |
0.00 |
0.16 |
1.00 |
| 1,4-Dihydroxybenzene |
1.00 |
1.00 |
1.16 |
0.60 |
0.83 |
| 1,4-Dimethylbenzene |
0.61 |
0.52 |
0.00 |
0.16 |
1.00 |
| 2-Aminophenol |
1.11 |
1.10 |
0.60 |
0.66 |
0.88 |
| 2-Bromoaniline |
1.07 |
0.98 |
0.31 |
0.39 |
0.99 |
| 2-Bromophenol |
1.04 |
0.90 |
0.35 |
0.31 |
0.95 |
| 2-Bromotoluene |
0.92 |
0.72 |
0.00 |
0.09 |
1.03 |
| 2-Chlorophenol |
0.85 |
0.88 |
0.32 |
0.31 |
0.90 |
| 2-Chlorotoluene |
0.76 |
0.65 |
0.00 |
0.07 |
0.98 |
| 2-Hydroxyacetophenone |
0.95 |
1.14 |
0.00 |
0.42 |
1.07 |
| 2-Hydroxybenzaldehyde |
0.96 |
1.15 |
0.11 |
0.31 |
0.93 |
| 2-Hydroxybenzamide |
1.14 |
1.50 |
0.59 |
0.53 |
1.03 |
| 2-Hydroxybenzonitrile |
0.92 |
1.33 |
0.78 |
0.34 |
0.93 |
| 2-Methoxyphenol |
0.84 |
0.91 |
0.22 |
0.52 |
0.98 |
| 2-Methylacetophenone |
0.78 |
1.00 |
0.00 |
0.51 |
1.16 |
| 2-Methylanisole |
0.73 |
0.75 |
0.00 |
0.30 |
1.06 |
| 2-Methylphenol |
0.84 |
0.86 |
0.52 |
0.30 |
0.92 |
| 2-Nitroaniline |
1.18 |
1.37 |
0.30 |
0.36 |
0.99 |
| 2-Nitrotoluene |
0.87 |
1.11 |
0.00 |
0.28 |
1.03 |
| 2-Phenyl-2-propanol |
0.85 |
0.85 |
0.32 |
0.65 |
1.20 |
| 2-Phenylethanol |
0.81 |
0.91 |
0.30 |
0.64 |
1.06 |
| 2-Phenylethyl bromide |
0.97 |
0.94 |
0.00 |
0.30 |
1.17 |
| 2-Phenylethyl chloride |
0.80 |
0.90 |
0.00 |
0.25 |
1.12 |
| 2-Phenylphenol |
1.55 |
1.40 |
0.56 |
0.49 |
1.38 |
| 2-Phenyltoluene |
1.33 |
0.88 |
0.00 |
0.26 |
1.47 |
| 2-Tolualdehyde |
0.87 |
0.96 |
0.00 |
0.40 |
1.01 |
| 2-Toluamide |
0.95 |
1.50 |
0.50 |
0.72 |
1.11 |
| 2-Toluidine |
0.97 |
0.92 |
0.23 |
0.59 |
0.96 |
| 2-Tolunitrile |
0.78 |
1.06 |
0.00 |
0.31 |
1.01 |
| 2,4-Dimethylphenol |
0.84 |
0.80 |
0.53 |
0.39 |
1.06 |
| 2,5-Dimethylphenol |
0.84 |
0.79 |
0.54 |
0.37 |
1.06 |
| 2,6-Dimethyl-4-nitrophenol |
1.12 |
1.64 |
0.79 |
0.26 |
1.23 |
| 3-Aminophenol |
1.13 |
1.15 |
0.65 |
0.78 |
0.88 |
| 3-Bromoaniline |
1.13 |
1.19 |
0.31 |
0.34 |
0.99 |
| 3-Bromophenol |
1.06 |
1.15 |
0.70 |
0.16 |
0.95 |
| 3-Bromotoluene |
0.90 |
0.75 |
0.00 |
0.09 |
1.03 |
| 3-Chlorophenol |
0.91 |
1.06 |
0.69 |
0.15 |
0.90 |
| 3-Chlorotoluene |
0.74 |
0.67 |
0.00 |
0.07 |
0.98 |
| 3-Hydroxyacetophenone |
0.98 |
1.35 |
0.72 |
0.55 |
1.07 |
| 3-Hydroxybenzaldehyde |
0.99 |
1.37 |
0.74 |
0.40 |
0.93 |
| 3-Hydroxybenzonitrile |
0.93 |
1.55 |
0.84 |
0.25 |
0.93 |
| 3-Methoxyphenol |
0.88 |
1.17 |
0.59 |
0.39 |
0.98 |
| 3-Methylacetophenone |
0.81 |
1.00 |
0.00 |
0.51 |
1.16 |
| 3-Methylanisole |
0.71 |
0.78 |
0.00 |
0.30 |
1.06 |
| 3-Methylphenol |
0.82 |
0.88 |
0.57 |
0.34 |
0.92 |
| 3-Nitroaniline |
1.20 |
1.71 |
0.40 |
0.35 |
0.99 |
| 3-Nitrobenzyl alcohol |
1.06 |
1.35 |
0.44 |
0.64 |
1.09 |
| 3-Nitrophenol |
1.05 |
1.57 |
0.79 |
0.23 |
0.95 |
| 3-Nitrotoluene |
0.87 |
1.10 |
0.00 |
0.25 |
1.03 |
| 3-Phenyl-1-propanol |
0.82 |
0.90 |
0.30 |
0.67 |
1.20 |
| 3-Phenyl-1-propene |
0.72 |
0.60 |
0.00 |
0.22 |
1.10 |
| 3-Phenyl-1-propionamide |
0.94 |
1.65 |
0.52 |
0.80 |
1.26 |
| 3-Phenyl-1-propionitrile |
0.77 |
1.35 |
0.00 |
0.51 |
1.15 |
| 3-Phenyl-1-propyl bromide |
1.08 |
1.00 |
0.00 |
0.27 |
1.30 |
| 3-Phenyl-1-propyl chloride |
0.79 |
0.90 |
0.00 |
0.24 |
1.26 |
| 3-Phenylphenol |
1.56 |
1.41 |
0.59 |
0.45 |
1.38 |
| 3-Phenyltoluene |
1.37 |
0.95 |
0.00 |
0.26 |
1.47 |
| 3-Tolualdehyde |
0.84 |
0.97 |
0.00 |
0.42 |
1.01 |
| 3-Toluamide |
0.99 |
1.50 |
0.49 |
0.63 |
1.11 |
| 3-Toluidine |
0.95 |
0.95 |
0.23 |
0.55 |
0.96 |
| 3-Tolunitrile |
0.76 |
1.08 |
0.00 |
0.34 |
1.01 |
| 4-Aminophenol |
1.15 |
1.20 |
0.65 |
0.80 |
0.88 |
| 4-Bromophenol |
1.08 |
1.17 |
0.67 |
0.20 |
0.95 |
| 4-Bromotoluene |
0.88 |
0.74 |
0.00 |
0.09 |
1.03 |
| 4-Chlorophenol |
0.92 |
1.08 |
0.67 |
0.20 |
0.90 |
| 4-Chlorotoluene |
0.71 |
0.74 |
0.00 |
0.05 |
0.98 |
| 4-Hydroxyacetophenone |
1.01 |
1.51 |
0.76 |
0.54 |
1.07 |
| 4-Hydroxybenzaldehyde |
1.01 |
1.54 |
0.85 |
0.37 |
0.93 |
| 4-Hydroxybenzonitrile |
0.94 |
1.63 |
0.80 |
0.29 |
0.93 |
| 4-Methoxyphenol |
0.90 |
1.17 |
0.57 |
0.48 |
0.98 |
| 4-Methylacetophenone |
0.84 |
1.00 |
0.00 |
0.52 |
1.16 |
| 4-Methylphenol |
0.82 |
0.87 |
0.57 |
0.31 |
0.92 |
| 4-Nitrobenzyl alcohol |
1.06 |
1.39 |
0.44 |
0.62 |
1.09 |
| 4-Nitrophenol |
1.07 |
1.72 |
0.82 |
0.26 |
0.95 |
| 4-Nitrotoluene |
0.87 |
1.11 |
0.00 |
0.28 |
1.03 |
| 4-Phenyl-1-butanol |
0.81 |
0.90 |
0.33 |
0.70 |
1.34 |
| 4-Phenyl-1-butyronitrile |
0.76 |
1.38 |
0.00 |
0.51 |
1.29 |
| 4-Phenyl-2-butanone |
0.75 |
1.14 |
0.00 |
0.65 |
1.30 |
| 4-Phenylphenol |
1.55 |
1.40 |
0.56 |
0.49 |
1.38 |
| 4-Phenyltoluene |
1.36 |
0.98 |
0.00 |
0.26 |
1.47 |
| 4-t-Butylphenol |
0.81 |
0.89 |
0.56 |
0.41 |
1.34 |
| 4-Tolualdehyde |
0.86 |
0.87 |
0.00 |
0.47 |
1.01 |
| 4-Toluamide |
0.99 |
1.50 |
0.49 |
0.65 |
1.11 |
| 4-Toluidine |
0.92 |
0.95 |
0.23 |
0.52 |
0.96 |
| 4-Tolunitrile |
0.74 |
1.10 |
0.00 |
0.34 |
1.01 |
| 5-Phenyl-1-pentanol |
0.80 |
0.90 |
0.33 |
0.72 |
1.48 |
| Acetophenone |
0.82 |
1.01 |
0.00 |
0.48 |
1.01 |
|
α-4-Dibromoacetophenone |
1.35 |
1.61 |
0.00 |
0.44 |
1.36 |
| Aniline |
0.96 |
0.96 |
0.26 |
0.50 |
0.82 |
| Anisole |
0.71 |
0.75 |
0.00 |
0.29 |
0.92 |
| Benzaldehyde |
0.82 |
1.00 |
0.00 |
0.39 |
0.87 |
| Benzamide |
0.99 |
1.50 |
0.49 |
0.67 |
0.97 |
| Benzene |
0.61 |
0.52 |
0.00 |
0.14 |
0.72 |
| Benzonitrile |
0.74 |
1.11 |
0.00 |
0.33 |
0.87 |
| Benzyl acetate |
0.80 |
1.06 |
0.00 |
0.65 |
1.21 |
| Benzyl alcohol |
0.80 |
0.87 |
0.39 |
0.56 |
0.92 |
| Benzyl bromide |
1.01 |
0.98 |
0.00 |
0.20 |
1.03 |
| Benzyl chloride |
0.82 |
0.82 |
0.00 |
0.33 |
0.98 |
| Benzyl cyanide |
0.75 |
1.15 |
0.00 |
0.45 |
1.01 |
| Biphenyle |
1.36 |
0.99 |
0.00 |
0.26 |
1.32 |
| Bromobenzene |
0.88 |
0.73 |
0.00 |
0.09 |
0.89 |
| Butyrophenone |
0.80 |
0.95 |
0.00 |
0.51 |
1.30 |
| Chlorobenzene |
0.72 |
0.65 |
0.00 |
0.07 |
0.84 |
| Dimethyl phthalate |
0.78 |
1.40 |
0.00 |
0.84 |
1.43 |
| Ethyl-3-phenylpropionate |
0.65 |
1.20 |
0.00 |
0.62 |
1.50 |
| Ethyl benzoate |
0.69 |
0.85 |
0.00 |
0.46 |
1.21 |
| Ethyl phenylacetate |
0.66 |
1.01 |
0.00 |
0.57 |
1.35 |
| Ethylbenzene |
0.61 |
0.51 |
0.00 |
0.15 |
1.00 |
| Heptanophenone |
0.72 |
0.95 |
0.00 |
0.50 |
1.72 |
| Hexanophenone |
0.72 |
0.95 |
0.00 |
0.50 |
1.58 |
| Isobutylbenzene |
0.58 |
0.47 |
0.00 |
0.15 |
1.28 |
| Isopropylbenzene |
0.60 |
0.49 |
0.00 |
0.16 |
1.14 |
| Methyl-2-hydroxybenzoate |
0.85 |
0.84 |
0.04 |
0.46 |
1.13 |
| Methyl-2-methylbenzoate |
0.77 |
0.87 |
0.00 |
0.43 |
1.21 |
| Methyl-3-hydroxybenzoate |
0.91 |
1.40 |
0.66 |
0.45 |
1.13 |
| Methyl-3-methylbenzoate |
0.75 |
0.88 |
0.00 |
0.47 |
1.21 |
| Methyl-3-phenylpropionate |
0.69 |
1.21 |
0.00 |
0.59 |
1.35 |
| Methyl-4-hydroxybenzoate |
0.90 |
1.37 |
0.69 |
0.45 |
1.13 |
| Methyl-4-methylbenzoate |
0.73 |
0.88 |
0.00 |
0.47 |
1.21 |
| Methyl-4-phenylbutyrate |
0.69 |
1.29 |
0.00 |
0.59 |
1.50 |
| Methyl benzoate |
0.73 |
0.85 |
0.00 |
0.46 |
1.07 |
| Methyl phenylacetate |
0.70 |
1.13 |
0.00 |
0.58 |
1.21 |
|
n-Butyl benzene |
0.60 |
0.51 |
0.00 |
0.15 |
1.28 |
|
N-Ethylaniline |
0.95 |
0.85 |
0.17 |
0.51 |
1.10 |
|
N-Methylbenzamide |
0.95 |
1.49 |
0.40 |
0.71 |
1.11 |
|
n-Propyl-4-hydroxybenzoate |
0.86 |
1.35 |
0.69 |
0.45 |
1.41 |
|
n-Propylbenzene |
0.60 |
0.50 |
0.00 |
0.15 |
1.14 |
|
N,N-Dimethylbenzamide |
0.95 |
1.40 |
0.00 |
0.98 |
1.26 |
| Nitrobenzene |
0.87 |
1.11 |
0.00 |
0.28 |
0.89 |
| Phenacyl bromide |
1.06 |
1.44 |
0.00 |
0.44 |
1.19 |
| Phenol |
0.81 |
0.89 |
0.60 |
0.30 |
0.78 |
| Phenylacetaldehyde |
0.76 |
0.70 |
0.00 |
0.64 |
1.01 |
| Phenylacetamide |
0.95 |
1.27 |
0.44 |
0.89 |
1.11 |
| Propiophenone |
0.80 |
0.95 |
0.00 |
0.51 |
1.16 |
|
s-Butylbenzene |
0.60 |
0.48 |
0.00 |
0.16 |
1.28 |
|
t-Butylbenzene |
0.62 |
0.49 |
0.00 |
0.18 |
0.13 |
| Thymol |
0.82 |
0.79 |
0.52 |
0.44 |
1.34 |
| Toluene |
0.60 |
0.52 |
0.00 |
0.14 |
0.86 |
| Valerophenone |
0.80 |
0.95 |
0.00 |
0.50 |
1.44 |
2.2. Computational methods
As noted above, the Bj constants are functions of an analyte's physico-chemical properties and the separation system under investigation. Analytes interact with the stationary and mobile phases through various dipole–dipole and hydrogen-bonding interactions. These interactions can be mathematically described using the Abraham solvation model. The basic model for solute transfer between two condensed phases is:| | | logk = c + eE + sS + aA + bB + vV | (2) |
where k is the retention factor, E is the excess molar refraction, S is dipolarity/polarizability of solute, A denotes the solute's hydrogen-bond acidity, B stands for the solute's hydrogen-bond basicity and V is the McGowan volume of the solute. In eqn (2) the coefficients c, e, s, a, b and v are the model constants (i.e. solvent's coefficients), which depend upon the solvent system under consideration. Numerical values of these coefficients have been reported for several water-to-organic solvent partition systems.17Eqn (2) was used for representing the retention factor of analytes in RPLC with a given solvent composition (mono-solvents or mixed solvents) as:| | | log k = c′+e′E+s′S+a′A+b′B + v′V | (3) |
in which the regressed parameters (i.e. c′, e′, s′, a′, b′ and v′) refer to the differences of stationary and mobile phases, e′ refer to the capability of interacting with analyte π and n-electron pairs, s′ dipolarity/polarizability, a′ hydrogen-bond basicity (an acidic analyte interacts with basic phase), b′ hydrogen-bond acidity and v′ hydrophobicity.12
The model constants of the Jouyban-Acree model could be correlated with the Abraham solvation parameters (of analytes and solvents) for building a generally trained version of the Jouyban-Acree model for predicting the retention factor of analytes in mixed solvent mobile phases. There are 2 kinds of model constants:
1) αi and βi, which denote the differences in the mobile phases (containing pure solvents) and solvated stationary phase capabilities to interact with the analyte, the larger the coefficient resulted from the linear regression, the larger the difference between the mobile and stationary phases with respect to the particular interactions. Also one can consider the first line of eqn (4) as modifier selector part of the model and the second line as solute behavior in pure aqueous mobile phase.
2) Wi, W′i and W′′i constants arising from the nature of the analytes and mobile phases of the analytical systems under investigation which is our main hypothesis. Another independent variable affecting these constants could be the nature of the solvated stationary phase, however we considered this variable as a constant since all data were collected using a single stationary phase. Therefore, the Jouyban-Acree model could be represented as eqn (4)in which α, β and W terms are the model constants. The numerical values of these terms could be computed by regressing log km against f1c1, f1e1E, f1s1S, f1a1A, f1b1B, f1v1V, f2c2, f2e2E, f2s2S, f2a2A, f2b2B, f2v2V, f1f2, f1f2(c1−c2)2, f1f2E(e1−e2)2, f1f2S(s1−s2)2, f1f2A(a1−a2)2, f1f2B(b1−b2)2, f1f2V(v1−v2)2, f1f2(f1−f2), f1f2(f1−f2)[(c1−c2)2], f1f2(f1−f2)[E(e1−e2)2], f1f2(f1−f2)[S(s1−s2)2], f1f2(f1−f2)[A(a1−a2)2], f1f2(f1−f2)[B(b1−b2)2], f1f2(f1−f2)[V(v1−v2)2], f1f2(f1−f2)2, f1f2(f1−f2)2[(c1−c2)2], f1f2(f1−f2)2[E(e1−e2)2], f1f2(f1−f2)2[S(s1−s2)2], f1f2(f1−f2)2[A(a1−a2)2], f1f2(f1−f2)2[B(b1−b2)2] and f1f2(f1−f2)2[V(v1−v2)2], using a no intercept least square analysis. It should be noted that the Abraham solvent coefficients used in our computations were taken from regression analysis of solubility data and infinite dilution activity coefficient data. The solvent coefficients represent only the mobile phase properties and no experimental chromatographic data are needed to compute these coefficients.
| | 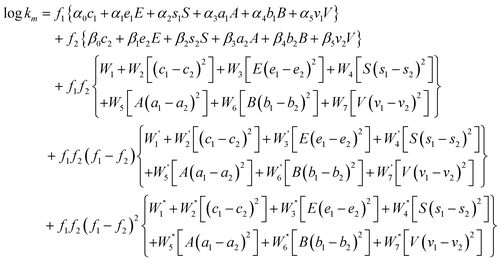 | (4) |
The predictive ability of the model was assessed in terms of the mean percentage deviation (MPD) of observed ((km)obs.) and calculated ((km)cal.) retention factors, defined by:
| | 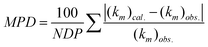 | (5) |
where
NDP is the number of data points. In addition, we also calculated the individual percentage deviation (
IPD):
| | 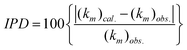 | (6) |
for each
retention factor data point.
3. Results and discussion
The available experimental km values collected from the literature were fitted to the proposed model and the constants with probability of < 0.05 were included in the model (eqn (7)).| | 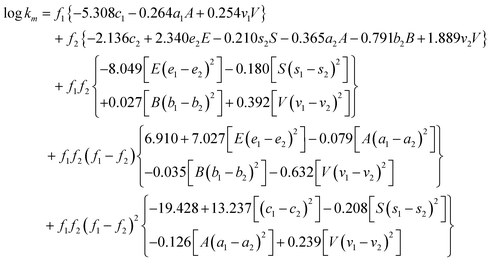 | (7) |
This correlation was significant at p < 0.0005, the F value of 1407 and the number of data points (NDP) fitted to the model was 1539. Solutes studied included both polar and nonpolar aromatic compounds, as well as aromatic compounds capable of hydrogen-bond formation. The solute descriptor range defined by the compounds studied would be: E = 0.58–1.55, S = 0.47–1.72, A = 0.00–1.16, B = 0.07–0.98 and V = 0.83–1.72.
The back-calculated km values were used to compute the MPDs and standard deviation values for the studied datasets. The details of the values were listed in Table 1 (see column 5). The overall MPD (± SD) was 20.9 (± 16.7) % and the number of data sets (NDS) was 292. When these values were analyzed considering a given organic modifier, the values were 16.5 (± 11.7) % and 25.3 (± 19.7) %, respectively for acetonitrile and methanol. Careful examination of the results revealed that a number of data sets produced very large MPD values and appeared to be possible outliers. We excluded the 10 data sets having the largest MPDs from the computations and in order to avoid any bias, the 10 data sets with the least MPDs were also excluded. The obtained overall MPDs for the back-calculated data using eqn (7) for remaining data sets was 19.2 (± 11.9) % (NDS = 272). The corresponding values for acetonitrile and methanol were 16.1 (± 8.8) % (NDS = 139) and 22.6 (± 13.6) % (NDS = 133), respectively. These MPD values are relatively high when compared with the corresponding values of the trained versions of the model for each analyte (8.1%) reported in a previous work.15 However, considering the proposed ab initio prediction method (without employing any experimental retention data of an analyte), the accuracy of the predictions could be considered acceptable. As it is evident from eqn (4) or (7), there is not an independent variable representing the properties of the stationary phases. Therefore, the model constants should be computed when other types of stationary phases are considered in the computations. As a more evident, eqn (4) was fitted to the km data of a number of analytes measured on five different stationary phases with aqueous mobile phases containing acetonitrile and methanol as organic modifiers.8 The obtained overall MPDs for these stationary phases were 26.1 (± 21.2) %, 20.6 (± 17.6) %, 29.1 (± 22.2) %, 20.3 (± 18.4) % and 18.4 (± 16.7) %, respectively for LiChrospher 100 RP-18e, LiChrospher 100 RP-8, Purospher RP-18e, SymmetrySheild RP-C18 and SymmeteryShield RP-C8 columns. Careful examination of these MPDs revealed that the proposed model could provide acceptable calculations for other types of stationary phases as the average of overall MPDs of these columns was 22.9%.
Fig. (1) shows the relative frequency of IPDs of the calculated km data listed in Table 1, sorted into four subgroups, i.e. IPD ≤ 15, 15–30, 30–45 and >45%. This result revealed that in more than 48% of the cases, the retention factor was predicted with an error less than 15%.
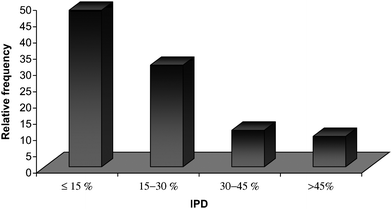 |
| | Fig. 1 The relative frequency of the individual percentage deviation (IPD), of the calculated retention factors (NDP = 1539), using eqn (7). | |
The first line of eqn (7) pertains to the organic modifier (acetonitrile or methanol) effect on retention factor. We found that the size (v1V) and hydrogen bond character (a1A) of the organic modifier (basicity) and solute (acidity) receive the most importance. In fact these parameters determine the kind of modifier which we should select for special retention factor. The second line of the equation denotes the behavior of solute in pure aqueous mobile phase. All solvation parameters have a role in retention factor which is in agreement with previous models that were developed for a single organic modifier (eqn (10) or (11)). The main difference between this part of the model and similar models is the impotence of polarizability parameter (E) which is larger than the size parameter (V). This finding is chemically reasonable because of the polarizable nature of water. The polarizability of the mobile phase versus stationary phase is an important consideration that one uses in selecting the best mobile phase needed to achieve a desired chromatographic separation. The remaining terms in the model denote the effect of mobile phase (hydro–organic) in retention factor. Solute polarizability and molecular size have high importance here, as expected based on the above discussion. The hydrogen-bonding character of the organic modifier is also a determining factor.
Eqn (4) was developed using the retention factors of various analytes in aqueous mobile phases containing acetonitrile and methanol as organic modifiers, the model could be reduced to represent the retention factor of various analytes in a single organic modifier system. In such cases, the accuracy of the model will be improved; however, the derived equation could only be applied to the data of the same organic modifier employed in training processes. When a single organic modifier is considered, the terms c1, e1, s1, a1, b1, v1, c2, e2, s2, a2, b2, v2, (c1−c2)2, (e1−e2)2, (s1−s2)2, (a1−a2)2, (b1−b2)2and (v1−v2)2 are constants and can be incorporated into the α, β and W terms. The trained model after excluding non-significant model constants (p > 0.05) for acetonitrile system was:
| |  | (8) |
and the corresponding model for
methanol was:
| | 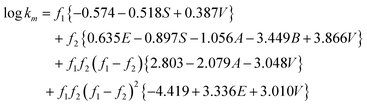 | (9) |
The overall MPD (± SD) for the back-calculated km values using eqn (8) and (9) were 13.1 (± 8.0) % (NDS = 139) and 16.1 (± 13.5) % (NDS = 133), respectively (for details of MPDs see column 7 of Table 1). The obtained models proved the previous findings about the importance of polarizability parameter in aqueous mobile phases. As it can be seen from eqn (8) and (9) the polarizability parameter is significant in the second line which is the retention factor in pure aqueous mobile phase. In other parts which the water solvation parameters were not entered the size parameter received the highest importance.
Similar models were reported in the literature13 to predict the retention factors of various analytes at different compositions of the mobile phase as for acetonitrile:
| | | log km = 1.679 + 0.198E − 0.455S − 0.485A − 1.214B + 1.291V − 4.328f1 + 1.672f21 | (10) |
and for
methanol:
| | | log km = 1.877 + 0.286E − 0.643S − 0.495A − 1.374B + 1.680V − 306f1 + 1.096f21 | (11) |
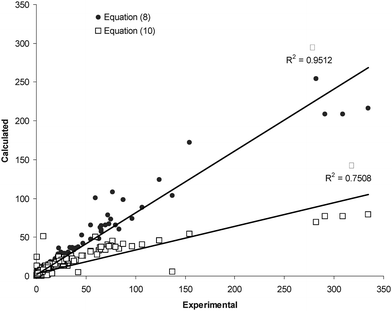
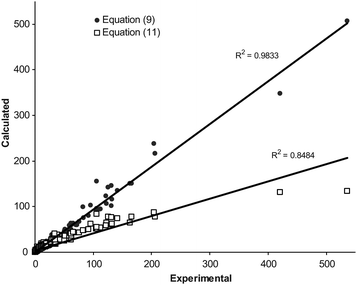
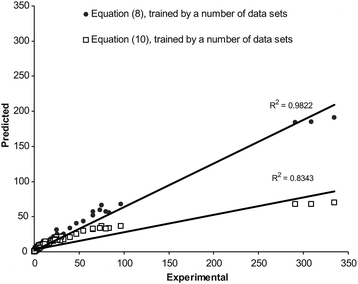
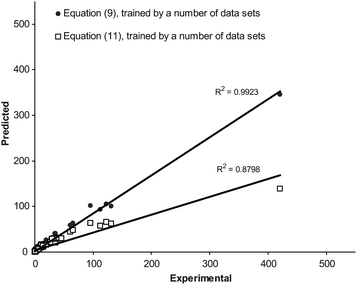
The overall MPD (± SD) for the back-calculated km values using eqn (10) and (11) were 25.3 (± 17.5) % (NDS = 139) and 26.4 (± 20.5) % (NDS = 133), respectively (for further details see column 8 of Table 1). There was significant reduction in MPD values when the pair similar equations (for acetonitrile and methanol) from this work and the previous work13 were compared (p < 0.0005). Fig. (2) and (3) show the linear plots of the calculated retention factors using the proposed and previous models against the corresponding experimental values. To show the prediction capabilities of the compared equations, they were trained using a number of data points (2/3 data sets) and then the rest of data (1/3 data sets) was predicted by using trained models. The results were shown in Fig. (4) and (5). Better scattering of the data around the regression line and also higher coefficients of determinations of the proposed models revealed that eqn (8) and (9) provide better predictions when compared with eqn (10) and (11). The same pattern has been observed when these models were trained using a number of data sets and the retention factor of prediction sets were considered (see Fig. (2)–(5)).The main advantage of eqn (8) and (9) over eqn (10) and (11) is that they provide more accurate calculations; whereas the major limitation is the larger number of curve-fit constants. Eqn (8) and (9) require more experimental retention data in the training process.
To validate the proposed method for predicting the retention factor of analytes, the experimental data of analytes reported in each reference was removed from the training process of the eqn (7). Then the km of the excluded data sets was predicted using the trained model, the MPD values were computed and listed in Table 1. The overall MPD (± SD) for this analysis was 19.1 (± 13.4) % (NDS = 272) and there was no significant difference between MPDs of this analysis and that of the back-calculated km values using eqn (7), i.e. 19.3 (± 11.9) % (paired t-test, p > 0.05). This finding confirmed that the proposed model is robust and could be used for predicting the retention factor of other analytes with C18 column and acetonitrile and/or methanol as organic modifier. Due to the variations of different C18 columns purchased from different manufacturers and/or batches, it is better to train the model using a column and then to use the trained model to predict the retention data on the same column. Developing the training model for the specific column being used should improve the model's predictive capability.
4. Conclusions
A generally trained model was proposed for predicting the retention factor of analytes in RPLC using different organic modifiers by combining the Jouyban-Acree and Abraham models. The constants of the proposed model could represent various interactions in the chromatographic system, and when their numerical values are computed for a given stationary phase, the model could be used to predict undetermined retention data, and therefore reduce the cost of the development process and also speed up the method development. The model has the advantage of modeling three variables, i.e. the analyte structure, the organic modifier type and the concentration of organic modifier in the mobile phase using a single model. To our knowledge, there is no such model reported in the literature to compare with the proposed one. It is obvious that the model is able to predict the effects of three mentioned variables on the retention of analytes and the other affecting variables usch as flow rate, pH of the buffer etc. should be fine tuned for achieving the best analytical conditions. The proposed model can be reduced to a simpler version to represent the effects of analyte structure and concentration of a given organic modifier. The accuracy of these versions was compared with two similar models taken from the literature and the results showed that the proposed models produce more accurate results.
Acknowledgements
The partial financial support (under grant No. 87-411) from the Drug Applied Research Center, Tabriz University of Medical Sciences was gratefully acknowledged.
References
- R. M. Smith and C. M. Burr, Retention prediction of analytes in reversed-phase high-performance liquid chromatography based on molecular structure: V. Cripes (chromatographic retention index prediction expert system), J. Chromatogr., A, 1989, 485, 325–340 CrossRef CAS.
- R. M. Smith and C. M. Burr, Retention prediction of analytes in reversed-phase high-performance liquid chromatography based on molecular structure: I. Monosubstituted aromatic compounds, J. Chromatogr., A, 1989, 475, 57–74 CrossRef CAS.
- R. M. Smith and C. M. Burr, Retention prediction of analytes in reversed-phase high-performance liquid chromatography based on molecular structure: III. Monosubstituted aliphatic compounds, J. Chromatogr., A, 1989, 481, 71–84 CrossRef CAS.
- R. M. Smith and C. M. Burr, Retention prediction of analytes in reversed-phase high-performance liquid chromatography based on molecular structure: II. Long term reproducibility of capacity factors and retention indices, J. Chromatogr., A, 1989, 475, 75–83 CrossRef CAS.
- R. M. Smith and C. M. Burr, Retention prediction of analytes in reversed-phase high-performance liquid chromatography based on molecular structure: IV. Branched and unsaturated alkylbenzenes, J. Chromatogr., A, 1989, 481, 85–95 CrossRef CAS.
- R. M. Smith and C. M. Burr, Retention prediction of analytes in reversed-phase high-performance liquid chromatography based on molecular structure: VI. Disubstituted aromatic compounds, J. Chromatogr., A, 1991, 550, 335–356 CrossRef CAS.
- A. Sandi and L. Szepesy, Characterization of various reversed-phase columns using the linear free energy relationship: I. Evaluation based on retention factors, J. Chromatogr., A, 1998, 818, 1–17 CrossRef CAS.
- A. Sandi and L. Szepesy, Evaluation and modulation of selectivity in reversed-phase high-performance liquid chromatography, J. Chromatogr., A, 1999, 845, 113–131 CrossRef CAS.
- A. Sandi, M. Nagy and L. Szepesy, Characterization of reversed-phase columns using the linear free energy relationship: III. Effect of the organic modifier and the mobile phase composition, J. Chromatogr., A, 2000, 893, 215–234 CrossRef CAS.
- J. R. Torres-Lapasió, M. C. García-Alvarez-Coque, M. Rosés and E. Bosch, Prediction of the retention in reversed-phase liquid chromatography using solute-mobile phase-stationary phase polarity parameters, J. Chromatogr., A, 2002, 955, 19–34 CrossRef CAS.
- F. Ruggieri, A. A. D'Archivio, G. Carlucci and P. Mazzeo, Application of artificial neural networks for prediction of retention factors of triazine herbicides in reversed-phase liquid chromatography, J. Chromatogr., A, 2005, 1076, 163–169 CAS.
- M. Vitha and P. W. Carr, The chemical interpretation and practice of linear solvation energy relationships in chromatography, J. Chromatogr., A, 2006, 1126, 143–194 CrossRef CAS.
- J. R. Torres-Lapasio, M. J. Ruiz-Ángel and M. C. García-Álvarez-Coque, Comparative study of solvation parameter models accounting the effects of mobile phase composition in reversed-phase liquid chromatography, J. Chromatogr., A, 2007, 1166, 85–96 CrossRef CAS.
- J. Hanaee, A. Jouyban, M. R. Rashidi, S. Esnaashari and W. E. Acree Jr., Correlation of retention factor of analytes in quaternary solvent mobile phases using the Jouyban-Acree model, Pharmazie, 2006, 61, 417–419 CAS.
- A. Jouyban, M. R. Rashidi, Z. Vaez-Gharamaleki, A. A. Matin and D. Djozan, Mathematical representation of analyte's capacity factor in binary solvent mobile phases using the Jouyban-Acree model, Pharmazie, 2005, 60, 827–829 CAS.
- A. Jouyban, Z. Vaez-Gharamaleki, A. A. Matin, D. Djozan and W. E. Acree Jr., Modelling of retention factors of analytes in chromatography with ternary solvent mobile phases, Chem. Anal. (Warsaw), 2005, 50, 981–989 CAS.
- D. M. Stovall, C. Givens, S. Keown, K. R. Hoover, E. Rodriguez, W. E. Acree Jr. and M. H. Abraham, Solubility of crystalline nonelectrolyte solutes in organic solvents: Mathematical correlation of 4-chloro-3-nitrobenzoic acid and 2-chloro-5-nitrobenzoic acid solubilities with the Abraham solvation parameter model, Phys. Chem. Liq., 2005, 43, 261–268 CrossRef CAS.
- J. R. Torres-Lapasio, M. C. García-Alvarez-Coque, M. Rosés, E. Bosch, A. M. Zissimos and M. H. Abraham, Analysis of a solute polarity parameter in reversed-phase liquid chromatography on a linear solvation relationship basis, Anal. Chim. Acta, 2004, 515, 209–227 CrossRef CAS.
-
http://www.pharma-algorithms.com/webboxes, 2008, Toronto, Canada.
|
| This journal is © The Royal Society of Chemistry 2010 |
Click here to see how this site uses Cookies. View our privacy policy here.