SERRS-based detection of dye-labeled DNA using positively-charged Ag nanoparticles†
Ron
Gill
* and
Gerald W.
Lucassen
Philips Research, High Tech Campus, 5656 AE, Eindhoven, The Netherlands. E-mail: ron.gill@philips.com; Fax: +31 40 2746321; Tel: +31 40 2747564
First published on 15th April 2010
Abstract
Dye-labeled DNA at nanomolar concentration can be used to aggregate positively charged silver nanoparticles, giving detectable, dye-dependent, Raman spectra. The kinetics of the aggregation procedure can be enhanced by addition of molecules with multiple carboxylic groups, such as citrate or ethylene diamine tetra acetate (EDTA) ions.
The use of surface enhanced resonance Raman scattering (SERRS) spectra, rather than fluorescence spectra from dyes used as labels, enable one to increase the multiplexing possibilities of homogeneous assays. Discrimination between 8 different dyes is known in the literature,1 and discrimination of up to 18 different dyes under a single excitation wavelength was demonstrated in our group.2 Most of the literature3,4 on multiplexed detection of dye-labeled DNA by SERRS includes protocols based on the commonly synthesized negatively charged silver nanoparticles (citrate or EDTA capped), with the need for a polyamine such as spermine to induce aggregation and to neutralize the charge of DNA which is also negative (Scheme 1(a)).
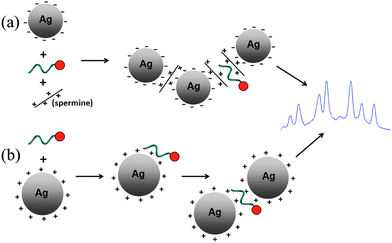 | ||
| Scheme 1 (a) Scheme of the commonly used protocol for SERRS detection of dye labeled DNA using negatively charged colloids and spermine. (b) Scheme of the protocol proposed in this article, in which the negatively charged backbone of the DNA induces the aggregation of CTAB-capped Ag nanoparticles. | ||
Here we report on the aggregation properties and SERRS signals generated using an alternative and simpler aggregation route, where positively charged silver particles are used for the generation of SERRS signals of dye-labeled DNA. As the negatively charged DNA is electrostatically attracted to the positively charged particles, no other chemicals are needed to induce aggregation (Scheme 1(b)). While other authors have used coulomb charge interactions to assemble (and detect) negatively charged ions onto a positively charged silver particles coated surface,5 or to assemble positively charged silver particles on DNA attached to a surface,6–8 the use of such interaction for homogenous assays of DNA is novel.
To study the possible use of positively charged particles for SERRS based detection of dye-labeled DNA, cetyl trimethyl ammonium bromide (CTAB)-coated Ag nanoparticles were synthesized according to the literature procedure.9 The average particle size was 15nm, as measured by TEM (Fig. S1, ESI†). The synthesized particles are very stable, with little precipitation occuring even after 6 months of storage at room temperature.
In a typical experiment, the Ag nanoparticles and R6G-labeled 29 nucleotides long single stranded DNA were mixed in a Tris-Tween buffer. SERRS signals were measured after different times of interaction using 532nm laser excitation. Fig. 1 shows the SERRS signals obtained with 8nM dye-labeled DNA. As can be seen, the aggregation of positively-charged nanoparticles around the dye-labelled DNA is a slow process, with the SERRS signal increasing slowly over a time scale of one hour. The nanoparticle aggregation process was confirmed by absorbance spectra (Fig. S2, ESI†). However, the adsorption of the negatively charged DNA on the positively charged nanoparticles is a very fast process, causing an observable, immediate, reduction of the fluorescence signal of R6G-labeled DNA upon the mixing of the DNA with the nanoparticles, due to quenching at the nanoparticle surface (Fig. S3, ESI†). The likely explanation for the slow aggregation of the nanoparticles is the electrostatic repulsion between particles, which remain positively charged even after DNA adsorption, as long as the DNA concentration is insufficiently high to form a monolayer on the surface. (Using an average diameter of 15 nm, and a concentration of 1 nM, even at a DNA concentration of 20 nM. there is still more than 35 nm2 of particle surface area available per 9 nm long ssDNA).
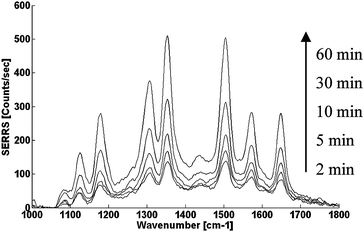 | ||
| Fig. 1 Time dependent SERRS spectra from CTAB-stabilized Ag nanoparticles/R6G-labeled DNA co-aggregation. The nanoparticles and DNA had a final concentration of 1 nM and 8 nM respectively in 10 mM Tris-HCl buffer pH = 7.5 supplemented with 0.01% Tween 20. | ||
Fig. 2 shows the effect of DNA concentration on the development of the SERRS signal with time (using the strongest SERRS peak at 1361 cm−1, which is assigned to the aromatic C–C stretch10). Upon increase of the concentration of DNA, not only the total signal increases, but also the signal per nM of dye labeled-DNA increases, as the electrostatic repulsion gets smaller when more and more DNA molecules adsorb on the surface. One can also see from the graph, that the slope increases with DNA concentration, which is to be expected, as particle aggregation becomes easier. These features are very different from those seen with the standard method of mixing the dye-labeled DNA with citrate-capped particles and spermine, where signals decrease with time because plasmon excitation becomes less efficient as bigger and bigger aggregates are formed (Fig. S4, ESI†).
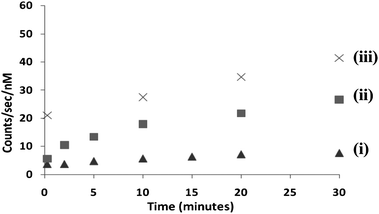 | ||
| Fig. 2 Time dependence of the SERRS peak value (from the 1361 cm−1 peak) using positively charged colloids (1 nM) for different concentrations of DNA: (i) 2 nM (ii) 8 nM (iii) 20 nM. | ||
In order to increase the rate at which the nanoparticles aggregate, the electrostatic repulsion between the CTAB-capped particles had to be reduced. In the case of the standard citrate-capped silver particles, polyamine containing molecules, such as spermidine (which has three primary amines) or spermine (which has four primary amines) are added for the reduction of electrostatic repulsion between the negatively charged particles. In a similar fashion, we tested molecules that contain multiple carboxyl functional groups for the reduction of electrostatic repulsion between our positively charged particles. We chose to study the effect of two molecules that are commonly used in biochemisty: citrate (which contains three carboxyl units) and EDTA (which containes four carboxyl units).
Fig. 3 compares the SERRS signal measured after mixing the DNA with the nanoparticles, in a buffer free of polycarboxyls to those measured in buffers containing citrate or EDTA ions. Unlike the previous results, when adding citrate or EDTA, the SERRS peak reached a maximum after only 10 min. At that point, a significant enhancement of the signal is observed for the solutions that included polycarboxyls.
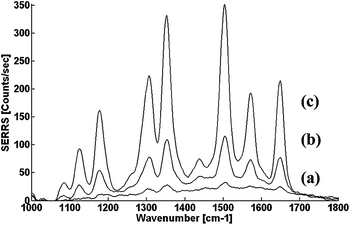 | ||
| Fig. 3 SERRS spectra from CTAB-stabilized Ag nanoparticles/R6G-labeled DNA co-aggregation for solutions which (a) do not contain any polyionic molecules. (b) contain 20 μM citrate ions. (c) contains 20 μM EDTA ions. All spectra were taken 10 min after mixing of DNA and nanoparticles. Final DNA concentration was 2 nM. | ||
Our results show that EDTA is more efficient than citrate, and this is ‘mirroring’ the fact that the tetra-amine was better than a tri-amine in the studies with negatively charged silver nanoparticles.11 The major difference between using a polyamine with negatively charged nanoparticles and a polycarboxyl-containing molecule with positively charged nanoparticles, is that the polyamine is also adsorbing on the DNA molecules, which are negatively charged, but the poly-carboxyl is only adsorbing on the positively charged particles without interacting with the DNA.
Finally, dyes with different charges were used in order to prove that this method is not significantly influenced by the charge of the dye at the end of the DNA. Previous reports on the standard citrate/spermine method showed that negative dyes do not give efficient SERRS unless special modifications to the DNA backbone were used.12Fig. 4 shows the SERRS spectra of different dye-labelled DNA obtained with the use of CTAB-coated Ag nanoparticles and EDTA as charge neutralization agent. Atto 532 has a charge of −1 after conjugation to DNA, Bodipy 558 is a neutral dye, and Atto 565 is a dye with a charge of +1 after conjugation. All three spectra are of the same magnitude, showing that the dye charge does not influence significantly the SERRS enhancement in this system. Having different dyes that give the same order of magnitude of the SERRS signal is important in assays where high multiplexing is used, where the mixed signal has to be deconvolved into its components.
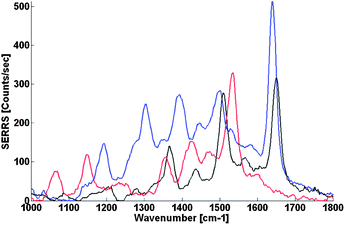 | ||
| Fig. 4 SERRS spectra from CTAB-stabilized Ag nanoparticles/dye-labeled DNA co-aggregation of Atto 532 dye-labelled DNA (blue), Atto 565 dye-labelled DNA (black) and Bodipy 558 dye-labelled DNA (red). Dye-labeled DNA concentration was 2 nM and the buffer contained 20 μM of EDTA in all measurements. | ||
In conclusion, a simplified, alternative route for aggregation of silver nanoparticles in the presence of dye-labeled DNA is shown. This method, using positively-charged CTAB-capped silver nanoparticles generates efficient SERRS signals irrespective of the charge of the dye label. The signal can be further enhanced by the addition of polyanionic ions. This can be used for the development of assays with higher multiplexing level than those possible using fluorescence detection.
The work of R. G. was supported by the European Commission (EC) through the Human Potential Programme within the 6th framework programme (Marie Curie ToK-DEV fellowship, MTKD-CT-2006-042410 LISA).
Notes and references
- L. Sun, C. X. Yu and J. Irudayaraj, Anal. Chem., 2007, 79, 3981–3988 CrossRef CAS.
- G. W. Lucassen, S. Neerken, K. Schmidt and A. Derome, unpublished work.
- D. Graham, W. E. Smith, A. M. T. Linacre, C. H. Munro, N. D. Watson and P. C. White, Anal. Chem., 1997, 69, 4703–4707 CrossRef CAS.
- D. Graham and K. Faulds, Chem. Soc. Rev., 2008, 37, 1042–1051 RSC.
- S. Tan, M. Erol, S. Sukhishvili and H. Du, Langmuir, 2008, 24, 4765–4771 CrossRef CAS.
- G. Wei, L. Wang, H. L. Zhou, Z. G. Liu, Y. H. Song and Z. A. Li, Appl. Surf. Sci., 2005, 252, 1189–1196 CrossRef CAS.
- L. Fabris, M. Dante, G. Braun, S. J. Lee, N. O. Reich, M. Moskovits, T.-Q. Nguyen and G. C. Bazan, J. Am. Chem. Soc., 2007, 129, 6086–6087 CrossRef CAS.
- J. Zhang, T. Boon Ping, N. R. Jana, G. Zhiqiang and J. Y. Ying, Small, 2009, 5, 1414–1417 CrossRef CAS.
- Z. M. Sui, X. Chen, L. Y. Wang, L. M. Xu, W. C. Zhuang, Y. C. Chai and C. J. Yang, Phys. E., 2006, 33, 308–314 CrossRef CAS.
- P. Hildebrandt and M. Stockburger, J. Phys. Chem., 1984, 88, 5935–5944 CrossRef CAS.
- in US Pat., 6127120, 2000.
- D. Graham, B. J. Mallinder and W. E. Smith, Angew. Chem., Int. Ed., 2000, 39, 1061–1063 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: TEM pictures of CTAB-stabilized particles, time-dependent absorption spectra of the CTAB-stabilized particles after the addition of dye-labelled DNA, fluorescence spectra of dye-labeled DNA before and after the addition of CTAB-stabilized particles, time-dependent SERRS spectra of dye labelled DNA using citrate-capped silver nanoparticles and spermine. See DOI: 10.1039/c0ay00190b |
| This journal is © The Royal Society of Chemistry 2010 |
