Automatic configuration of optimized sample-weighted least-squares support vector machine by particle swarm optimization for multivariate spectral analysis
Hai-Yan
Fu
,
Hai-Long
Wu
*,
Hong-Yan
Zou
,
Li-Juan
Tang
,
Lu
Xu
,
Chen-Bo
Cai
,
Jin-Fang
Nie
and
Ru-Qin
Yu
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Hunan University, Changsha, 410082, P. R. China. E-mail: hlwu@hnu.cn; Fax: +86-731-88821818
First published on 25th January 2010
Abstract
Due to the high dimensionality and complexity of multivariate spectral data space and the uncertainty involved in the sampling process, the representation of training samples in the whole sample space is difficult to evaluate and selection of representative training samples for conventional multivariate calibration depends largely on experiential methods. If the training samples fail to represent the sample space, sometimes the prediction of new samples can be degraded. To circumvent this problem, in this paper, a new optimized sample-weighted least-squares support vector machine (OSWLS-SVM) multivariate calibration method is proposed by incorporating the concept of weighted sampling into LS-SVM, where the complexity and predictivity of the model are considered simultaneously. A recently suggested global optimization technique base on particle swarm optimization (PSO) is invoked to simultaneously search for the best sample weights and the hyper-parameters involved in OSWLS-SVM optimizing the training of a calibration set and the prediction of an independent validation set. The implementation of PSO achieves complete automatization of the OSWLS-SVM modeling process and high efficiency in convergence to a desired optimum. Three real multivariate spectral data sets including two public data sets and an experimental data set are investigated and the results are compared favorably with those of PLS and LS-SVM to demonstrate the advantages of the proposed method. The stability and efficiency of OSWLS-SVM is also surveyed, the results reveal that the proposed method can obtain desirable results within moderate PSO cycles.
Introduction
With the increasing practical need for fast and reliable quantification of sought-for components in complex samples and the development of modern spectrometers, many classical multivariate calibration methods such as principal component regression (PCR),1 multiple linear regression (MLR),2 partial-least squares (PLS)3 and support vector machine (SVM)4–6 have been developed successfully and widely utilized for the processing of multivariate spectral data. The main role of these methods is to establish a calibration model relating the measured spectral signals to certain properties of samples for predicting the same properties of new samples not involved in the calibration set. Whether the properties can be predicted with satisfactory accuracy depends to a great extent on the performance of the applied multivariate calibration method. However, in some practical multivariate spectral data analyses, these methods may encounter some disadvantages. For example, PLS, as a popular modeling method, might still be susceptible to overfitting when uninformative variables and uncertain nonlinear factors exist in data. Though SVM is less vulnerable to overfitting for such data, it is difficult to find the final SVM model owing to solving a set of nonlinear equations (quadratic programming). Recently, as a simplification of the approach, least-squares SVM (LS-SVM) was proposed and introduced successfully into multivariate calibration by many investigators for its attractive features and promising empirical performance applied in many studies,7–14 it not only encompasses similar advantages as SVM which has the capability of dealing with linear and nonlinear multivariate calibration, but also only requires solving a set of linear equations (linear programming) thereby reducing the computation burden of the SVM model. However, like SVM, the inherent limitation of LS-SVM is the issue of choosing the right kernel function, importance of parameters tuning (cost and epsilon), etc. Conventional LS-SVM is performed firstly by adjusting the parameters in regularization item and kernel function called hyper-parameters, which play an important role in the algorithm performance. The existing methods for adjusting the hyper-parameters in LS-SVM can be summarized into two kinds: one is based on analytical techniques which are computationally intractable to optimize these parameters.15,16 The other is based on heuristic searches including genetic algorithms (GA), simulated annealing algorithms (SA), evolution algorithm (EA) and other evolutionary strategies,17–20 which are the tricky tasks since a suitable sampling step varies from kernel to kernel and the grid interval may not be easy to determine without a prior knowledge of the problem. Moreover, when there are more than two hyper-parameters, the manual model selection may become intractable. To avoid deficiency of these methods, a stochastic global optimization technique, particle swarm optimization (PSO) method21,22 is recommended in the work. Unlike the analytical techniques mentioned above, PSO needs no time-consuming validation of possible models. Compared with GA, SA and EA, PSO is conceptually simpler and requires lower computation costs, it has no evolution operators and does not need any prior knowledge on the analytic property of the generalization performance measure and can be used to determine multiple hyper-parameters at the same time.The performance of multivariate calibration models is strongly dependent upon the homogeneity of the model errors and the uniformity of the data sampling. Reasonable sampling can be as an expectation for further improving performance of models. In analytical chemistry, one of the most important goals of sampling is to select samples that are as representative as possible for the purpose of drawing general conclusions about the population.23–25 Usually, we can follow some obvious and intuitive principles, such as experiences and experimental designs26,27 to select representative samples for calibration. But some unknown negative factors and some uncontrollable conditions28–32 may occur in the distribution of whole sample space and high dimensional spectral space, sampling process and feature computation and so on, which would make the representation of the samples in a calibration set become uncertain to some extent. In cases where the training samples are singularly distributed into clusters and the model errors are highly heterogeneous, performing an experimental design or outlier diagnosis can not necessarily ensure a representative training set. To obtain representative training samples more effectively, it is reasonable that the samples in the calibration set should not be treated without discrimination. That is, samples with high representation should be attached more importance to, while the ones with poor representation should be discounted. Representation of the samples in the calibration set should be rescaled by their contribution to the model. Xu et al.33 have proposed sample weighting with PLS for spectra data analysis, but it can not perform well when non-linear factors exist. Zhou et al.34 have developed a boosting support vector regression method for QSAR studies, but the process of the parameter selection is complex and the computation is slow. Wen et al.35 have combined outlier detection and adaptive weighted least square support vector machine for QSAR studies, but computation of weights is complicated, and the weights and parameters involved in model can not be optimized simultaneously, which means the final solution could not be a global optimization and the validation of possible models would be time-consuming.
In this paper, the concept of weighted sampling is introduced to LS-SVM and a new intelligentized algorithm called optimized sample-weighted least-squares support vector machine (OSWLS-SVM) is developed. In the proposed methodology, a new calibration set is constructed by sampling the individuals in the original calibration set for different times, where samples in the original calibration set are weighted with different non-negative values. Automatic configuration of the best sample weights and the hyper-parameters involved in LS-SVM are performed simultaneously by the particle swarm optimization (PSO) technique optimizing the training of a calibration set and the prediction of an independent validation set. The details are described in the next parts of the paper. The proposed OSWLS-SVM algorithm is applied to multivariate spectral data analysis including two public data sets and an experimental data set, the results show satisfactory performance in training and prediction for three real data sets. The stability and efficiency of OSWLS-SVM is also studied, the results demonstrate that the proposed method can obtain desirable results within moderate PSO cycles.
Theory
Optimized sample-weighted least-squares support vector regression algorithm
The theory of PLS, SVM and LS-SVM has been described clearly in original literatures and some papers with their applications.3–14 Thus only a detailed description of OSWLS-SVM is given in this paper.Consider an n × p matrix X including p predictor variables for n samples and an n × 1 vector y including the corresponding dependent variable for the n samples, for simplicity and without loss of generality, both X and y are column centered. Not like in a conventional LS-SVM model, when the calibration set is determined to build the model describing the relationship between the dependent variables and the predictor variables, all samples are considered to make the same contribution to the model, each sample in a calibration set is essentially weighted with the same value 1. This is only really a special case of sample weighting, where the difference of representation between the samples is ignored. In OSWLS-SVM, the idea of the difference of representation between the samples with different weighted values is integrated into LS-SVM. OSWLS-SVM is expected to be more flexible and rational. The key issue is how to sample or weight the original samples to form a representative calibration set. Now conceive a new calibration set Xn of the size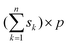 and the corresponding new dependent variable yn of the size
and the corresponding new dependent variable yn of the size 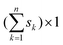 , Xn and yn can be formed in such a way that the kth sample in the original calibration set X is sampled for sk times or given a weight sk. The samples appearing in the new calibration set Xn and yn have been column-centered in the original calibration set X and y. It is very clear that a sample in the original calibration set with higher representation should be given a bigger weight and vice versa. However, according to the theory of sampling, a problem arises that whether a sample is representative or not cannot be determined by any kind of inspection or characterization of the sample itself and the recognition of representative samples requires a full qualification of the sampling process. Hereby, to obtain a reasonable new calibration set, a sampling design should be based on such considerations that a good OSWLS-SVM calibration model with a set of highly representative samples can lead to a good prediction of the samples in the whole sample space. Thus, the goal of OSWLS-SVM is to automatically find a non-negative vector of sample weights s = [s1, s2, s3,…, sn]T and hyper-parameters to optimize the calibration of the original training set and the prediction of an independent validation set. The PSO algorithm is simultaneously implemented for the sample weights and hyper-parameters training. A predefined fitness function is used to evaluate the performance of each particle in OSWLS-SVM, whose minimization would be obtained. In this paper, in addition to an n × 1 vector with all the elements being 1, 99 non-negative vectors are randomly generated to form 100 initial feasible solutions. The following objective function is minimized in the optimization process:
, Xn and yn can be formed in such a way that the kth sample in the original calibration set X is sampled for sk times or given a weight sk. The samples appearing in the new calibration set Xn and yn have been column-centered in the original calibration set X and y. It is very clear that a sample in the original calibration set with higher representation should be given a bigger weight and vice versa. However, according to the theory of sampling, a problem arises that whether a sample is representative or not cannot be determined by any kind of inspection or characterization of the sample itself and the recognition of representative samples requires a full qualification of the sampling process. Hereby, to obtain a reasonable new calibration set, a sampling design should be based on such considerations that a good OSWLS-SVM calibration model with a set of highly representative samples can lead to a good prediction of the samples in the whole sample space. Thus, the goal of OSWLS-SVM is to automatically find a non-negative vector of sample weights s = [s1, s2, s3,…, sn]T and hyper-parameters to optimize the calibration of the original training set and the prediction of an independent validation set. The PSO algorithm is simultaneously implemented for the sample weights and hyper-parameters training. A predefined fitness function is used to evaluate the performance of each particle in OSWLS-SVM, whose minimization would be obtained. In this paper, in addition to an n × 1 vector with all the elements being 1, 99 non-negative vectors are randomly generated to form 100 initial feasible solutions. The following objective function is minimized in the optimization process:
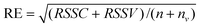 | (1) |
The steps of OSWLS-SVM can be described as follows:
Step 1. With the 100 initial feasible solutions of sample weighting vector s and two hyper-parameters (kernel widths and the relative weight of the error term) in LS-SVM. A LS-SVM model between predictor matrix Xn and dependent variable yn is built.
Step 2. For different numbers of generations, PSO is used to search for s and two hyper-parameters that minimize the objective function RE defined in eq. (1).
Step 3. With the best optimized sample weighting vector s and hyper-parameters determined in Step 2, an OSWLS-SVM model is built and then applied to unknown samples.
Data sets
Public data sets
Experimental data set
Results and discussion
Due to the poor selectivity of NIR absorption bands, broad and serious overlapping and high complexity of the measured NIR data, data preprocessing should be considered to remove noise, baseline and other interfering factors. So, for two NIR data sets including Grass data and Tablets data, prior to calibration modeling, multiple scatter correction (MSC)38 is used to preprocess the spectra. Since Monte Carlo cross-validation (MCCV)39,40 can mitigate the probability of overfitting, it is used to determine latent variables of PLS model and hyper-parameters of conventional LS-SVM in this paper. For three data sets, the parameters in MCCV are all such selected that in each randomly sampling process, 40% of the training samples are left out and the number of sampling times is 100. The results of all models based on PLS, LS-SVM and OSWLS-SVM for three real multivariate data are listed in Table 1.| Data (analytes) | PLS | LS-SVM | OSWLS-SVM | |||
|---|---|---|---|---|---|---|
| RMSEC | RMSEP | RMSEC | RMSEP | RE | RMSEP | |
| Grass (nitrogen) | 0.24 | 0.27 | 0.09 | 0.25 | 0.07 | 0.15 |
| Fuel (saturates) | 0.82 | 0.91 | 0.80 | 0.79 | 0.56 | 0.64 |
| Fuel (monoaromatics) | 0.64 | 0.85 | 0.65 | 0.82 | 0.50 | 0.65 |
| Fuel (diaromatics) | 0.17 | 0.18 | 0.14 | 0.14 | 0.09 | 0.09 |
| Fuel (polyaromatics) | 0.04 | 0.15 | 0.03 | 0.05 | 0.03 | 0.04 |
| Tablet (paracetamol) | 2.34 | 3.80 | 2.67 | 2.86 | 1.52 | 1.76 |
For the grass data, there are 1050 spectral channels in the spectra in all and the wavelengths are not released, so the wavelengths are just referred to as wavelength variable 1 to 1050. The total 141 samples are split into a calibration set with 70 samples, a validation set with 34 samples and a test set with 37 samples by the DUPLEX method.41 The 104 samples in the calibration set and validation set form the training set for PLS. The number of PLS latent variables is determined to be 9. The test set is used for prediction, the root mean squared error of calibration (RMSEC) and root mean squared error of prediction (RMSEP) by PLS are 0.24 and 0.27, respectively. As to LS-SVM, the calibration set and validation set are also combined as the training set for the training model. RMSEC and RMSEP is 0.09 and 0.25 respectively. The results show that the LS-SVM model obtains slight better prediction ability than the PLS model, but the root mean squared error of the LS-SVM model for the test set is much larger than the training set, indicating the LS-SVM model exhibits an overfitting problem. For OSWLS-SVM, the 34 samples picked are used as a validation set independent of the training set for training the OSWLS-SVM model. A 300-cycle PSO is carried out to search for the sample weights of the calibration set and hyper-parameters minimizing the objective function defined RE, the corresponding sample weights are shown in Fig. 1. Where each sample in the original calibration set is weighted differently to account for its representativeness, all weighted samples in the calibration set can more effectively represent the sample space thereby avoiding the degradation of prediction ability for new samples. The OSWLS-SVM model gives an RMSEP for the prediction set of 0.15. The RMSEP is reduced by 45% compared with that of the PLS model and reduced by 40% compared with that of the LS-SVM model. This data set revealed that the proposed OSWLS-SVM offered substantially improved performance of the LS-SVM algorithm.
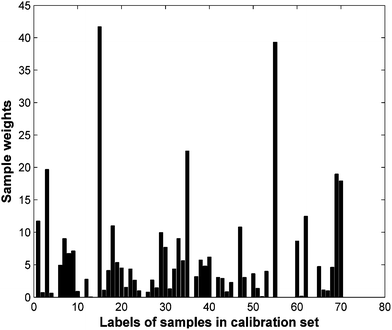 | ||
| Fig. 1 Sample weights obtained by a 300-cycle PSO search for grass data. | ||
As to the fuel data, the 114 samples are split into a calibration set with 56 samples, a validation set with 28 samples and a test set with 30 samples by the DUPLEX method. The 84 samples in the calibration set and validation set form the training set for PLS. For four kinds of analytes of fuel samples including saturates, monoaromatics, diaromatics and polyaromatics in the fuel data, the number of each PLS model optimum latent variables is determined as 7, 7, 6, and 6 by MCCV, respectively. A LS-SVM model is also built on 84 samples containing the calibration set and the validation set. 28 samples picked are used as a validation set independent of the training set for training the OSWLS-SVM model. For each of the four components in the fuel data, a 300-cycle PSO is carried out to search for the weights of 56 samples in the calibration set and hyper-parameters minimizing the objective function defined RE. Each sample in the original calibration set is weighted with different non-negative values and each sample can bring into play itself effect in the OSWLS-SVM model (figures not shown). As shown in Table 1, the OSWLS-SVM provides better results for the four analytes than those of PLS and LS-SVM. These results verify that the proposed algorithm has the ability to model nonlinearity more effectively than the PLS and is more capable of resisting the overfitting problem than LS-SVM. This might be on account of the strategy of optimizing the sample weighting being capable of mitigating the overfitting to singular sample distribution and heterogeneous errors.
For again checking the proposed OSWLS-SVM algorithm as a competitive alternative to conventional multivariate calibration approaches, it was applied to an experiment data, and the PLS and LS-SVM are also tested as comparisons. The 60 cold tablet samples were split into a calibration set with 30 samples, a validation set with 14 samples and a test set with 16 samples by the DUPLEX method. A PLS model is built on 44 samples containing the calibration set and the validation set. The number of PLS model latent variables was determined as 9. The test set is used for prediction, the root mean squared error of calibration (RMSEC) and root mean squared error of prediction (RMSEP) is 2.34 and 3.80, respectively. A LS-SVM model is also built on 44 samples containing the calibration set and the validation set. The RMSEC and RMSEP obtained by LS-SVM is 2.67 and 2.86, respectively. Results of LS-SVM are better than that of PLS. This might be attributed to contamination of some nonlinear factor in this data set. For OSWLS-SVM, 14 samples picked are used as a validation set independent of the training set for training the OSWLS-SVM model. A 400-cycle PSO is carried out to search simultaneously for the optimum sample weight vectors of the calibration set and hyper-parameters minimizing the objective function defined RE. The corresponding sample weights are depicted in Fig. 2. It can be seen some samples are highly weighted and some samples have small weights. Difference of representation between the samples can be rescaled with different weighted values, each sample in the calibration set makes a different contribution to training of the model for prediction. Weights of sample 16 and 27 are very low. They are not representative and seem like outliers. When the two samples are left, the LS-SVM algorithm gives a root mean squared error (RMSE) of 2.05 for the training set and 2.31 for the test set. It shows better performance than that of LS-SVM with all samples in calibration set. But when the OSWLS-SVM algorithm is used, no outliers are taken out and better results are obtained with the all weighted samples in the calibration set. The OSWLS-SVM model gives the most satisfactory results, with a RE value 1.52 and a RMSEP value 1.76. These results verify that the integration of weighted sampling with LS-SVM offers the possibility of improving the performance of the calibration model, this might be due to the fact that it solves the representativeness of samples to some extent.
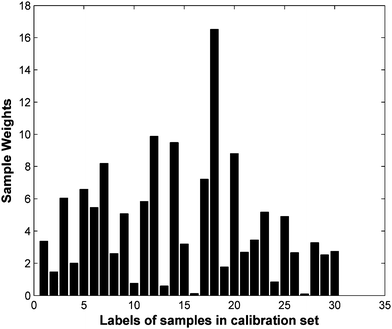 | ||
| Fig. 2 Sample weights obtained by a 400-cycle PSO search for cold tablet data. | ||
Based on the results for the three real data sets, it is demonstrated adequately that the OSWLS-SVM method yielded superior performance of model training and prediction to the conventional LS-SVM and PLS method. This is due to that reasonable construction of new calibration set in OSWLS-SVM model solves the problem of the representation of samples to some extent and is more effective to represent the sample space, model errors can be mitigated when data may encounter some negative impacts including non-uniformly distribution of samples, heterogeneous noises and so on. Compared with some sample weighting methods in literatures, such as OSWPLS, BSVR and AWLS-SVM,33–35 the present method will have some advantages. Compared with OSWPLS, which weights the training samples by PSO for linear models, the proposed method will extend the sample-weighting strategy to nonlinear calibration models and proves to be successful. Compared with BSVR and AWLS-SVM, which weight the training samples by other criterions, the present method obtains the global optimum of sample weights and model parameters by minimizing a reasonable objective function considering both training and predicting performances. Since the representativeness of samples can not be determined by inspecting the samples themselves, for SVM, it is of importance to weight training samples by considering the model parameters. Therefore, by simultaneously optimizing the samples weights and model parameters, the influences of sample representativeness on calibration models are fully considered in our method.
Stability investigation of the intelligentized OSWLS-SVM model
OSWLS-SVM as an intelligentized algorithm is achieved by invoking the PSO technique, the stability and efficiency of the particle swarm optimization (PSO) in OSWLS-SVM needs to be studied. Hundreds-cycle PSO have been carried out for the three real data sets, for each cycle, the training error RE defined in eq. (1) and the root mean squared error of prediction (RMSEP) for each analyte are recorded, respectively. All the results obtained are demonstrated in Fig. 3, where training error, prediction error of OSWLS-SVM model for all analytes in PSO cycles are represented by curve 1 and curve 2, respectively. It can be seen that the trends of variation of curves 1 and 2 for every analyte seem to be parallel to each other and might finally turn out to be identical, indicating no sign of overfitting. The trend is very clear that RE and RMSEP decreases with the increasing of PSO cycles, though there are slight fluctuations of RE and RMSEP during the PSO search. The time required to run the OSWLS-SVM is satisfactory. For the grass data sets, OSWLS-SVM converges to the optimal stable solution and offers significant improvements of prediction ability by a search of 110 or more cycles, it takes less than 2 min for our computer (Celeron(R) CPU 2.29 GHz, and 1.00 GB memory) to calculate the OSWLS-SVM model. As to convergence curves of OSWLS-SVM model to every analyte in fuel data sets, a search of 100 or more cycles offers significant improvements and the solution is stable for saturates. For the other analytes in the fuel data, more cycles (less than 200) of PSO search are required. For the cold tablet data, a search of 150 or more cycles gives a steady solution and obtains a desirable result. All the results demonstrate that the optimization process is stable and effective, satisfactory prediction results can be obtained within a reasonable number of PSO cycles.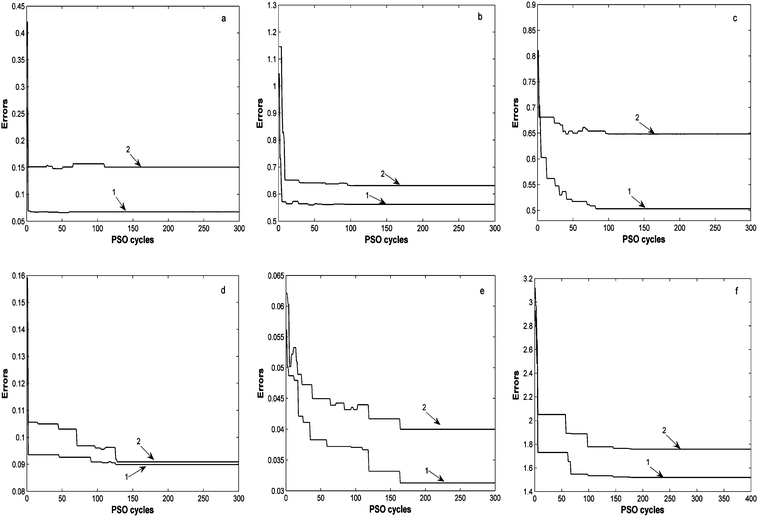 | ||
| Fig. 3 The convergence curves of the OSWLS-SVM model for all analytes, curve 1 indicates the values RE of the objective function defined in eq. (1), curve 2 represents the RMSEP values of prediction of the test set (a) for grass data during 300 PSO cycles; (b), (c), (d) and (e) for analytes in fuel data during 300 PSO cycles, respectively; (f) for cold tablet data during 400 PSO cycles. | ||
Conclusions
In conventional multivariate calibration, each sample in the calibration set is treated equally to give the same contribution to the model, where the difference of representativeness among the samples is seldom considered, which might lead to poor predictions in some cases. This paper developed an intelligentized algorithm called optimized sample-weighted LS-SVM (OSWLS-SVM) by incorporating weighted sampling into LS-SVM for solving the problem of the representation of samples to some extent. PSO is used to search for multiple parameters of the OSWLS-SVM model including the best sample-weighting vectors and hyper-parameters to optimize calibration of the original training set and prediction of an independent validation set. From the results of three real data sets, OSWLS-SVM performs better than PLS and LS-SVM in terms of both training and prediction performances. The stability of the OSWLS-SVM model has also been investigated. The RMSEP value is reduced significantly by PSO and despite the slight fluctuations of RMSEP during the search, satisfactory prediction results can be obtained within a reasonable number of PSO cycles. The advantages of the OSWLS-SVM method compared with some sample weighting methods in literatures are also mentioned in this work. The overall conclusion is that OSWLS-SVM is a promising multivariate calibration method for more practical applications, especially when the data may encounter some factors including non-uniformly distributed samples, heteroscedastic noises and non-composition-related factors and so on.Acknowledgements
The authors gratefully acknowledge the National Nature Science Foundation of China (Grant No. 20775025), the National Basic Research Program (No. 2007CB216404) and the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT) for financial support.References
- P. Geladi and B. R. Kowalski, Anal. Chim. Acta, 1986, 185, 1–17 CrossRef CAS.
- M. Grotti, R. Leardi and R. Frache, Anal. Chim. Acta, 1998, 376, 293–304 CrossRef CAS.
- M. Sjostrom, S. Wold, W. Lindberg, J. A. Persson and H. Martens, Anal. Chim. Acta, 1983, 150, 61–70 CrossRef.
- V. N. Vapnik, in The Nature of Statistical Learning Theory, ed. M. Jordan, S. L. Lauritzen, J. F. Lawless and V. Nair, Springer-Verlag, New York, 2nd edn, 1995, pp. 138–279 Search PubMed.
- V. N. Vapnik, in Statistical Learning Theory, ed. S. Haykin, John Wiley and Sons, New York, 1st edn, 1998, pp. 401–567 Search PubMed.
- A. I. Belousov, S. A. Verzakov and J. V. Frese, J. Chemom., 2002, 16, 482–489 CrossRef CAS.
- J. A. K. Suykens and J. Vandewalle, Neural Process. Lett., 1999, 9, 293–300 CrossRef.
- J. A. K. Suykens, T. V. Gestel, J. D. Brabanter, B. D. Moor and J. Vandewalle, Least Squares Support Vector Machines, World Scientific, Singapore, 1st edn, 2002, pp. 71–239 Search PubMed.
- A. Borin, M. F. Ferrão, C. Mello, D. A. Maretto and R. J. Poppi, Anal. Chim. Acta, 2006, 579, 25–32 CrossRef CAS.
- M. F. Ferrão, S. C. Godoy, A. E. Gerbase, C. Mello, J. C. Furtado, C. L. Petzhold and R. J. Poppi, Anal. Chim. Acta, 2007, 595, 114–119 CrossRef CAS.
- U. Thissen, B. Ustun, W. J. Melssen and L. M. C. Buydens, Anal. Chem., 2004, 76, 3099–3105 CrossRef CAS.
- J. Z. Li, H. X. Liu, X. J. Yao, M. C. Liu, Z. D. Hu and B. T. Fan, Chemom. Intell. Lab. Syst., 2007, 87, 139–146 CrossRef CAS.
- A. Niazi, S. Sharifi and E. Amjadi, J. Electroanal. Chem., 2008, 623, 86–92 CrossRef CAS.
- H. Y. Yu, X. Y. Niu, H. J. Lin, Y. B. Ying, B. B. Li and X. X. Pan, Food Chem., 2009, 113, 291–296 CrossRef CAS.
- S. J. An, W. Q. Liu and S. Venkatesh, Pattern Recognit., 2007, 40, 2154–2162 CrossRef.
- M. W. Chang and C. J. Lin, Neural Comput., 2005, 17, 1188–1222 CrossRef.
- J. Caballero, L. Fernandez, M. Garriga, J. I. Abreu, S. Collina and M. Fernández, J. Mol. Graphics Modell., 2007, 26, 166–178 CrossRef CAS.
- F. Friedrichs and C. Igel, Neurocomputing, 2005, 64, 107–117 CrossRef.
- H. Frohlich, O. Chapelle and B. Scholkopf, Proceedings of the 15th IEEE International Conference on Tools with Artificial Intelligence, Washington, 2003 Search PubMed.
- D. R. Eads, D. Hill, S. Davis, S. J. Perkins, J. Ma, R. B. Porter and J. P. Theiler, Applications and Science of Neural Networks, Fuzzy Systems, and Evolutionary Computation V. Proceedings of the SPIE, Seattle, 2002 Search PubMed.
- J. Kennedy and R. Eberhart, Proceedings of the IEEE International Conference on Neural Networks, Perth, 1995 Search PubMed.
- Q. Shen, J. H. Jiang, C. X. Jiao, S. Y. Huan, G. L. Shen and R. Q. Yu, J. Chem. Inf. Comput. Sci., 2004, 44, 2027–2031 CrossRef CAS.
- P. M. Gy, Sampling for Analytical Purposes, Wiley, New York, 1st edn, 1998, pp. 1–131 Search PubMed.
- L. Petersen, P. Minkkinen and K. H. Esbensen, Chemom. Intell. Lab. Syst., 2005, 77, 261–277 CAS.
- P. J. Worsfold, L. J. Gimbert, U. Mankasingh, O. N. Omaka, G. Harahan, P. C. F. C. Gardolinskic, P. M. Haygarth, B. L. Turner, M. J. Keith-Roacha and I. D. McKelvie, Talanta, 2005, 66, 273–293 CrossRef CAS.
- H. Mark, Anal. Chem., 1986, 58, 2814–2819 CrossRef CAS.
- O. Devos, B. Fanget, A. I. Saber, L. Paturel, E. Naffrechoux and J. Jarosz, Anal. Chem., 2002, 74, 678–683 CrossRef CAS.
- O. Yamamoto and M. Yanagisawa, Anal. Chem., 1970, 42, 1463–1465 CrossRef CAS.
- A. O. Grady, A. C. Dennis, D. Denvir, J. J. M. Garvey and S. E. J. Bell, Anal. Chem., 2001, 73, 2058–2065 CrossRef CAS.
- M. Faber, D. L. Duewer, S. J. Choquette, T. L. Green and S. N. Chesler, Anal. Chem., 1998, 70, 2972–2982 CrossRef CAS.
- C. D. Brown, Anal. Chem., 2004, 76, 4364–4373 CrossRef CAS.
- C. E. Efstathiou, Talanta, 2000, 52, 711–715 CrossRef CAS.
- L. Xu, J. H. Jiang, W. Q. Lin, Y. P. Zhou, H. L. Wu, G. L. Shen and R. Q. Yu, Talanta, 2007, 71, 561–567 CrossRef CAS.
- Y. P. Zhou, J. H. Jiang, W. Q. Lin, H. Y. Zou, H. L. Wu, G. L. Shen and R. Q. Yu, Eur. J. Pharm. Sci., 2006, 28, 344–353 CrossRef CAS.
- W. T. Cui and X. F. Yan, Chemom. Intell. Lab. Syst., 2009, 98, 130–135 CrossRef CAS.
- http://kerouac.pharm.uky.edu/asrg/cnirs/cnirs.html .
- P. D. Wentzell, D. T. Andrews, J. M. Walsh, J. M. Cooley and P. Spencer, Can. J. Chem., 1999, 77, 391–400 CrossRef CAS.
- H. Martens, J. P. Nielsen and S. B. Engelsen, Anal. Chem., 2003, 75, 394–404 CrossRef CAS.
- Q. S. Xu and Y. Z. Liang, Chemom. Intell. Lab. Syst., 2001, 56, 1–11 CrossRef CAS.
- S. Gourvénec, J. A. F. Pierna and D. L. Massart, Chemom. Intell. Lab. Syst., 2003, 68, 41–51 CrossRef CAS.
- R. D. Snee, Technometrics, 1977, 19, 415–428.
| This journal is © The Royal Society of Chemistry 2010 |
