Impedimetric approach for monitoring bacterial cultures based on the changes in the magnitude of the interface capacitance
Xavier
Muñoz-Berbel
*a,
Núria
Vigués
b,
Montserrat
Cortina-Puig
a,
Roger
Escudé
a,
Cristina
García-Aljaro
a,
Jordi
Mas
b and
Francesc Xavier
Muñoz
a
aCentre Nacional de Microelectrònica (IMB-CSIC), Campus Univ. Autònoma de Barcelona, Esfera UAB, 08193,, Bellaterra, Barcelona, Spain. Fax: +34 93 580 14 96; E-mail: fxmber@gmail.com; Fax: +34 93 594 77 00
bGrup de Microbiologia Ambiental, Universitat Autònoma de Barcelona, Campus UAB, E-08193, , Bellaterra, Spain
First published on 28th May 2010
Abstract
A previously reported methodology is applied to monitor the concentration of bacterial suspensions from an incubator using impedance spectroscopy and platinum electrodes. The interface capacitance, commonly fitted as a constant phase element, CPEi, was found sensitive to the suspended bacteria concentration after short exposure times in a wide range of concentrations (from 102 to 107 colony forming units per mL, CFU mL−1) with a limit of detection of 10 CFU mL−1. The effect of the substances released during bacterial growth (enzymes, toxins, exopolysaccharide, etc.) in the CPEi magnitude was found to be negligible and samples only containing cells washed with saline solutions showed results comparable to those obtained using aliquots directly extracted from the bacterial incubator. This impedimetric approach showed correlation with classical microbiological methods for measuring bacterial concentration such as plating onto agar, optical density and fluorescence microscopy. The impedimetric approach was simplified to a single frequency analysis by selecting the optimal frequency for the measurement.
1. Introduction
Bacterial species can be found adapted to live elsewhere (air, water or soil) as free cells (planktonic cells) or in communities, such as biofilms.1 The demands on cell counting are dependent upon the field or industry which requires the method,2 being especially relevant in the fermentation industries where a strict control of the cells’ activity in the incubator is necessary during the fermentation process. The presence of bacteria, even non-pathogenic ones, is very restricted in other areas. Some examples include the environmental monitoring, the food and the beverage industries and the clinical chemistry. For instance, the presence of non-pathogenic bacteria is limited to 102 colony forming units per mL (CFU mL−1) in drinking water.3 Such strict controls in vital areas require fast and simple detection methods for the real-time quantification of planktonic bacteria in liquids, especially at low concentrations.Conventional methods for the detection of viable bacteria typically rely on the culture-based assays with excessive sample treatment and long incubation times. With the advent of modern molecular biological techniques, many new approaches have been investigated for this purpose, such as bioluminescent assays,4 Fluorescent In Situ Hybridization (FISH),5 optical tweezers,6 nucleic acid amplification method (Polymerase Chain Reaction, PCR),7 Reverse Transcription-Polymerase Chain Reaction (RT-PCR)8,9 and Nucleic Acid Sequence-Based Amplification (NASBA).10 Although these methods can offer high sensitivity, they are either time-consuming or rely on laboratory facilities, which limit their application for rapid operation and on-site analysis. In recent years, a number of new methodologies have been applied to the detection of planktonic bacteria. These include voltamperometry,2 micromechanical oscillators11 and Quartz Crystal Microbalances (QCMs).12 These methods demonstrate good sensitivities (around 102-103 CFU mL−1) with short measurement times. However, the complexity of the assay, which frequently requires the manipulation of fragile or biologically delicate materials, renders this method vulnerable to error.
Recently in our group, a new approach for monitoring bacterial concentration has been developed.13,14 The detection scheme is based on the changes produced in the electrode-solution interface by bacterial attachment in the initial stage of the electrode surface colonization (early reversible bacteria attachment). Previous works of the group showed that bacterial attachment to platinum electrodes could be used for suspended bacteria quantification. Next, this approach was probed to be able to quantify different bacterial species (Pseudomonas aeruginosa and Staphylococcus aureus) and even yeasts (Saccharomyces cerevisiae) following the same measurement protocol.15 In combination with an artificial neural network, this approach was found to be selective enough to distinguish between binary mixtures of microorganisms after a suitable training process.
In this paper, the approach previously described is applied to the monitoring of real bacterial samples from an incubator where Escherichia coli (E. coli) are growing under optimal experimental conditions. Before its application to the control of the bacterial concentration of real samples extracted from the incubator, the influence of the products released during the bacterial metabolism (secreted proteins, such as enzymes or toxins, exopolysaccharide, lipopolysaccharide, cell debris, peptidoglycan or waste products) in the impedimetric measurement is investigated. The impedimetric approach is compared with classical microbiological methods, such as plating onto agar (the classical method for counting bacteria, very accurate but extremely tedious),1 epifluorescence microscopy counting (a standard method, faster than plating but also tedious)16 and optical density (the most employed method for the on-line monitoring of bacterial concentration).1 This approach was also simplified to a single frequency analysis.
2. Experimental
2.1 Electrochemical cell
The platinum sensors were fabricated using photolithographic processes. A platinum disk working electrode surrounded by a platinum counter electrode of 1.4 mm2 was integrated on a silicon nitride substrate of 9 mm2 (3 mm length per 3 mm width) using electron beam metallization.17 The electrodes were 0.08 mm apart, ensuring near-homogeneous polarization of the working electrode.18 The working electrode/counter electrode (WE/CE) chip production was carried out in-house at the Centre Nacional de Microelectrònica de Barcelona (CNM-IMB, CSIC). The measurement systems comprised the WE/CE chip and an external Ag|AgCl reference electrode (Crison, Barcelona, Spain).2.2 Microbiological preparation
E. coli (CGSC 5073 K12) was grown overnight at 37 °C in AB Minimal Medium (ABMM) containing glucose.19 The bacterial concentration was measured using plating on (Luria-Bertani) LB medium containing 1.5% agar, with stock concentration at around 109 CFU mL−1. Bacteria were centrifuged and re-suspended in sterile ABMM. This process was repeated 3 times to ensure the complete removal of the substances secreted during bacterial growth. The suspension was then serially diluted down to 101 CFU mL−1 in decade steps. A dilution bank ranging from 0 to 107 CFU mL−1 was prepared for the calibration of the impedimetric approach.For the monitoring of the bacterial growth, 1 mL of the suitable bacterial suspension was inoculated into a 1.5 L water-jacketed glass reactor containing ABMM (under sterile conditions) to achieve the initial concentration of interest. During bacterial growth, the reactor was thermostatically kept at 37 °C with constant agitation [MR 2000 (Heidolph, Germany)] and aeration [BioFlo (New Brunswick Scientific, New Jersey, USA)]. Under these conditions, E. coli grew aerobically with a duplication time experimentally found between 30 and 40 min. After the inoculation, no nutrients were added into the reactor (batch process).
Using the aseptic acquisition system, aliquots of 20 mL were extracted from the incubator at 30 min intervals and directly measured using impedance spectroscopy, optical density and fluorescence microscopy. After 20 min of centrifugation at 4388 × g [Sigma 4–10 centrifuge (Sigma, Switzerland)] the secreted substances (supernatant containing secreted proteins, such as enzymes or toxins, exopolysaccharide, lipopolysaccharide, cell debris, peptidoglycan or waste products from the metabolism) and the suspended cells (pellet cleaned and re-suspended in sterile ABMM) were separately measured using impedance spectroscopy. Biological samples were stored at 4 °C to slow growth until measurement. All of the manipulations were performed under sterile conditions.
2.3 Impedance measurements
Impedance measurements were performed following the protocol detailed in previous publications of the group.14 Briefly, the impedance analyser AUTOLAB PGSTAT 12 (EcoChemie, BV, The Netherlands) containing a FRA-2 module was used. Impedance spectra were recorded with the FRA-2 software (EcoChemie, BV, The Netherlands). Experimentally, 25 mV AC potential was applied at the cell open circuit potential (+0.26 ± 0.05 V vs. Ag|AgCl) over a frequency range between 100 kHz and 10 Hz recording 17 points per frequency decade with a total measurement time of around 1 min. All measurements given in this work are in relation to the Ag|AgCl reference electrode.Bacterial suspensions were introduced to the electrochemical cell, which was thermostatically kept at 4 °C. Impedance measurements were made after 15 s of exposure for the calibration solutions, direct (non-centrifuged samples), secreted substances or suspended cells samples. After the measurement, the electrochemical cell, including electrodes, was cleaned with water and sterilized with ethanol (Panreac, Spain). Ethanol residues were eliminated by washing another time with water.
2.4 Optical density measurements
The magnitude of the optical density at 550 nm (OD550) was used to follow the bacterial concentration of the culture. OD550 measurements were made with the spectrophotometer Ultrospec 1100 pro (Biochrom, Cambridge, UK). OD550 values were interpolated in a calibration curve previously obtained for E. coli growing under the same experimental conditions, which was used as a calibration curve (data not shown).2.5 Fluorescence microscopy measurements
The bacteria concentration in the incubator was monitored with time using fluorescence microscopy for correlation with impedance measurements. Bacteria cells of each aliquot were fixed with formaldehyde (CH2O, Sigma) and retained in 0.2 μm pore size GTBP filters (Millipore, Billerica, Massachusets, USA). Bacteria were stained with 20 μg mL−1 4′-6-diamidino-2-phenylindole (DAPI) (Merk, Germany) for 5 min and then rinsed in phosphate buffered saline (PBS) immediately prior to imaging. An Olympus BH Fluorescence Microscope (Olympus, California, USA) was used.3. Results and discussion
3.1 Fitting and interpretation of impedance spectra
Impedance spectra were fitted using the Z-View software to the equivalent circuit shown in Fig. 1. This circuit was composed of the solution resistance, RS, the interface capacitance, which was modelled with a constant phase element to improve the fitting, CPEi, and an extra capacitance of small magnitude (experimentally found to be below 5 nF) associated with the presence of an external reference electrode, Cref, which was not sensitive to the bacteria concentration. The electrical elements of the circuit showed fitting errors smaller than 5% in all cases. The use of the CPEi instead of a conventional double layer capacitance was discussed in previous works of the group.13–15 The reason of this change is currently under study but the roughness of the electrode appears to play a relevant role on it.20 | ||
| Fig. 1 Admittance complex plane plot for bacterial suspensions extracted from the incubator 30 and 270 min after the inoculation of the bacterial starter suspension. Impedance data were fitted using the equivalent circuit shown inset. The experimental impedance spectra (points in the plot) and the ideal impedance spectra from the fitting (line in the plot) are shown. Also the spectra corresponding to the culture medium without bacteria is added in each plot as control (red line). Below, the calculated values and errors of each element from the fitting are detailed. | ||
Fig. 1 shows the admittance Nyquist plots and the magnitude and errors of each parameter of the electrical equivalent circuit for bacterial suspensions extracted from the incubator 30 and 270 min after the inoculation of the stock bacterial suspension used as starter. Regarding the fitting, from the elements of the equivalent circuit only the magnitude of the CPEi (Ki-T in the figure) was found to change with time as a consequence of bacterial growth in the incubator. The fact that the other parameter of the electrical equivalent circuit related to the CPEi, Ki-P, did not change with bacterial concentration was not surprising since, although this parameter depends on several factors, the roughness of the electrode seems to be one important aspect,20 changing from 1 for ideally flat electrodes to 0.5 for very rough ones.
3.2 Monitoring of bacteria concentrations extracted from an incubator using impedance spectroscopy: Evaluation of the influence of cells and metabolites in the impedance magnitude
Impedance measurements were made and fitted as previously shown. The most relevant results are shown in Fig. 2. It should be noted that after the fitting, the measured CPEi magnitude, Ki-T, was normalized using eqn (1). Ki-T(it) is the value of CPEi at any incubation time and Ki-T(m) is the value of CPEi of the culture medium in absence of bacteria (before inoculation): | (1) |
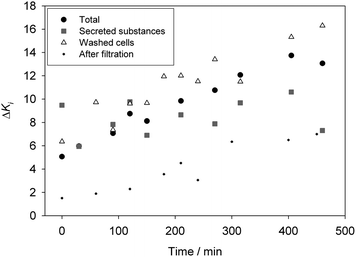 | ||
| Fig. 2 Representation of the normalized Kiversus the incubation time for total samples containing both cells and released substances (total samples), samples only containing released substances (secreted substance samples), samples containing only cells (washed cell samples) and secreted substance samples after filtration (after filtration). | ||
A constant RS value was obtained with time for total samples (samples containing both suspended cells and secreted substances), cells samples (samples containing re-suspended bacteria) and secreted molecule samples (samples mainly containing secreted proteins, such as enzymes or toxins, exopolysaccharide, lipopolysaccharide, cell debris, peptidoglycan or waste products from the metabolism) with a magnitude of 445 ± 19 Ω. Sample conductivity was measured separately with a conductance meter and found to be sample invariant, which confirmed that, under the experimental conditions previously exposed, the conductivity of the medium did not vary with the E. coli growth (or with the secretion of molecules and wasting products from the metabolism).
K i -T changed with time in total and cells samples. The increase in CPEi coincided with that reported in previous works of the group where Ki-T was found to correlate very well with the concentration of washed cells between 102 and 107 CFU mL−1.13,14 Briefly, the attachment of bacteria, considered to be behaving as simple charged colloidal particles during the very early attachment stage, was thought to modify the structure of the double layer at the electrode interface. Particularly, bacteria attachment to the electrode surface may decrease the Debye length at the electrode double layer, causing the increase in CPEi. However, the correlation of this change with time, and thus with bacteria growth, was slightly better in the total samples. This is likely due to the centrifugation step used to prepare the cell samples where some bacteria may be lost.
When considering samples only containing secreted molecules, a random oscillation in the CPEi magnitude was recorded, probably caused by the presence of bacteria since the variation decreased after filtration in 0.2 μm pore size GTBP filters. Thus, substances produced and secreted by E. coli during bacteria growth under the experimental conditions discussed herein did not modify the CPEi magnitude either.
3.3 Evaluation of the global behaviour of the impedimetric approach in the determination of bacteria concentration
The global behaviour of the new impedimetric approach in the determination of bacteria concentration for total and cell samples was evaluated by plotting the predicted values (from impedance spectroscopy) against the expected ones (from plating onto agar). First of all, impedance data from total and cells samples were converted into concentration values using the calibration curve shown in Fig. 3 where the Ki-T shows a linear relationship with the logarithm of the bacteria concentration. The calibration curve was obtained by measuring the samples described in section 2.2, following the measurement protocol detailed in section 2.3. It has to be emphasized that both experiments (calibration curves and impedance data from total and cells samples) were comparable since both of them were made using the same bacteria, culture medium, equipment, experimental conditions and identical platinum electrodes. | ||
| Fig. 3 Representation of the normalized Kiversus the logarithm of the suspended concentration of bacteria. This calibration curve was used for converting capacitance values into bacteria concentrations. | ||
Results from the comparison of both approaches are plotted in Fig. 4. A good method should display comparative lines with high correlation and a slope equal to one with zero intercept. As illustrated in Fig. 4, in both cases impedance spectroscopy measurements showed good correlation with comparison lines practically indistinguishable from the theoretical values. However, better results were obtained in the case of total samples directly extracted from the incubator. As discussed above, this is likely due to the centrifugation step. Some cells remained in suspension after centrifugation, introducing variability in the impedance measurement.
 | ||
| Fig. 4 Representation of the predicted bacteria concentration magnitude (obtained from total (A) and cells samples (B) when using impedance spectroscopy) against the expected ones (from plating onto agar). The dotted line represents the theoretical comparison line y = x. | ||
Finally, the Student's t-test for paired samples was used for checking whether there were significant differences between the predicted and the expected values, 95% confidence value being considered significant. The tabulated values of the t (ttab), 2.26 and 2.20 for total and cells samples respectively, were always found to be bigger than the calculated ones (0.21 and 0.85 for total and cells samples, respectively). Thus, no significant differences between the predicted and the expected values were obtained. Again, better results were achieved when using total samples directly extracted from the incubator.
3.4 Comparison of the impedimetric approach with standard methods: Epifluorescence microscopy counting and optical density
Bacteria concentration values from impedance spectroscopy were compared with those obtained from optical density and epifluorescence microscopy counting measurements. Optical density is the most popular method for the on-line monitoring of bacterial growth in the industry with commercially available equipment based on it.1 The results supplied for the impedimetric approach were compared with those obtained using this traditional method. Below 105–106 CFU mL−1, the limit of detection of the optical density method, impedance measurements were compared with another standard method widely used in microbiology: epifluorescence microbiology counting.16Impedance, optical density and epifluorescence microscopy measurements were made as described in Section 2. Impedance spectroscopy suspended concentration values were obtained by correlating the response magnitude with the calibration curve shown in Fig. 3. Optical density suspended concentration values were similarly obtained by correlation with an appropriate calibration curve (data not shown).
The impedance spectroscopy approach showed great fidelity when compared to epifluorescence microscopy (Fig. 5) in a wide range of concentrations (from 102 to 106 CFU mL−1). However, it could not be compared with the optical density method since the linear range of both approaches did not coincide, they were found complementary. The impedimetric approach is faster, simpler and less tedious than the epifluorescence microscopy method, although epifluorescence microscopy is much more accurate (since it is directly proportional to the bacteria concentration while the impedance approach is proportional to the logarithm of the concentration) and has a better limit of detection.
 | ||
| Fig. 5 Representation of the variation of the concentration values from impedance spectroscopy, epifluorescence microscopy and optical density measurements with the incubation time. | ||
3.5 Single frequency recording: Determination of the optimal frequency for the impedimetric approach
In order to reduce the measurement time, to simplify the approach and to facilitate the data interpretation, the impedance spectra were analyzed to find out the most sensitive frequency. Fig. 6 shows the variation of the phase angle with the frequency for a bacterial sample extracted after 60 min of incubation. | ||
| Fig. 6 Representation of the variation of the phase angle (ϕ) with the frequency for a bacterial sample extracted after 60 min of incubation. The slope and the intercept of the comparison plots comparing the predicted bacteria concentration (from impedance spectroscopy) against the expected ones (from plating onto agar) for each frequency under study are also included. | ||
The phase angle changed from a value close to 0 (at high frequencies) to almost −80 (at low frequencies) with a minimum value at 10 Hz. Considering that the interface capacitance was found sensitive to the bacteria concentration, the ideal frequency for this application would be that one with a phase angle closer to −90. The small differences in the phase angle value in the region from 1 kHz to 10 Hz suggested a deeper analysis. The interface capacitance magnitude at a single frequency was calculated considering that this behaved as an ideal capacitance by using the imaginary magnitude of the impedance at that frequency (Z′′) and the angular frequency (ω) as follows:
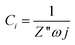 | (2) |
4. Conclusions
Recent investigations of our group demonstrate that the study of the changes produced in the electrode-solution interface though the CPEi magnitude could be used to analyze the initial steadies of bacterial colonization on metallic surfaces.13 At the very early bacterial attachment, a linear relationship between the magnitude of the interface capacitance and the number of suspended bacteria was found in a wide range of concentrations for washed bacterial cells.14Staphylococcus aureus and Saccharomyces cerevisiae as examples of respectively Gram-positive bacteria and yeasts also showed correlation between the number of suspended cells and the CPEi,15 enhancing the range of application of this approach. In this paper, the methodology previously developed has been used to monitor the concentration of real bacteria samples directly extracted from an incubator containing E. coli growing at optimal conditions. The magnitude of the interface capacitance, the parameter of the electrical equivalent circuit capable to monitor the bacterial concentration, has been found to be insensitive to the presence of secreted substances in the medium and only dependent on bacteria concentration. After interpolation in a calibration curve, the bacteria concentration values obtained from impedance measurements have shown good correlation with classical microbiological methods for the determination of bacteria concentration such as plating onto agar and epifluorescence microscopy counting in a wide range of concentrations (from 102 to 106 CFU mL−1), and it has been found complementary to optical density. The impedimetric approach was simplified to a single frequency analysis by selecting the most sensitive frequency. 10 Hz was found to be the optimal frequency to monitor bacterial concentration under these experimental conditions.Recently in our group, a new approach for monitoring bacterial concentration has been developed.13,14 The detection scheme is based on the changes produced in the electrode–solution interface by bacterial attachment in the initial stage of the electrode surface colonization (early reversible bacteria attachment). Previous works of the group showed that bacterial attachment to platinum electrodes could be used for suspended bacteria quantification. Next, this approach was probed to be able to quantify different bacterial species (Pseudomonas aeruginosa and Staphylococcus aureus) and even yeasts (Saccharomyces cerevisiae) following the same measurement protocol.15 In combination with an artificial neural network, this approach was found selective enough to distinguish between binary mixtures of microorganisms after a suitable training process.
Acknowledgements
The authors would like to acknowledge funding through the FPU program and the MICROBIOTOX project. Part of the work was supported by grants CSD2006-00044 TRAGUA (CONSOLIDER-INGENIO2010) and TEC2006-12109-C03-02/MIC from the Spanish Ministry of Education and Science.References
- M. T. Madigan, J. M. Martinko, J. Parker, Brock Biology of Microorganisms, Prentice Hall International, Hertfordshire, U.K., 1997 Search PubMed.
- H. Tang, W. Zhang, P. Geng, Q. Wang, L. Jin, Z. Wu and M. Lou, Anal. Chim. Acta, 2006, 562, 190–196 CrossRef CAS.
- P. Lebaron, A. Henry, A. S. Lepeuple, G. Pena and P. Servais, Mar. Pollut. Bull., 2005, 50, 652–659 CrossRef CAS.
- J. Lee and R. A. Deininger, Luminescence, 2004, 19, 209–211 CrossRef.
- T. Garcia-Armisen and P. Servais, J. Microbiol. Methods, 2004, 58, 269–279 CrossRef CAS.
- M. Ericsson, D. Hanstorp, P. Hagberg, J. Enger and T. Nystrom, J. Bacteriol., 2000, 182, 5551–5555 CrossRef CAS.
- T. B. Tims and D. V. Lim, J. Microbiol. Methods, 2003, 55, 141–147 CrossRef CAS.
- G. Bleve, L. Rizzotti, F. Dellaglio and S. Torriani, Appl. Environ. Microbiol., 2003, 69, 4116–4122 CrossRef CAS.
- M.d.M. Lleo, S. Pierobon, M. C. Tafi, C. Signoretto and P. Canepari, Appl. Environ. Microbiol., 2000, 66, 4564–4567 CrossRef CAS.
- S. A. Simpkins, A. B. Chan, J. Hays, B. Pöpping and N. Cook, Lett. Appl. Microbiol., 2000, 30, 75–79 CrossRef CAS.
- K. Y. Gfeller, N. Nugaeva and M. Hegner, Biosens. Bioelectron., 2005, 21, 528–533 CrossRef CAS.
- G. Yu, J. Niu, M. Shen, H. Shao and L. Chen, Clin. Chim. Acta, 2006, 366, 281–286 CrossRef CAS.
- X. Muñoz-Berbel, N. Vigués, J. Mas, A. T. A. Jenkins and F. J. Muñoz, Electrochem. Commun., 2007, 9, 2654–2660 CrossRef CAS.
- X. Muñoz-Berbel, N. Vigués, A. T. A. Jenkins, J. Mas and F. J. Muñoz, Biosens. Bioelectron., 2008, 23, 1540–1546 CrossRef CAS.
- X. Muñoz-Berbel, N. Vigués, J. Mas, M. del Valle, F. J. Muñoz and M. Cortina-Puig, Biosens. Bioelectron., 2008, 24, 958–962 CrossRef CAS.
- K. Alef, P. Nannipieri, Methods in Applied Soil Microbiology and Biochemistry, Academic Press, New York, 1st edn, 1995 Search PubMed.
- M. A. Alonso-Lomillo, J. Gonzalo-Ruiz and F. J. Muñoz-Pascual, Anal. Chim. Acta, 2005, 547, 209–214 CrossRef CAS.
- J. E. Harrar and I. Shain, Anal. Chem., 1966, 38, 1148–1158 CrossRef CAS.
- D. Balestrino, J. A. J. Haagensen, C. Rich and C. Forestier, J. Bacteriol., 2005, 187, 2870–2880 CrossRef CAS.
- H. Fricke, Philos. Mag., 1932, 7, 310–318.
| This journal is © The Royal Society of Chemistry 2010 |
