The biosyntheticgene cluster of zorbamycin, a member of the bleomycin family of antitumor antibiotics, from Streptomyces flavoviridis ATCC 21892†
Ute
Galm
a,
Evelyn
Wendt-Pienkowski
a,
Liyan
Wang
a,
Nicholas P.
George
b,
Tae-Jin
Oh
a,
Fan
Yi
a,
Meifeng
Tao
a,
Jane M.
Coughlin
c and
Ben
Shen
*abcd
aDivision of Pharmaceutical Sciences, Division of Pharmaceutical Sciences, School of Pharmacy, University of Wisconsin-Madison, 777 Highland Ave., Madison, Wisconsin 53705, USA. E-mail: bshen@pharmacy.wisc.edu; Fax: +1 608 262-5345; Tel: +1 608 263-2673
bMicrobiology Doctoral Training Program, Division of Pharmaceutical Sciences, School of Pharmacy, University of Wisconsin-Madison, 777 Highland Ave., Madison, Wisconsin 53705, USA
cDepartment of Chemistry, Division of Pharmaceutical Sciences, School of Pharmacy, University of Wisconsin-Madison, 777 Highland Ave., Madison, Wisconsin 53705, USA
dUniversity of Wisconsin National Cooperative Drug Discovery Group, Division of Pharmaceutical Sciences, School of Pharmacy, University of Wisconsin-Madison, 777 Highland Ave., Madison, Wisconsin 53705, USA
First published on 12th November 2008
Abstract
The biosyntheticgene cluster for the glycopeptide-derived antitumor antibiotic zorbamycin (ZBM) was cloned by screening a cosmid library of Streptomyces flavoviridis ATCC 21892. Sequence analysis revealed 40 ORFs belonging to the ZBM biosyntheticgene cluster. However, only 23 and 22 ORFs showed striking similarities to the biosyntheticgene clusters for the bleomycins (BLMs) and tallysomycins (TLMs), respectively; the remaining ORFs do not show significant homology to ORFs from the related BLM and TLM clusters. The ZBM gene cluster consists of 16 nonribosomal peptide synthetase (NRPS) genes encoding eight complete NRPS modules, three incomplete didomain NRPS modules, and eight freestanding single NRPS domains or associated enzymes, a polyketide synthase (PKS) gene encoding one PKS module, six sugarbiosynthesisgenes, as well as genes encoding other biosynthesis and resistance proteins. A genetic system using Escherichia coli–Streptomyces flavoviridis intergeneric conjugation was developed to enable ZBM gene cluster boundary determinations and biosynthetic pathway manipulations.
Introduction
Zorbamycin (ZBM) belongs to the bleomycin (BLM)1–3 family of glycopeptide antitumor antibiotics and also encompasses the closely related BLMs (e.g.BLM A2 and B2), the phleomycins (PLMs, e.g.PLM D1),1,2,4 and the tallysomycins (TLMs, e.g.TLM S10B)5,6 (Fig. 1). The structure of the intact ZBM molecule has only recently been fully established7 and its hybrid peptide–polyketide backbone is thought to be biosynthesized in part by nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs), as has been shown for the BLMs8 and TLMs.9 Under the trade name Blenoxane® the BLMs are currently used clinically in combination with a number of other agents for the treatment of testicular cancer, certain types of lymphoma, and numerous other malignancies.10,11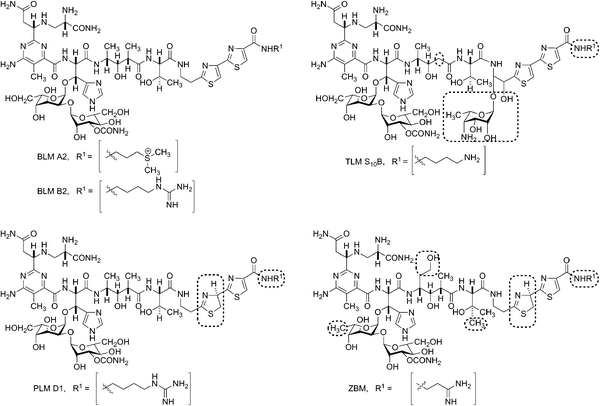 | ||
| Fig. 1 Structures of selected members of the bleomycin (BLM) family of antitumor antibiotics: BLM A2 and B2, tallysomycin (TLM) S10B, phleomycin (PLM) D1, and zorbamycin (ZBM). Structural differences between BLMs and other members of this family are highlighted by boxes. | ||
Nonribosomal peptide synthetases (NRPSs) and modular polyketide synthases (PKSs) are multifunctional proteins catalyzing natural product formation by sequential condensation of amino acids and short carboxylic acids, respectively. They are usually organized into modules consisting of a minimal set of essential domains for chain elongation and optional domains for additional chain modifications. Recently, a growing number of biosyntheticgene clusters was found to violate this unwritten rule comprising freestanding (sometimes partial) modules or isolated domains acting in trans to complement the functionality of the multimodular NRPSs and PKSs.12,13 In the biosynthetic pathway for the nonribosomal peptidesyringomycin, the last in cisNRPS module was found to lack the amino acid activating adenylation (A) domain, and a freestanding adenylation (A)-peptidyl carrier protein (PCP) didomain, amended by a halogenase and an acyltransferase, catalyzes the formation of nonproteinogenic 4-Cl-L-Thr and its subsequent transfer onto the PCP domain of the incomplete NRPS module.14
The study of biosynthetic pathways and generation of new analogs by combinatorial biosynthesis requires fulfillment of three primary criteria: (i) the gene cluster of the target natural product has to be cloned, (ii) the producing strain has to be genetically amenable, and (iii) the natural product has to be produced in quantities sufficient for detection, isolation, and structural elucidation. During our ongoing research on hybrid peptide–polyketidenatural productbiosynthesis, we cloned and characterized the gene clusters for BLMbiosynthesis from Streptomyces verticillus ATCC150038,15–17 and TLM biosynthesis from Streptoalloteichus hindustanus E465-94 ATCC31158,9 unveiling novel hybrid NRPS-PKS machineries for both. Although the yield of BLMs (10–12 mg L−1) and TLMs (18–20 mg L−1) from the wild-type strains are suitable for laboratory scale fermentation, exhaustive efforts to develop an expedient genetic system for the BLM producer S. verticillus17 or the TLM producer S. hindustanus9 have thus far achieved only limited success due to slow growth rates, poor sporulation, inefficient introduction of plasmidDNA into these organisms, and intrinsic low homologous recombination activity in both. The recently reported ZBM overproducer Streptomyces flavoviridis SB9001 represents a useful alternative organism in which to produce new BLM analogs via the application of combinatorial biosynthesis strategies.7
Here we present (i) the cloning and DNA sequence analysis of the ZBM cluster, (ii) boundary determination by gene inactivation and mutant complementation in S. flavoviridis, and (iii) functional assignments of the ZBM gene products, affording the proposed ZBM biosynthetic pathway.
Results
Cloning of a thiazole-forming NRPS module from S. flavoviridis ATCC 21892 by PCR and mapping of the zbmgene cluster
Two cyclization (Cy) domains and one oxidation (Ox) domain were PCR -amplified with degenerate primers from S. flavoviridis ATCC 21892 and one Cy domain was used to identify the first of four overlapping cosmids covering the entire ZBM biosyntheticgene cluster.With the previously described degenerate PCR primers Cy-FP and Cy-RP18 and S. flavoviridis ATCC 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 892 genomic DNA as the template, a distinct band with the expected size of 1.1 kb for Cy domains was readily amplified. Nucleotide sequences obtained from 10 randomly selected clones of the PCR product could be grouped into two populations, pBS9000 (eight clones) and pBS9001 (two clones), with high similarity to the NRPS-1-Cy and NRPS-0-Cy domains, respectively, of BlmIV from the BLMbiosyntheticgene cluster.8 Two sets of PCR primers were designed using the end-sequences of both Cy fragments (Fig. S1). No product was obtained with Cy0-FP and Cy1-RP, but Cy1-FP and Cy0-RP, yielded a 2.5 kb fragment, confirming that the pBS9000 insert was located upstream of the pBS9001 insert. Sequencing of this 2.5 kb fragment (pBS9002) revealed the presence of one A and one PCP domain, flanked by the two Cy domains (Fig. S1† ). One complete thiazole forming NRPS module was PCR amplified as a distinct band with the predicted size of 3.0 kb using the Cy0-FP and the previously described degenerate Ox-RP18 primers with cosmidDNA (pBS9005) as template. DNA sequencing of the cloned fragment (pBS9003) revealed that it encoded one A and one PCP domain flanked, on the upstream end, by the NRPS-0-Cy domain and on the downstream end by an Ox domain with similarity to NRPS-0 of BlmIII (Fig. S1† ).
892 genomic DNA as the template, a distinct band with the expected size of 1.1 kb for Cy domains was readily amplified. Nucleotide sequences obtained from 10 randomly selected clones of the PCR product could be grouped into two populations, pBS9000 (eight clones) and pBS9001 (two clones), with high similarity to the NRPS-1-Cy and NRPS-0-Cy domains, respectively, of BlmIV from the BLMbiosyntheticgene cluster.8 Two sets of PCR primers were designed using the end-sequences of both Cy fragments (Fig. S1). No product was obtained with Cy0-FP and Cy1-RP, but Cy1-FP and Cy0-RP, yielded a 2.5 kb fragment, confirming that the pBS9000 insert was located upstream of the pBS9001 insert. Sequencing of this 2.5 kb fragment (pBS9002) revealed the presence of one A and one PCP domain, flanked by the two Cy domains (Fig. S1† ). One complete thiazole forming NRPS module was PCR amplified as a distinct band with the predicted size of 3.0 kb using the Cy0-FP and the previously described degenerate Ox-RP18 primers with cosmidDNA (pBS9005) as template. DNA sequencing of the cloned fragment (pBS9003) revealed that it encoded one A and one PCP domain flanked, on the upstream end, by the NRPS-0-Cy domain and on the downstream end by an Ox domain with similarity to NRPS-0 of BlmIII (Fig. S1† ).
The NRPS-1-Cy domain was recovered as a 1.1 kb EcoRI fragment from pBS9000 and used as a probe (Fig. 2A, probe 1) to screen an S. flavoviridis genomic library. Of the approximately 8000 colonies screened, 6 positive clones were confirmed by Southern analysis to contain the same fragment as the NRPS-1-Cy probe. Chromosomal walking from this locus was carried out with probe 2 and probe 3 (Fig. 2A), eventually leading to the localization of an 84.8 kb contiguous DNA region covered by overlapping cosmids as represented by pBS9004, pBS9005, pBS9006, and pBS9007 (Fig. 2A).
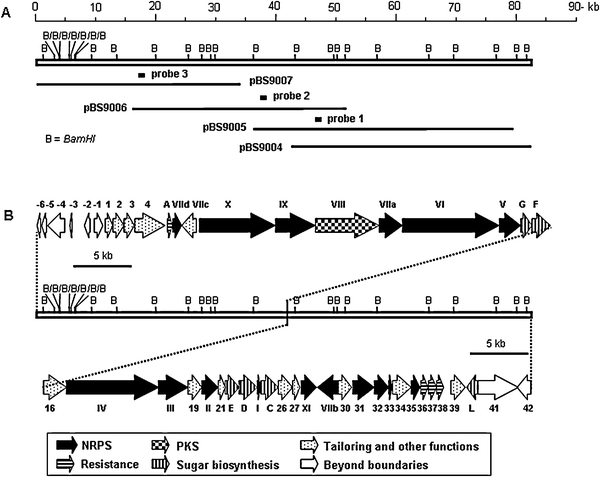 | ||
| Fig. 2 (A) Restriction map of the 84.8 kb DNA region from S. flavoviridis ATCC 21892 as represented by the four overlapping cosmids pBS9007, pBS9006, pBS9005, and pBS9004 and (B) genetic organization of the ZBM biosyntheticgene cluster. Proposed functions for individual ORFs are summarized in Table 1. B, BamHI. | ||
The overall GC content of the sequenced region is 72.2%, characteristic of StreptomycesDNA,19 and bioinformatic analysis revealed 48 open reading frames (ORFs) (Fig. 2B). Functional assignments to individual ORFs as summarized in Table 1 were made by comparing the deduced gene products with proteins of known function in the database and by comparison to the BLM8 and TLM9biosyntheticgene clusters.
| Gene | Length (amino acids) | Proposed functiona | Sequence homologsb | ZBM-BLM Comparisonc | ZBM-TLM Comparisonc |
|---|---|---|---|---|---|
| a Abbrevations for NRPS and PKS domains are: A, adenylation; ACP, acyl carrier protein; AL, acyl CoA ligase; AT, acyltransferase; C and C′, condensation; Cy, cyclization; KR, ketoreductase; KS, ketosynthase; Ox, oxidation; PCP, peptidyl carrier protein . b Protein accession numbers are given in parentheses. c Amino acid comparison of homologs identified from the TLM and BLM clusters as expressed in % identity/%similarity (calculated by AlignX in the Vector NTI Advance™ 10 program from Invitrogen for full length protein alignments). | |||||
| orf(−6)–(−1) | ORFs that are beyond the zbm cluster boundary | ||||
| Predicted upstream boundary of the ZBM gene cluster | |||||
| orf1 | 209 | Hypothetical protein | S. ambofaciens (CAJ88624) | — | — |
| orf2 | 326 | Putative dehydrogenase | S. ambofaciens (CAJ88623) | — | — |
| orf3 | 276 | Putative hydrolase | S. ambofaciens (CAJ88622) | — | — |
| orf4 | 833 | Acylase | S. sp. M664 AhlM (AAT68473) | — | — |
| zbmA | 132 | ZBM binding protein | BlmA (AAB00458)/TlmA (ABL74956) | 49/60 | 55/67 |
| zbmVIId | 269 | Acyltransferase | S. tubercidicus, TeLB (AAT45287) | — | — |
| zbmVIIc | 416 | Cytochrome P450 | S. tubercidicus, CypLB (AAT45286) | — | — |
| zbmX | 2131 | NRPS (C/A/PCP/C/A/PCP) | BlmX (AAG02355) (NRPS9-8)/TlmX (ABL74936) | 51/61 | 50/62 |
| zbmIX | 1105 | NRPS (C/A/PCP) | BlmIX (AAG02356) (NRPS7)/TlmIX (ABL74937) | 44/55 | 43/53 |
| zbmVIII | 1811 | PKS (KS/AT/MT/KR/ACP) | BlmVIII (AAG02357) (PKS)/TlmVIII (ABL74938) | 42/53 | 37/47 |
| zbmVIIa | 643 | NRPS (C/PCP) | BlmVII (AAG02358) (NRPS6)/TlmVII (ABL74939) | 19/24 | 20/25 |
| zbmVI | 2714 | NRPS (AL/ACP/C/A/PCP/C/A) | BlmVI (AAG02359) (NRPS5-4-3)/TlmVI (ABL74940) | 49/59 | 48/58 |
| zbmV | 597 | NRPS (PCP/C’) | BlmV (AAG02360) (NRPS3CT)/TlmV (ABL74941) | 35/47 | 40/52 |
| zbmG | 320 | NAD-dependent sugar epimerase | BlmG (AAG02361)/TlmG (ABL74942) | 64/71 | 63/71 |
| zbmF | 518 | Glycosyl transferase and hydroxylase | BlmF (AAG02362)/TlmF (ABL74943) | 46/55 | 39/49 |
| orf16 | 637 | Gln-dependent amino transferase | Blm-Orf18 (AAG02363)/Tlm-Orf21 (ABL74944) | 52/63 | 51/63 |
| zbmIV | 2639 | NRPS (C/A/PCP/Cy/A/PCP/Cy) | BlmIV (AAG02364) (NRPS2-1)/TlmIV (ABL74945) | 47/57 | 50/61 |
| zbmIII | 847 | NRPS (A/PCP/Ox) | BlmIII (AAG02365) (NRPS0)/TlmIII (ABL74946) | 38/50 | 41/54 |
| orf19 | 381 | Oxygenase | Blm-Orf15 (AAG02366)/Tlm-R3 (ABL74947) | 49/57 | 47/58 |
| zbmII | 470 | NRPS (C) | BlmII (AAG02367)/TlmII (ABL74948) | 31/39 | 28/39 |
| orf21 | 203 | Unknown | Blm-Orf13 (AAG02368)/Tlm-Orf16 (ABL74949) | 51/60 | 53/65 |
| zbmE | 386 | Glycosyl transferase | BlmE (AAG02369)/TlmE (ABL74950) | 53/62 | 52/63 |
| zbmD | 542 | Carbamoyltransferase | BlmD (AAG02370)/TlmD (ABL74951) | 67/77 | 65/76 |
| zbmI | 88 | Type II PCP | BlmI (AAD42077)/TlmI (ABL74952) | 43/56 | 44/61 |
| zbmC | 497 | NTP-sugar synthase | BlmC (AAG02371)/TlmC (ABL74953) | 52/63 | 50/63 |
| orf26 | 424 | SAM-dependent oxidase or methyl transferase | Blm-Orf8 (AAG02372)/Tlm-Orf11 (ABL74954) | 67/76 | 65/75 |
| orf27 | 206 | Ankyrin-like protein | Blm-Orf3 (AAB00459)/Tlm-Orf8 (ABL74957) | 36/46 | 40/51 |
| zbmXI | 449 | NRPS (C) | BlmXI (AAG02354) | 11/16 | — |
| zbmVIIb | 591 | NRPS (A/PCP) | Salinispora tropica (CP000667) | — | — |
| orf30 | 314 | α-Ketoglutarate-dependent hydroxylase | Blm-Orf1 (AAB00457)/Tlm-Orf10 (ABL74955) | 59/68 | 57/68 |
| orf31 | 609 | NRPS (C) | Bacillus licheniformis, BA2 (BAA36755) | — | — |
| orf32 | 423 | Type II PCP | Brevibacillus parabrevis, TycC (O30409) | — | — |
| orf33 | 88 | Type II PCP | Stigmatella aurantiaca, MxcG (AAG31130) | — | — |
| orf34 | 532 | Acyl-CoA synthetase | Rhizobium etli (YP_472634) | — | — |
| orf35 | 259 | NRPS (TE) | Melittangium lichenicola, MelG (CAD89778) | — | — |
| orf36 | 202 | ABC transporter | Dinoroseobacter shibae DFL 12 (ZP_01587238) | — | — |
| orf37 | 226 | ABC transporter | Saccharopolyspora erythraea (YP_001103442) | — | — |
| orf38 | 199 | ABC transporter | Saccharopolyspora erythraea (YP_001103441) | — | — |
| orf39 | 398 | Biotin synthase | S. coelicolor (NP_625532) | — | — |
| zbmL | 337 | GDP-mannose-4,6-dehydratase | S. hygroscopicus, Hyg5 (ABC42542) | — | — |
| Downstream boundary of the ZBM gene cluster | |||||
| orf41–42 (partial) | ORFs that are beyond the zbm cluster boundary | ||||
Confirmation of the identity of the zbmbiosynthetic locus by gene replacement
Establishment of a genetic system for S. flavoviridis ATCC21892 and its recently described improved derivative S. flavoviridis SB90017 allowed the generation of a zbmXgene replacement mutant that, by its ZBM non-producing phenotype , confirmed the identity of the ZBM biosynthetic locus.Standard procedures for protoplast transformation and intergeneric conjugation were applied,19 and for plasmid introduction as well as homologous recombination both methods resulted in frequencies comparable to those obtained for Streptomyces lividans under similar conditions.19,20 A slightly modified version of the literature protocol was chosen for conjugations between E. coli S17-1 and S. flavoviridis SB9001. The pSET151 based plasmid pBS9013 (Fig. 3A) was introduced into wild-type S. flavoviridis SB9001 and a double crossover mutant, SB9002, selected (Fig. 3B). This mutant strain completely lost its ability to produce ZBM (Fig. 3C, trace II), confirming that the NRPS encoding zbmX is required for ZBM biosynthesis.
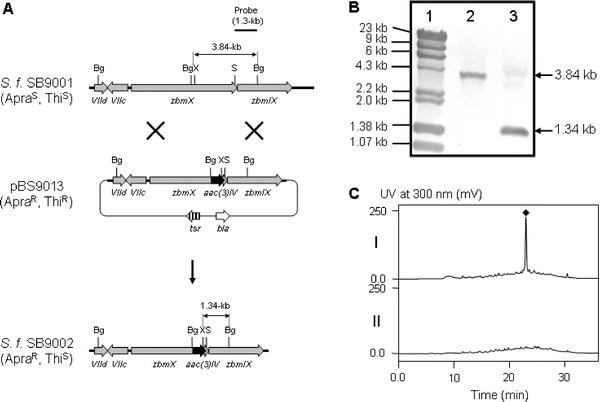 | ||
| Fig. 3 Inactivation of zbmX by gene replacement. (A) Construction of the zbmXgene replacement mutant and restriction map of S. flavoviridis SB9001 wild-type and SB9002 mutant strains showing fragment sizes upon BglII-XhoI digestion. Bg, BglII; S, StuI; X, XhoI; ApraR, apramycin resistant; ApraS, apramycin sensitive; ThiR, thiostrepton resistant; ThiS, thiostrepton sensitive. (B) Southern analysis of SB9001 (lane 2) and SB9002 (lane 3) genomic DNA digested with BglII and XhoI using a 1.3-kb BglII-StuI fragment as a probe. Lane 1, molecular weight marker. (C) HPLC analysis of ZBM (◆) production in wild-type SB9001 (I) and recombinant strain SB9002 (II). | ||
Determination of the zbmgene cluster boundaries by systematic gene inactivations
Through inactivation of selected genes (zbm-orf(−1), zbm-orf2, zbm-orf4, zbmVIId, zbmL, zbm-orf41) residing at distal ends of the sequenced region (Fig. 2B and Table 1), the upstream and downstream zbm cluster boundaries were assigned to reside between zbm-orf(−1) and zbm-orf1 and between zbmL and zbm-orf41, respectively.At the upstream end of the cluster, gene replacements of zbm-orf2, zbm-orf4, and zbmVIId encoding proteins with similarities to dihydroorotate dehydrogenases, aculeacin acylases, and type II thioesterases, respectively, yielded the corresponding mutant strains SB9008, SB9006, and SB9005, respectively, that completely lost the ability to produce ZBM (Fig. 4, traces IV, V and VI). Inactivation of the further upstream zbm-orf(−1) gene encoding another protein with similarity to type II thioesterases afforded the SB9007 mutant strain that showed greatly reduced ZBM production (Fig. 4, trace II). However, ZBM production by the complemented strain SB9010, failed to exceed that of the parent strain SB9007 (Fig. 4, traces III and II). These data revealed the involvement of Zbm-Orf2, Zbm-Orf4, and ZbmVIId in ZBM biosynthesis and suggested that the zbm-orf(−1) gene product is dispensible for ZBM production.
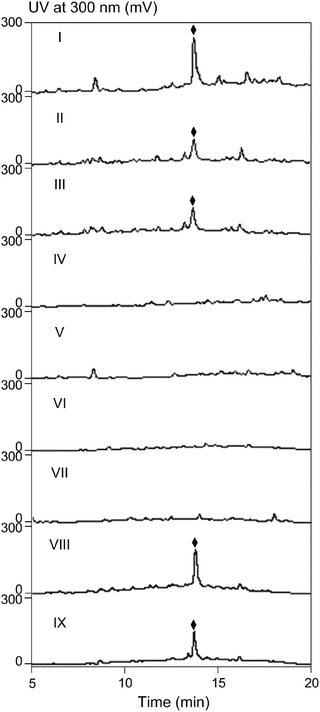 | ||
| Fig. 4 HPLC analysis of ZBM (◆) production in wild-type SB9001 (I), recombinant strains SB9007 (II), SB9008 (IV), SB9006 (V), SB9005 (VI), SB9003 (VII), SB9004 (IX), and complemented strains SB9010 (III) and SB9009 (VIII). | ||
At the downstream boundary, zbmL encodes a putative GDP mannose-4, 6-dehydratase and was thought to be the last gene within the zbm cluster. No function in ZBM biosynthesis was apparent for the adjacent ORF, zbm-orf41, encoding an NRPS harboring one condensation (C) and one adenylation (A) domain. Inactivation of zbm-orf41 had a marginal effect on ZBM production (∼60% of the wild-type SB9001 strain; Fig. 4, traces IX and I) whereas the zbmL deficient mutant strain showed complete abolishment of ZBM production (Fig. 4, trace VII). Introduction of the integrative zbmL complementation construct, pBS9019, into the zbmL deficient SB9003 mutant strain restored ZBM production to ∼70% of previous production levels (Fig. 4, trace VIII).
Sequence analysis and organization of the zbmbiosyntheticgene cluster
The deduced functions of the sequenced region (Table 1), together with the experiments described above, show that the zbmgene cluster spans 74.5-kb of DNA, encompassing 40 ORFs from zbm-orf1 to zbmL (Fig. 2B). Among them, 16 are biosyntheticgenes encoding NRPSs and associated enzymes (zbmVIId, zbmX, zbmIX, zbmVIIa, zbmVI, zbmV, zbmIV, zbmIII, zbmII, zbmI, zbmXI, zbmVIIb, zbm-orf31, zbm-orf32, zbm-orf33, and zbm-orf35), one gene encodes a polyketide synthase (PKS) (zbmVIII), six deduced gene products are similar to sugarbiosynthesis enzymes (zbmC, zbmD, zbmE, zbmF, zbmG, and zbmL), and five genes encode other tailoring enzymes (zbmVIIc, zbm-orf16, 19, 26, and 30). Four genes (zbmA, zbm-orf36, 37, and 38) are thought to confer self-resistance to the ZBM producing organism and the remaining eight ORFs (zbm-orf1, 2, 3, 4, 21, 27, 34, and 39) are genes whose function could not be predicted by sequence comparison alone.All of the ORFs belonging to the zbmgene cluster except for three (zbmVIIc, zbmVIIb, and zbmL) are transcribed in the same direction. The 40 ORFs are thought to be organized into 11 transcriptional units with as many as 20 ORFs forming a single 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 444 bp operon. Possible promoter regions might be located in front of zbm-orf1, zbm-orf4, zbmA, zbmVIIc, zbmX, zbmXI, zbmVIIb, zbm-orf30, zbm-orf31, zbm-orf39, and zbmL. Direct homologs for 22 ORFs were found in the BLM and TLM biosyntheticgene clusters, for one additional ORF (zbmXI), the corresponding homologous gene was found in the BLM cluster only.
444 bp operon. Possible promoter regions might be located in front of zbm-orf1, zbm-orf4, zbmA, zbmVIIc, zbmX, zbmXI, zbmVIIb, zbm-orf30, zbm-orf31, zbm-orf39, and zbmL. Direct homologs for 22 ORFs were found in the BLM and TLM biosyntheticgene clusters, for one additional ORF (zbmXI), the corresponding homologous gene was found in the BLM cluster only.
The zbmNRPS-PKS megasynthase genes
The zbmX, zbmIX, zbmVIII, zbmVIIa, zbmVI, zbmV, zbmIV, and zbmIIIgenes are organized in the same order and direction as in the BLM and TLM clusters and the deduced gene products, which are thought to constitute the ZBM hybrid NRPS-PKS megasynthase, show high similarities to their respective BLM (19–51% identity, 24–61% similarity) and TLM (20–50% identity, 25–62% similarity) homologs.
DNA and the deduced amino acid sequence analyses of the zbmVIgene suggested that it is not transcribed from one of the typically observed ATG or GTG start codons,19 but from an uncommon CTG start codon. Three possible start codons (a CTG at bp 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 650, an ATG at bp 30
650, an ATG at bp 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 818, and a GTG at bp 30
818, and a GTG at bp 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 821) for the respective protein coding region were found. For all three gene products (2714 aa, 2658 aa, and 2657 aa) BLAST analyses returned acceptable alignments with BlmVI (2675 aa) and TlmVI (2742 aa), but the results for the longest ORF seemed most reasonable, especially since the predicted CTG start codon was the only one preceded by an apparent ribosomal binding site.
821) for the respective protein coding region were found. For all three gene products (2714 aa, 2658 aa, and 2657 aa) BLAST analyses returned acceptable alignments with BlmVI (2675 aa) and TlmVI (2742 aa), but the results for the longest ORF seemed most reasonable, especially since the predicted CTG start codon was the only one preceded by an apparent ribosomal binding site.
Based on the specificity conferring code21,22 extracted by the online analysis tool NRPS predictor,23 the A domains of ZbmVI (NRPS-4), ZbmVI (NRPS-3), ZbmX (NRPS-9), ZbmX (NRPS-8), ZbmIV (NRPS-2), and ZbmIV (NRPS-1) are predicted to activate L-Ser, L-Asn, L-Asn, L-His, β-Ala, and L-Cys, respectively (Table 2 and Fig. 5A), which is in perfect agreement with the chemical structure of ZBM and the predictions made for BLM8 and TLM9biosynthesis. The ZbmIII (NRPS-0) A domain is proposed to be inactive since the essential aspartate (D235) and lysine (K517) moieties are replaced by different amino acids; similar amino acid substitutions have also been reported for the BlmIII (NRPS-0) domain, which has been confirmed biochemically to be non-functional,24 and the homologous TlmIII (NRPS-0) A domain.9 The ten specificity conferring amino acids of the ZbmIX (NRPS-7) A domain resemble the only known D-lysergic acid activating A domain contained in the ergopeptine NRPS, ps2, from Claviceps purpurea.25 The NRPS-6 module encoded by zbmVIIa was found to be incomplete since ZbmVIIa (643 aa) is considerably shorter than a full C-A-PCP NRPS module (∼1000 aa), and only a C and PCP domain were detected. The zbmVIIIgene encodes a PKS module consisting of a ketoacyl synthase (KS), an acyltransferase (AT), a methyltransferase (MT), a ketoreductase (KR), and an acyl carrier protein (ACP) domain. The exact same domain organization is also found in BlmVIII and thought to be responsible for the formation of the methylated polyketide unit in the BLM structure.8
![(A) A linear model for the ZBM hybrid NRPS-PKS templated assembly of the ZBM aglycone from nine amino acids and one acetate. Abbrevations for NRPS and PKS domains are: A, adenylation; ACP, acyl carrier protein; AL, acyl CoA ligase; AT, acyltransferase; C and C’, condensation; Cy, cyclization; KR, ketoreductase; KS, ketosynthase; MT, methyltransferase; Ox, oxidation; PCP, peptidyl carrier protein. (B) Proposed pathway for ZBM biosynthesis. [?] indicates a step whose enzyme activity could not be identified within the sequenced ZBM cluster. While all intermediates for ZBM biosynthesis are hypothetical, the analogous compounds, except the ones in brackets, have been identified in BLMbiosynthesis from S. verticillus fermentation as the corresponding free acids.51](/image/article/2009/MB/b814075h/b814075h-f5.gif) | ||
| Fig. 5 (A) A linear model for the ZBM hybrid NRPS-PKS templated assembly of the ZBM aglycone from nine amino acids and one acetate. Abbrevations for NRPS and PKS domains are: A, adenylation; ACP, acyl carrier protein; AL, acyl CoA ligase; AT, acyltransferase; C and C’, condensation; Cy, cyclization; KR, ketoreductase; KS, ketosynthase; MT, methyltransferase; Ox, oxidation; PCP, peptidyl carrier protein . (B) Proposed pathway for ZBM biosynthesis. [?] indicates a step whose enzyme activity could not be identified within the sequenced ZBM cluster. While all intermediates for ZBM biosynthesis are hypothetical, the analogous compounds, except the ones in brackets, have been identified in BLMbiosynthesis from S. verticillus fermentation as the corresponding free acids.51 | ||
| Domain | 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | 517 | Similarity (%)a |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a Similarity is calculated by AlignX in the Vector NTI Advance™ 10 program from Invitrogen. b X indicates a variable amino acid within the determined code. In similarity calculations, X is not recognized as an arbitrary amino acid, hence similarity values appear to be lower than calculated manually (100% for ZbmIV (NRPS2) compared to the β-Ala code; 100% for ZbmX (NRPS8) compared to the L-His code). | |||||||||||
| L-Cys(2) | D | L | Y | N | L | S | L | I | W | K | |
| ZbmIII (NRPS0) | E | R | Y | S | A | S | L | I | W | R | 70 |
| ZbmIV (NRPS1) | D | L | Y | N | L | S | L | I | W | K | 100 |
| β-Ala | V | D | Xb | V | I | S | Xb | G | D | K | |
| ZbmIV (NRPS2) | V | D | A | L | V | S | L | A | D | K | 80 |
| L-Asn | D | L | T | K | L | G | E | V | G | K | |
| ZbmVI (NRPS3) | D | L | T | K | V | G | E | V | G | K | 100 |
| ZbmX (NRPS9) | D | F | T | K | V | G | E | V | G | K | 90 |
| L-Ser | D | V | W | H | L | S | L | I | D | K | |
| ZbmVI (NRPS4) | D | V | W | H | L | S | L | I | D | K | 100 |
| L-Val (1) | D | A | F | W | I | G | G | T | F | K | |
| ZbmVIIb (NRPS6b) | D | A | F | W | L | G | G | T | F | K | 100 |
| D-Lyserg | D | V | F | S | V | G | L | Y | M | K | |
| ZbmIX (NRPS7) | D | V | F | S | N | G | L | T | H | K | 70 |
| ps2 (Q8J0L6, D-Lyserg) | D | V | F | S | V | G | L | Y | M | K | 100 |
| FUSS A (AAT28740, L-Homoser) | D | M | T | F | S | A | G | I | I | K | 60 |
| L-His | D | S | Xb | L | Xb | A | E | V | Xb | K | |
| ZbmX (NRPS8) | D | S | V | L | T | A | E | V | W | K | 70 |
Genes encoding other NRPS and associated enzymes
Several genes located up- and downstream of the ZBM megasynthase genes encode fragments of typical NRPS modules such as an A-PCP didomain (zbmVIIb), freestanding PCP (zbmI, zbm-orf32, zbm-orf33), C (zbmII, zbmXI, zbm-orf31), and thioesterase (TE) (zbm-orf35) domains as well as an NRPS associated type II thioesterase (zbmVIId).The zbmVIIbgene product is similar to other A-PCP didomains in the database, e.g. from Salinispora tropica (accession number CP000667; 39% identity and 52% similarity) and the A domain of ZbmVIIb is predicted to activate L-valine. The zbmIgene encodes a discrete protein homologous to individual PCP domains of modular NRPSs, e.g. to blmI (43% identity and 56% similarity) and tlmI (44% identity and 61% similarity). BlmI has been characterized as a type II PCP,26 but its role in BLMbiosynthesis remains unknown. The gene product of zbm-orf33 shows homology to PCP domains of large multimodular NRPSs, especially to MxcG from Stigmatella aurantiaca (accession number AAG31130; 39% identity and 57% similarity), its size of only 88 aa, however, indicates that is belongs to the aforementioned family of type II PCPs. The zbm-orf32gene encodes a 423 aa protein showing similarity to several A (25–27% identity and 37–40% similarity) and PCP (28–33% identity and 44–51% similarity) domains of TycC (accession number O30409) from Brevibacillus parabrevis. The A domain is considered inactive due to the lack of all but one (A1) of the commonly found ten core motifs.21,22 The C terminal ∼100 aa of Zbm-Orf32, however, comprise an intact PCP domain, which may be involved in ZBM biosynthesis.
The zbmII, zbmXI, and zbm-orf31genes encode discrete C domains of type I NRPSs. Direct homologs of zbmII were found in both the BLM (blmII, 31% identity and 39% similarity) and TLM (tlmII, 28% identity and 39% similarity) biosyntheticgene clusters. BlmII and its homolog TlmII are thought to catalyze the coupling reaction between the respective NRPS-bound full length peptide intermediate and the terminal amines,8,9 and a similar function is proposed for ZbmII (Fig. 5A). The respective homolog of zbmXI was only detected in the BLM cluster (blmXI, 11% identity and 16% similarity), but ZbmXI (449 aa) was found to be considerably shorter than BlmXI (688 aa); bioinformatic analyses of both proteins does not allow any predictions whether they are required for biosynthesis of the respective compound. For zbm-orf31 no corresponding gene could be found in the related clusters. The 609 aa Zbm-Orf31 protein contains a C domain in the ∼300 aa N terminal region showing similarity to the bacitracin synthetase 2 from Bacillus licheniformis (accession number BAA36755; 21% identity and 36% similarity; C domain alignment only). So far, no function in ZBM biosynthesis could be assigned to Zbm-Orf31.
The zbmVIIdgene product is highly homologous to known type II TEs, such as LnmN from Streptomyces atroolivaceus (accession number AAN85527; 30% identity and 43% similarity). Zbm-Orf35 (259 aa) in contrast resembles TE domains located at the C terminal end of many NRPS assembly lines, like the MelG TE domain from Melittangium lichenicola (accession number CAD89778; 21% identity and 33% similarity; TE domain only). TE domains usually control the release of the final peptide product by cleaving the thioester bond between the PCP domain of the last NRPS module and the full length peptide generating either a linear or a cyclic peptide product.27 Since no such genes for peptide release were detected in the BLM and TLM clusters, the function of Zbm-Orf35 in ZBM biosynthesis is unclear.
Sugar biosynthesis genes
Six genes, zbmC, zbmD, zbmE, zbmF, zbmG, and zbmL, encode proteins that are thought to be responsible for 6-deoxy-L-gulose-3-O-carbamoyl-D-mannose biosynthesis and attachment.ZbmC, ZbmD, ZbmE, ZbmF, and ZbmG exhibit significant similarity on amino acid level to their respective BLM (46–67% identity and 55–76% similarity) and TLM (39–65% identity and 49–77% similarity) counterparts. They are expected to catalyze the corresponding reactions in the formation of the 6-deoxy-L-gulose-3-O-carbamoyl-D-mannose disaccharide and its attachment to the ZBM aglycone as proposed for the L-gulose-3-O-carbamoyl-D-mannose disaccharide in the BLM8 and TLM9biosynthetic pathways (Fig. 5B). The gene product of zbmL exhibits high similarity to GDP-mannose 4,6-dehydratases such as Hyg5 from Streptomyces hygroscopicus (accession number ABC42542; 59% identity and 69% similarity) reported to be involved in hygromycinbiosynthesis.28
Genes encoding tailoring enzymes or resistance proteins
The ZbmVIIc, Zbm-Orf19, Zbm-Orf30, Zbm-Orf16, and Zbm-Orf26 proteins are proposed to carry out hydroxylation, transamination, and methylation reactions on various ZBM intermediates, while the zbmA, zbm-orf37, and zbm-orf38gene products are thought to confer self-resistance to the producing organism by drug binding and transport mechanisms.The deduced gene product of zbmVIIc shows significant homology to known cytochrome P-450 hydroxylases, such as CypLB from Streptomyces tubercidicus (accession number AAT45286; 43% identity and 56% similarity) and Ecm12 from Streptomyces lasaliensis (accession number BAE98161; 34% identity and 49% similarity). The deduced gene product of zbm-orf19 shows significant similarity to Blm-Orf15 (49% identity and 57% similarity), TlmR3 (47% identity and 58% similarity) and to α-ketoglutarate-dependent oxygenases. The zbm-orf30gene product and its homologs, Blm-Orf1 (59% identity and 68% similarity), Tlm-Orf10 (57% identity and 68% similarity), and TlmH (40% identity and 53% similarity) also resemble α-ketoglutarate-dependent oxygenases that were recently shown to generate β-hydroxylated amino acids.29 Blm-Orf18,30 as well as Tlm-Orf109 and TlmH9 have been proposed to catalyze such β-hydroxylation reactions in their respective pathways and Zbm-Orf19 or Zbm-Orf30 are therefore thought to carry out the corresponding His hydroxylation in ZBM biosynthesis (Fig. 5B). The zbm-orf16gene encodes a putative glutamine-dependent transaminase highly similar to Blm-Orf18 (52% identity and 63% similarity) and Tlm-Orf21 (51% identity and 63% similarity), which were previously proposed to be involved in biosynthesis of the pyrimidoblamic acid moiety8,9 (Fig. 5B). Zbm-Orf26 is closely related to Blm-Orf8 (67% identity and 76% similarity) and Tlm-Orf11 (65% identity and 75% similarity), belonging to the superfamily of radical SAM dependent enzymes. In analogy to Blm-Orf88 and Tlm-Orf11,9 Zbm-Orf26 is suggested to carry out methylation or oxidation reactions in the ZBM biosynthetic pathway (Fig. 5B).
The zbmAgene encodes a protein with high homology to BlmA (49% identity and 60% similarity) and TlmA (55% identity and 67% similarity).8,9 BlmA and TlmA have long been known to confer resistance to the BLM family of antibiotics by drug sequestering,30,31 and ZbmA is expected to fulfil the same function in the ZBM producer. Zbm-Orf36, Zbm-Orf37 and Zbm-Orf38, belong to the family of ATP dependent transporters. Zbm-Orf36 is related to BioY from Dinoroseobacter shibae (accession number ZP_01587238; 45% identity and 66% similarity), Zbm-Orf37 closely resembles a transport protein from Saccharopolyspora erythraea (accession number YP_001103442; 58% identity and 70% similarity), and Zbm-Orf38 shows significant similarity to a cobalt transport system permease protein from Saccharopolyspora erythraea (accession number YP_001103441; 36% identity and 52% similarity). Drug transport is a common resistance mechanism found in antibiotic producing microorganisms,32 and Zbm-Orf36, Zbm-Orf37 and Zbm-Orf38 may constitute a transport system that provides some degree of ZBM resistance to S. flavoviridis by transporting the drug out of the cells.
Other genes
The remaining eight ORFs, zbm-orf1, zbm-orf2, zbm-orf3, zbm-orf4, zbm-orf21, zbm-orf27, zbm-orf34, and zbm-orf39, encode proteins whose function cannot be predicted by sequence comparison alone.The zbm-orf1gene product exhibits similarity to a conserved hypothetical protein from Streptomyces ambofaciens (accession number CAJ88624; 43% identity and 55% similarity). The zbm-orf2gene encodes a protein highly homologous to dihydroorotate dehydrogenases from Streptomyces ambofaciens (accession number CAJ88623; 54% identity and 65% similarity) and from various other microorganisms. The zbm-orf3 product is related to creatinine amidohydrolases from Streptomyces ambofaciens (accession number CAJ88622; 52% identity and 66% similarity) and other microorganisms. Zbm-Orf4 shows significant similarity to aculeacin and other acylases, e.g. from Sigmatella aurantiaca (accession number ZP_01463795; 41% identity and 55% similarity). The gene product of zbm-orf21, like the product of blm-orf13 (51% identity and 60% similarity) and tlm-orf16 (53% identity and 65% similarity), shows similarity to MbtH, a protein of unknown function whose homologs are found in many antibioticbiosyntheticgene clusters, especially the ones encoding NRPSs,33,34 The precise role of MbtH and its homologs in antibiotic biosynthesis, however, remains unkown. The zbm-orf27gene, is proposed to start with a highly unusual CTG start codon and its 206 aa gene product was found to return reasonable BLAST alignments with its homologs from the BLM and TLM biosyntheticgene clusters. Zbm-Orf27, like Blm-Orf3 (36% identity and 46% similarity) and Tlm-Orf8 (40% identity and 51% similarity), resembles ankyrin-like proteins which are thought to play a role in protein-protein interaction.35 The zbm-orf34gene product shows considerable similarity to individual acyl-CoA synthetases, e.g. from Rhizobium etli (accession number YP_472634; 34% identity and 52% similarity), and to acyl-CoA ligase domains of type I PKSs, such as HbmAI from Streptomyces hygroscopicus (accession number AAY28225; 23% identity and 35% similarity, domain alignment only). Zbm-Orf39 shows very high sequence similarity to biotin synthases, e.g. from Streptomyces coelicolor (accession number NP_625532; 89% identity and 91% similarity) and its function in ZBM biosynthesis is not clear.
Discussion
The presence of a thiazolinyl-thiazole moiety in the molecular structure of ZBM inspired us to screen the S. flavoviridis ATCC 21892 chromosome for Cy and Ox domains,18 resulting in the identification of the ZBM biosynthetic locus. The incorporation and cyclization of the amino acidcysteine, leading to the formation of thiazoline moieties in nonribosomal peptides, is typically catalyzed by Cy domains.36 Conversion of these thiazolinyl into thiazole moieties by dehydration is then carried out by Ox domains generally located in the same NRPS module as the respective Cy domain.36,37 As expected from the molecular characteristics of ZBM and in analogy to both the BLM8 and TLM9biosyntheticgene clusters, two Cy domains, presumably responsible for the incorporation of two cysteine moieties, were amplified from S. flavoviridis genomic DNA. An entire NRPS module comprising both a Cy and Ox domain was next identified from the ZBM producer. The Ox domain contained in this module was later found to be the only Ox domain present in the entire ZBM biosyntheticgene cluster, which is consistent with the presence of a thiazolinyl-thiazole moiety in ZBM (Fig. 1 and Fig. 5B).Establishment of a genetic system for the ZBM producer S. flavoviridis ATCC21892 allowed for multiple gene knockout experiments to confirm the identity of the cloned biosynthetic locus and to experimentally define the ZBM cluster boundaries. Although the biosyntheticgene clusters of BLM and TLM have been known for some time, thorough exploitation of their biosynthetic pathways has thus far been hampered by the severely limited amenability of S. verticillus ATCC15003 and S. hindustanus E465-94 ATCC31158, respectively, to genetic manipulation.8,9,17 In contrast, the ZBM producer S. flavoviridis proved easily accessible by intergeneric conjugation or protoplast transformation and could therefore constitute a promising host for the engineered production of novel BLM related antitumor antibiotics in the future. Seven gene inactivation experiments illustrated the involvement of zbmX, zbm-orf2, zbm-orf4, zbmVIId, and zbmL in ZBM biosynthesis and expanded the predicted cluster boundaries by an additional ∼5 kb.
The ZBM cluster harbours several novel features to distinguish the ZBM from the related BLM and TLM biosynthetic pathways, e.g. the GDP-mannose-4,6-dehydratase encoding zbmL, the supposedly L-homoserine activating A domain of ZbmIX, and the zbmVIIa, VIIb, VIIc, and VIIdgenes presumably involved in L-OH-valine biosynthesis and incorporation. The additional sugarbiosynthesisgene, zbmL, at the downstream end of the ZBM cluster encodes a GDP-mannose-4,6-dehydratase. Only one extra enzymatic step, the dehydration of NDP-D-mannose to NDP-4-keto-6-deoxy-D-mannose, is expected to distinguish between the pathways for NDP-6-deoxy-L-gulose formation in ZBM (Fig. 5B) and NDP-L-gulose formation in BLM and TLM biosynthesis. The enzymatic function of a GDP-mannose-4,6-dehydratase fits perfectly into this model of ZBM biosynthesis. Furthermore, these results support the hypothesis that the disaccharide for ZBM is biosynthesized from D-mannose-phosphate, possibly using GDP as the nucleotide, which was also proposed for BLM and TLM biosynthesis.
The NRPS-7 A domain in ZbmIX deviates from the L-alanine specificity conferring code found in BlmIX8 and TlmIX.9 Based on a single database entry only,23 it is predicted to activate D-lysergic acid (Table 2), which is inconsistent with the chemical structure of ZBM that contains L-homoserine at the respective position in the peptide backbone.7 Only one L-homoserine activating A domain, FUSS A from the fungus Fusarium moniliforme, has been reported in literature so far,38 however, its specificity conferring region showed only 40% similarity with the corresponding region of ZBM NRPS-7. As reported for Streptomyces sp. NRRL 5331,39 the non-proteinogenic amino acidL-homoserine may be supplied by the L-threoninebiosynthetic pathway of S. flavoviridis.
The current paradigm for hybrid peptide–polyketide biosynthesis would predict the NRPS-6 module to have a C-A-PCP domain organization with the A domain loading either L-OH-valine or its precursor L-valine. Sequence analysis of the ZBM gene cluster, however, revealed that the ZBM NRPS-6 module completely lacks the expected L-OH-valine or L-valine activating A domain, exhibiting a C-PCP domain organization. Instead, we identified a gene (zbmVIIb) downstream of the ZBM megasynthase encoding a discrete NRPS A-PCP didomain protein with predicted L-valine specificity that might substitute in trans for the missing part of the NRPS-6 module (Table 2). After priming of the ZbmVIIb PCP domain, ZbmVIIc most likely hydroxylates the PCP bound L-valine to afford the non-proteinogenic amino acid β-hydroxy-L-valine (Fig. 5B), as has been outlined for L-tryptophane hydroxylation by Ecm12 in echinomycinbiosynthesis.40 This enzymatic modification could be performed (i) at the stage of the free amino acid, (ii) while L-valine is tethered to a PCP domain, or (iii) after the peptide-polyketide backbone is completed. Although examples for all three mechanisms have been reported,8,9,41,42 the involvement of aminoacyl-S-enzyme intermediates in β-hydroxylations of amino acids seems to be by far the most common.42 The β-hydroxy-L-valine then needs to be transferred onto the NRPS-6 PCP domain of the ZBM megasynthase complex. Two scenarios can be envisioned for this transfer reaction, and examples for both have been published in the current literature. In a mechanistic analogy to SyrB1, SyrB2 and SyrE of the syringomycinbiosynthetic pathway,14L-valine may be loaded onto the PCP domain of ZbmVIIb, hydroxylated by ZbmVIIc, and subsequently transferred onto the NRPS-6 PCP domain (ZbmVIIa) by the predicted type II thioesterase ZbmVIId (Fig. 5A and S3A). The alternative route reflects the reaction sequence in coronamic acidbiosynthesis43 catalyzed by CmaA, CmaE, CmaD, and CmaB, with L-valine being loaded onto the PCP domain of ZbmVIIb, transferred onto either one of the discrete PCP proteins, Zbm-Orf32 or Zbm-Orf33, by one of the two predicted thioesterases ZbmVIId or Zbm-Orf35, hydroxylated by ZbmVIIc, and finally transferred onto the NRPS-6 PCP domain (ZbmVIIa) by ZbmVIId or Zbm-Orf35 (Fig. S3B). BarC, another potential acyltransferase from the barbamidebiosynthetic pathway was proposed to either directly transfer trichloroleucine from BarA onto the PCP domain of BarE or to release trichloroleucine by a thioesterase-like mechanism.44 The active-site cysteine of SyrC (C224)14 and CmaE (C105)43 have been shown to be directly involved in aminoacyltransfer. Amino acid alignments of the critical active site residues of all three enzymes, SyrC,14 CmaE,43 and BarC,44 with the corresponding residues of ZbmVIId, and Zbm-Orf35 clearly split into two groups: SyrC (C224), CmaE (C105), and ZbmVIId (C90) comprise a cysteine residue in the active site, while BarC (S95) and Zbm-Orf35 (S87), like ordinary thioesterases, contain a serine residue at the corresponding positions (Fig. S3C). These facts suggest that ZbmVIId is the better candidate for the amino acid shuttling reaction and that the syringomycin model requiring only one such transfer reaction is more likely to be true.
With more and more isolated NRPS or PKS domains, incomplete or freestanding modules and shuttling acyltransferases showing up in literature,12,13 a new view on their utilization for combinatorial biosynthesis in the especially difficult to manipulate modular NRPS and PKSbiosynthetic pathways of small molecules arises. Although many of these freestanding domains and partial modules simply seem to be superfluous, quite a few were shown to be essential for natural productbiosynthesis14,43–45 and some of the corresponding acyltransferases were characterized in vitro.14,43,45 In the ZBM biosynthetic pathway, nature apparently has deleted one A domain from the NRPS-6 module and replaced it in trans by a set of two proteins, the incomplete A-T didomain module of ZbmVIIb and the acyltransferase ZbmVIId, to create diversity at this position of the peptide backbone. Provided that the respective acyltranferases show sufficient substrate flexibility regarding their acyl and PCP substrates, this natural strategy may be a very useful way for the artificial creation of new unnatural natural products with various activities.
Conclusions
The biosynthetic pathway of the hybrid peptide-polyketideantibiotic ZBM involves novel biochemistry, such as the formation of the rarely seen 6-deoxy-L-gulose sugar involving a GDP-mannose-4,6-dehydratase, an A domain supposedly exhibiting L-homoserine specificity, and formation as well as incorporation of the nonproteinogenic amino acid hydroxy-L-valine. Although the BLM and TLM biosyntheticgene clusters have been cloned and characterized previously from S. verticillus and S. hindustanus, respectively, genetic engineering of the biosynthetic machinery for this family of important antitumor antibiotics has been severely hampered by the lack of an efficient genetic system for these organisms. The availability of the ZBM gene cluster and the genetic amenability of S. flavoviridis now provide a platform to decipher these enigmas, expand the repertoire of NRPS/PKS systems for combinatorial biosynthesis of new compounds, especially new BLM family derivatives, and develop new antitumor drugs with similarly high activity and reduced toxicity.Experimental
Bacterial strains and culture conditions
Streptomyces flavoviridis ATCC21892 (American Type Culture Collection, Rockville, MD, USA), S. flavoviridis SB9001,7 and recombinant strains generated in this study were routinely grown at 28 °C in R2YE or TSB liquid medium.19 ISP4 medium supplemented with 0.05% yeast extract, 0.1% tryptone, and MgCl2 to a final concentration of 20 mM was used to prepare S. flavoviridisspores and to plate out conjugation mixtures. For ZBM production, S. flavoviridis SB9001 and recombinant strains were cultured in 250 mL-baffled flasks containing 50 mL of the previously described seed medium7 for 2 days at 28 °C and 250 rpm. Then, 5 mL of the seed cultures were inoculated into 250 mL baffled flasks containing 50 mL of the production medium (6.4% millet jelley, 0.5% glucose, 3.5% soybean meal, 0.75% cornsteep solid, 0.2% NaNO3, 0.3% NaCl, and 0.1% K2HPO4, with the pH adjusted to 7.2 with 1.0 N HCl) and fermented at 28 °C and 250 rpm for 12 days. Escherichia coli XL1 Blue MR (Stratagene, La Jolla, CA, USA) and E. coli DH5α46 were used for routine subcloning, plasmid preparations, and cosmid library preparation. E. coli S17-119 was the donor in E. coli–S. flavoviridis SB9001 conjugation and E. coli BW25113/pIJ790 was the host for λRED-mediated mutagenesis using the REDIRECT Technology kit.47Plasmids and biochemicals
The pGEM series of vectors (Promega, Madison, WI) and Litmus 28, (New England Biolabs, Beverly, MA) were from commercial sources, pIJ773,47 pANT841 (accession number AF438749),48 pSET152,49 pSET151,49 pWHM1250,50 and the aac(3)IVapramycin resistance cassette19 were described previously. A modified version of SuperCos 1 (Stratagene) that lacks the kanamycin resistance marker was used for S. flavoviridiscosmid library preparation. Common biochemicals, chemicals, media, restriction enzymes, and other molecular biology reagents were from standard commercial sources. Ampicillin (150 μg mL−1), apramycin (50 μg mL−1), and chloramphenicol (25 μg mL−1) were used for selection in E. coli. Apramycin (50–100 μg mL−1) and thiostrepton (50–100 μg mL−1) were used for selection of S. flavoviridis recombinants. Nalidixic acid (50 μg mL−1) was used to select against the E. coli conjugal donor after conjugation.Isolation and analysis of ZBM
ZBM and intermediates produced by the respective recombinant strains were isolated and analyzed as described previously.7 The identity of ZBM was confirmed by LC-ESI mass spectrometry on an Agilent 1100 HPLC-MSD SL quadrupole mass spectrometer or an Agilent 1100 HPLC-MSD SL ion trap mass spectrometer.DNA isolation and manipulation
DNA isolations and manipulations in E. coli46 and Streptomyces19 were carried out according to standard procedures. For Southern analysis, digoxigenin labeling of DNA probes, hybridization, and detection were performed according to the protocols provided by the manufacturer (Roche Diagnostics Corp., Indianapolis, IN, USA).Cloning, isolation and sequencing of the ZBM biosyntheticgene cluster
Amplification of putative NRPS Cy domains from S. flavoviridis genomic DNA was carried out as described previously.18 The obtained PCR product was cloned into pGEM-T and sequenced, yielding plasmid populations pBS9000 and pBS9001. PCR primers Cy1-FP (5′-TCA TCG GTG ACT TCA CCT CC-3′, at the 3′ end of the pBS9000 insert), Cy1-RP (5′-ACA TAG CCG TAG GTC GAG ACG-3′, at the 5′ end of the pBS9000 insert), Cy0-FP (5′-CGA CAT CGA TCA GGT GAT CG-3′, at the 3′ end of the pBS9001 insert), and Cy0-RP (5′-GCT CTT CCA GAT CGA GTT CG-3′, at the 5′ end of the pBS9001 insert), were designed (Fig. S1). The PCR product obtained with Cy1-FP and Cy0-RP was cloned into pGEM-T Easy to yield pBS9002. The entire NRPS module was amplified from the S. flavoviridiscosmid pBS9005 using primers Cy0-FP and Ox-RP16 and cloned into pGEM-T to afford pBS9003.A cosmid library (Stratagene) of S. flavoviridis chromosomal DNA was constructed by partial digestion with Sau3AI, dephosphorylation and ligation into the BamHI site of a modified version of SuperCos1. The ligation mixture was packaged with Gigapack III XL packaging extract and transduced into E. coli XL1 Blue MR cells. The genomic library was screened by colony hybridization with the digoxigenin-labeled 1.1 kb EcoRI fragment from pBS9000 as a probe (Fig. 2A, probe 1) and positive clones were confirmed by Southern hybridization. The 2.7 kb EcoRI-KpnI fragment excised from the upstream end of pBS9005 (probe 2), and the 2.2-kb EcoRI-KpnI fragment derived from the upstream end of pBS9006 (probe 3), were used as probes for chromosomal walking to isolate overlapping cosmids covering the entire cluster (Fig. 2A).
DNA sequencing and computer assisted sequence analysis
Sequencing of two cosmids, pBS9005 and pBS9007, and subclones thereof was performed on both strands by the dideoxynucleotide chain termination method using M13 universal primers or primers designed for the cosmid vector or by primer walking. A third cosmid, pBS9006 was chosen to fill-in the 2700 bp gap between pBS9007 and pBS9005, and a fourth cosmid, pBS9004, was used to obtain the 3121 bp sequence downstream of pBS9005. Sequencing reactions were run using Big Dye Terminator mix (Applied Biosystems, Foster City, CA), cleaned using CleanSeq magnetic beads (Agencourt Biosciences, Beverly, MA), and sequenced by the University of Wisconsin Biotechnology Center. Samples were electrophoresed on an Applied Biosystems 3700 automated DNA sequencing instrument, and data were analyzed using PE-Biosystems version 3.7 for Sequence Analyses. ORF assignments were made with the assistance of Codon Preference (GCG package, Madison, WI) and BLAST analyses. Sequence alignments, identity and similarity calculations were carried out by AlignX in the Vector NTI Advance™ 10 program from Invitrogen, the amino acid specificity of NRPS A domains was predicted using NRPS predictor.23Genetic manipulation of S. flavoviridis
E. coli–S. flavoviridis conjugation was carried out using standard literature conditions19 with minor modifications. In brief, approximately 108S. flavoviridis SB9001 spores were heat shocked in 500 μL of TSB medium supplemented with 0.4% glycine and 10.0% sucrose at 50 °C for 10 min, followed by incubation at 30 °C for 3–4 h as S. flavoviridis recipients. E. coli S17-1 containing the respective plasmid was grown in LB medium to an OD600 of 0.4–0.6. Cells from 2 mL of culture were pelleted, washed twice with LB medium, and resuspended in 500 μL of LB medium as the E. coli donors. For conjugation, the donors (500 μL) and recipients (500 μL, ∼108spores) were mixed and distributed onto ISP4 plates supplemented with 0.05% yeast extract, 0.1% tryptone, and MgCl2 to a final concentration of 20 mM. The plates were incubated at 30 °C for 16–20 h, overlaid with 1 mL of water containing final concentrations of 50 μg mL−1nalidixic acid to select against E. coli and either 100 μg mL−1apramycin or 100 μg mL−1thiostrepton to select for S. flavoviridis exconjugants. Incubation was continued at 30 °C until exconjugants appeared.Generation of the integrative vector pBS9010
The blaampicillin resistance gene46 was cloned as a 1 kb BspHI-SspI-fragment into the NcoI-Ecl136 II sites of Litmus 28 to yield pBS9008. The tsrthiostrepton resistance gene19 was isolated as a 1 kb BclI fragment from pWHM1250 and cloned behind the blagene into the BamHI site of pBS9008 to create pBS9009 (tsrgene in the same orientation as the blagene). A 2 kb EcoRI (Klenow treated)-StuI fragment was then moved from pBS9009 into the Ecl136 II sites of pSET152 yielding pBS9010, with the bla and tsrgenes facing the opposite direction as the replaced aac(3)IVapramycin resistance gene.Inactivation of zbmX (NRPS 9-8)
A 13.5 kb EcoRI-SbfI DNA fragment containing zbmX was cloned into the EcoRI-PstI sites of pGEM3Zf to yield pBS9011. A 2.5 kb ClaI-StuI internal fragment was then replaced with the aac(3)IVgene on a ClaI-EcoRV fragment to afford pBS9012 and the inactivated zbmX was moved as a 12 kb SspI-HindIII fragment into the BamHI (Klenow treated)-HindIII sites of pSET151 to yield pBS9013.Inactivation and complementation of zbmL (4,6-GDP-mannose dehydratase)
A 2.4-kb BamHI-NcoI DNA fragment containing zbmL was Klenow treated and cloned into the Ecl136 II-HincII sites of pGEM3Zf with the blunt ended NcoI site ligating to the HincII site to afford pBS9014. The aac(3)IVgene was then inserted as a KpnI-BamHI fragment in the same orientation as zbmL into the KpnI-BglII sites of pBS9014 yielding pBS9015. The inactivated zbmL was transferred as a ∼3.4 kb EcoRI-PstI fragment into the same sites of pSET151 to yield pBS9016.The ErmE* promoter19 was isolated from pWHM125050 as an SstI-BamHI fragment and cloned into Litmus 28 at the same sites to yield pBS9017. The zbmLgene was then inserted into the EcoRI-NsiI sites of pBS9017 behind the promoter as an EcoRI-PstI fragment from pBS9014 to form pBS9018. The entire insert was moved as a ∼2.7 kb Ecl136II-SpeI fragment from pBS9018 into pBS9010 at EcoRI (Klenow treated)-XbaI to yield the zbmL expression plasmid pBS9019.
Inactivation of zbm-orf41 (NRPS)
A 2.7 kb NcoI DNA fragment (blunt ended by Klenow treatment) containing most of the zbm-orf41gene was cloned into the Ecl136II-HincII sites of pGEM3Zf to create pBS9020 with the upstream NcoI site of the insert ligated to the Ecl136II site. A 0.6 kb internal BamHI-EcoRV fragment of pBS9020 was then replaced with the aac(3)IVgene on a BamHI-EcoRI (Klenow treated) fragment to afford pBS9021. The inactivated zbm-orf41 was moved as a ∼3.2 kb EcoRI-PstI fragment into the same sites of pSET151 to afford pBS9022.Inactivation of zbmVIId (thioesterase)
The zbmVIIdgene was replaced by the aac(3)IV-oriT cassette from pIJ773 viaλRED-mediated PCR -targeting47 using oligonucleotides zbm5-f1 (5′-cgaggcggccgaaacgacccggacggttcgttgagcgtgATTCCGGGGATCCGTCGACC-3′) and zbm5-r1 (5′-cccgtccgggtccgcgtgtcccctcacgccgccgagttcTGTAGGCTGGAGCTGCTTC-3′), with lower case letters representing DNA sequence originating from S. flavoviridis and upper case letters representing DNA sequence flanking the aac(3)IV-oriT cassette from pIJ773, to yield pBS9023.Inactivation of zbm-orf4 (aculeacin acylase)
A 4.3 kb XhoI-BglII fragment containing zbm-orf4 was cloned into the XhoI-BamHI sites of pGEM7Zf, yielding pBS9024. A ∼1.4 kb NsiI-EcoRV fragment containing the aac(3)IVgene was then cloned into the PstI-EcoRV sites of pBS9024, replacing a 1.2 kb internal fragment of zbm-orf4, to afford pBS9025. The inactivated zbm-orf4 was moved as a 4.1 kb XbaI-NsiI fragment into the XbaI and PstI sites of pSET151 to yield pBS9026.Inactivation and complementation of zbm-orf(−1) (thioesterase)
A 5 kb NdeI-HindIII fragment containing the zbm-orf(−1) gene was cloned into the same sites of pANT841,48 yielding pBS9027. A 1.3-kb PstI-EcoRV fragment from pBS9027 was subcloned into pGEM5Zf to form pBS9028, where a 210 bp KpnI-BamHI internal portion of zbm-orf(−1) was replaced with the aac(3)IVgene affording pBS9029. The 1.2 kb NdeI-PstI fragment from pBS9027 was inserted into the same sites in pBS9029 in front of zbm-orf(−1) to create pBS9030. The entire insert of pBS9030 was moved as ∼3.3 kb NsiI-EcoRV fragment, together with the 2.5-kb EcoRV-HindIII fragment from pBS9027 into the PstI and HindIII sites of pSET151 in a three piece ligation, resulting in pBS9031.The 1270 bp PstI-EcoRV insert of pBS9028 was cloned as a PstI-NcoI fragment into Litmus28 and then transferred as a BglII-XbaI fragment into pBS9010 along with the ErmE* promoter on an EcoRI-BamHI fragment to create the zbm-orf(−1) expression plasmid pBS9032.
Inactivation of zbm-orf2 (dihydroorotate dehydrogenase)
The zbm-orf2gene was replaced by the aac(3)IV-oriT cassette from pIJ773 via λRED-mediated PCR -targeting47 using oligonucleotides zbm1-f1 (5′-cgacgccgtgctcgccgtcaccgagcagccggcgtgaggATTCCGGGGATCCGTCGACC-3′) and zbm1-r1 (5′-acccaccaggtcccgggtctccgccagaatgcgctcgagTGTAGGCTGGAGCTGCTTC-3′), with lower case letters representing DNA sequence originating from S. flavoviridis and upper case letters representing DNA sequence flanking the aac(3)IV-oriT cassette from pIJ773, to yield pBS9033.Generation of the mutant and complementation strains and analysis of their secondary metabolite profile
The inactivation constructs pBS9013, pBS9016, pBS9022, pBS9023, pBS9026, pBS9031, and pBS9033 were introduced into the wild-type S. flavoviridis SB9001 strain by intergeneric conjugation. Exconjugants were selected either by using the apramycin-resistant and thiostrepton sensitive phenotype or, for S. flavoviridis SB9005 and SB9008, by a combination of the apramycin-resistant phenotype and PCR screening using oligonucleotide pairs C-zorf5-f1 (5′-GGATCGAGGTCCTCGGCTTC-3′) and C-zorf5-r1 (5′-ACCCTGCCGCACACGATGTC-3′) as well as C-zorf1-f1 (5′-CGTACTCGTCACCGAACAGC-3′) and C-zorf1-r1 (5′-AGTGGCCGTTGACCACGACG-3′), respectively, affording the mutant strains S. flavoviridis SB9002, SB9003, SB9004, SB9005, SB9006, SB9007, and SB9008 (Fig. 3A and S2† ). The genotypes of all apparent double crossover mutant strains were further confirmed by Southern blot analyses (Fig. 3B and S2† ). The mutants were cultured and examined for ZBM production with the wild-type S. flavoviridis SB9001 strain as a control (Fig. 3C and 4, traces I, VII, IX, VI, V, II, IV). The isolated yields of ZBM from the SB9004 and SB9007 mutant strains were 4–6 mg L−1 (∼60% of the wild-type SB9001 strain) and 2–4 mg L−1 (∼40% of the wild-type SB9001 strain), respectively.The complementation constructs pBS9019 and pBS9032 were introduced into SB9003 and SB9007 mutant strains, respectively, by intergeneric conjugation. Maintenance of pBS9019 and pBS9032 in the resultant S. flavoviridis SB9009 and SB9010 strains, respectively, was verified by PCR analysis using oligonucleotides O.127 (5′-CGACGTGTACTCAGCGACACG-3′), O.108 (5′-GGTCCTGCATCGGCACTCC-3′), O.851 (5′-GCGACCGTCGAAGGGCTCGC-3′), and O.2353 (5′-AGCGCCGTTCTTTGCGTTTTCTGT-3′) (data not shown). The complementation strains were cultured and investigated for ZBM production with the wild-type S. flavoviridis SB9001 and the parent SB9003 and SB9007 strains as controls (Fig. 4, traces I, VII, VIII, II, and III). The isolated yields of ZBM from the SB9009 and SB9010 complementation strains were ∼8–10 mg L−1 (∼70% of the wild-type SB9001 strain) and 2–4 mg L−1 (∼40% of the wild-type SB9001 strain), respectively.
Nucleotide sequence accession number
The nucleotide sequence reported in this study is available in the GenBank database under accession no. EU670723.Acknowledgements
We thank the Analytical Instrumentation Center of the School of Pharmacy, UW-Madison, for support in obtaining LC-MS data, the John Innes Center, Norwich, UK, for providing the REDIRECT Technology kit, and Dr Scott R. Rajski for critical reading of the manuscript. This work was supported in part by the NIH grant CA94426. N.P.G. is supported in part by NIH grant T32 GM08505 and U.G. is a Postdoctoral Fellow of the Deutsche Forschungsgemeinschaft (DFG).References
- T. Takita, Y. Muraoka, A. Fujii, H. Ito and K. Maeda, The structure of the sulfur-containing chromophore of phleomycin, and chemical transformation of phleomycin to bleomycin, J. Antibiot., 1972, 25(3), 197–199 CAS.
- T. Takita, Y. Muraoka, T. Nakatani, A. Fujii, Y. Umezawa, H. Naganawa and H. Umezawa, Chemistry of bleomycin. XIX Revised structures of bleomycin and phleomycin, J. Antibiot., 1978, 31(8), 801–804 CAS.
- H. Umezawa, K. Maeda, T. Takeuchi and Y. Okami, New antibiotics, bleomycin A and B, J. Antibiot., 1966, 19(5), 200–209 CAS.
- K. Maeda, H. Kosaka, K. Yagashita and H. Umezawa, A new antibiotic, phleomycin, J. Antibiot., 1956, 9(2), 82–85 CAS.
- H. Kawaguchi, H. Tsukiura, K. Tomita, M. Konishi, K. Saito, S. Kobaru, K. Numata, K. Fujisawa, T. Miyaki, M. Hatori and H. Koshiyama, Tallysomycin, a new antitumor antibiotic complex related to bleomycin. I. Production, isolation and properties, J. Antibiot., 1977, 30(10), 779–788 CAS.
- M. Konishi, K. Saito, K. Numata, T. Tsuno, K. Asama, H. Tsukiura, T. Naito and H. Kawaguchi, Tallysomycin, a new antitumor antibiotic complex related to bleomycin. II. Structure determination of tallysomycins A and B, J. Antibiot., 1977, 30(10), 789–805 CAS.
- L. Wang, B. S. Yun, N. P. George, E. Wendt-Pienkowski, U. Galm, T. J. Oh, J. M. Coughlin, G. Zhang, M. Tao and B. Shen, Glycopeptide antitumor antibiotic zorbamycin from Streptomyces flavoviridis ATCC 21892: strain improvement and structure elucidation, J. Nat. Prod., 2007, 70(3), 402–406 CrossRef CAS.
- L. Du, C. Sanchez, M. Chen, D. J. Edwards and B. Shen, The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase, Chem. Biol., 2000, 7(8), 623–642 CrossRef CAS.
- M. Tao, L. Wang, E. Wendt-Pienkowski, N. P. George, U. Galm, G. Zhang, J. M. Coughlin and B. Shen, The tallysomycin biosynthetic gene cluster from Streptoalloteichus hindustanus E465-94 ATCC 31158 unveiling new insights into the biosynthesis of the bleomycin family of antitumor antibiotics, Mol. Biosyst., 2007, 3(1), 60–74 RSC.
- S. M. Hecht, Bleomycin: new perspectives on the mechanism of action, J. Nat. Prod., 2000, 63(1), 158–168 CrossRef.
- J. Chen and J. Stubbe, Bleomycins: towards better therapeutics, Nat. Rev. Cancer, 2005, 5(2), 102–112 CrossRef CAS.
- C. R. Hutchinson, Polyketide and non-ribosomal peptide synthases: falling together by coming apart, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(6), 3010–3012 CrossRef CAS.
- T. Nguyen, K. Ishida, H. Jenke-Kodama, E. Dittmann, C. Gurgui, T. Hochmuth, S. Taudien, M. Platzer, C. Hertweck and J. Piel, Exploiting the mosaic structure of trnas-acyltransferase polyketide synthases for natural product discovery and pathway dissection, Nat. Biotechnol., 2008, 26(2), 225–233 CrossRef CAS.
- G. M. Singh, F. H. Vaillancourt, J. Yin and C. T. Walsh, Characterization of SyrC, an aminoacyltransferase shuttling threonyl and chlorothreonyl residues in the syringomycin biosynthetic assembly line, Chem. Biol., 2007, 14(1), 31–40 CrossRef CAS.
- B. Shen, L. Du, C. Sanchez, D. J. Edwards, M. Chen and J. M. Murrell, The biosynthetic gene cluster for the anticancer drug bleomycin from Streptomyces verticillus ATCC15003 as a model for hybrid peptide-polyketide natural product biosynthesis, J. Ind. Microbiol. Biotechnol., 2001, 27(6), 378–385 CrossRef CAS.
- B. Shen, L. Du, C. Sanchez, D. J. Edwards, M. Chen and J. M. Murrell, Cloning and characterization of the bleomycin biosynthetic gene cluster from Streptomyces verticillus ATCC15003, J. Nat. Prod., 2002, 65(3), 422–431 CrossRef CAS.
- U. Galm, L. Wang, E. Wendt-Pienkowski, R. Yang, W. Liu, M. Tao, J. M. Coughlin and B. Shen, In vivo manipulation of the bleomycin biosynthetic gene cluster in Streptomyces verticillus ATCC15003 revealing new insights into its biosynthetic pathway, J. Biol. Chem., 2008, 283(42), 28236–28245 CrossRef CAS.
- Y. Q. Cheng, G. L. Tang and B. Shen, Identification and localization of the gene cluster encoding biosynthesis of the antitumor macrolactam leinamycin in Streptomyces atroolivaceus S-140, J. Bacteriol., 2002, 184(24), 7013–7024 CrossRef CAS.
- T. Kieser, M. J. Bibb, M. J. Buttner, K. F. Chater and D. A. Hopwood, Practical Streptomyces Genetics, John Innes Foundation, Norwich, UK, 2000 Search PubMed.
- F. Flett, V. Mersinias and C. P. Smith, High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes, FEMS Microbiol. Lett., 1997, 155(2), 223–229 CrossRef CAS.
- T. Stachelhaus, H. D. Mootz and M. A. Marahiel, The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases, Chem. Biol., 1999, 6(8), 493–505 CrossRef CAS.
- G. L. Challis, J. Ravel and C. A. Townsend, Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains, Chem. Biol., 2000, 7(3), 211–224 CrossRef CAS.
- C. Rausch, T. Weber, O. Kohlbacher, W. Wohlleben and D. H. Huson, Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs), Nucleic Acids Res., 2005, 33(18), 5799–5808 CrossRef CAS.
- L. Du, M. Chen, Y. Zhang and B. Shen, BlmIII and BlmIV nonribosomal peptide synthetase-catalyzed biosynthesis of the bleomycin bithiazole moiety involving both in cis and in trans aminoacylation, Biochemistry, 2003, 42(32), 9731–9740 CrossRef CAS.
- T. Correia, N. Grammel, I. Ortel, U. Keller and P. Tudzynski, Molecular cloning and analysis of the ergopeptine assembly system in the ergot fungus Claviceps purpurea, Chem. Biol., 2003, 10(12), 1281–1292 CrossRef CAS.
- L. Du and B. Shen, Identification and characterization of a type II peptidyl carrier protein from the bleomycin producer Streptomyces verticillus ATCC 15003, Chem. Biol., 1999, 6(8), 507–517 CrossRef CAS.
- J. W. Trauger, R. M. Kohli, H. D. Mootz, M. A. Marahiel and C. T. Walsh, Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase, Nature, 2000, 407(6801), 215–218 CrossRef CAS.
- N. Palaniappan, S. Ayers, S. Gupta, E.-S. Habib and K. A. Reynolds, Production of hygromycin A analogs in Streptomyces hygroscopicus NRRL 2388 through identification and manipulation of the biosynthetic gene cluster, Chem. Biol., 2006, 13(7), 753–764 CrossRef CAS.
- M. Strieker, F. Kopp, C. Mahlert, L. O. Essen and M. A. Marahiel, Mechanistic and structural basis of stereospecific Cbeta-hydroxylation in calcium-dependent antibiotic, a daptomycin-type lipopeptide, ACS Chem. Biol., 2007, 2(3), 187–196 CrossRef CAS.
- M. J. Calcutt and F. J. Schmidt, Gene organization in the bleomycin-resistance region of the producer organism Streptomyces verticillus, Gene, 1994, 151(1–2), 17–21 CrossRef CAS.
- A. Gatignol, H. Durand and G. Tiraby, Bleomycin resistance conferred by a drug-binding protein, FEBS Lett., 1988, 230(1–2), 171–175 CrossRef CAS.
- C. Mendez and J. A. Salas, ABC transporters in antibiotic-producing actinomycetes, FEMS Microbiol. Lett., 1998, 158(1), 1–8 CrossRef CAS.
- S. Lautru, D. Oves-Costales, J. L. Pernodet and G. L. Challis, MbtH-like protein-mediated cross-talk between non-ribosomal peptide antibiotic and siderophore biosynthetic pathways in Streptomyces coelicolor M145, Microbiology, 2007, 153(Pt 5), 1405–1412 CrossRef CAS.
- M. Wolpert, B. Gust, B. Kammerer and L. Heide, Effects of deletions of mbtH-like genes on clorobiocin biosynthesis in Streptomyces coelicolor, Microbiology, 2007, 153(Pt 5), 1413–1423 CrossRef CAS.
- V. Bennett, Ankyrins. Adaptors between diverse plasma membrane proteins and the cytoplasm, J. Biol. Chem., 1992, 267(13), 8703–8706 CAS.
- C. T. Walsh, H. Chen, T. A. Keating, B. K. Hubbard, H. C. Losey, L. Luo, C. G. Marshall, D. A. Miller and H. M. Patel, Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines, Curr. Opin. Chem. Biol., 2001, 5(5), 525–534 CrossRef CAS.
- T. L. Schneider, B. Shen and C. T. Walsh, Oxidase domains in epothilone and belomycin biosynthesis: thiazoline to thiazole oxidation during chain elongation, Biochemistry, 2004, 42(32), 9722–9730.
- D. O. Rees, N. Bushby, R. J. Cox, J. R. Harding, T. J. Simpson and C. L. Willis, Synthesis of [1,2-13C215N]-L-homoserine and its incorporation by the PKS-NRPS system of Fusarium moniliforme into the mycotoxin fusarin C, Chembiochem, 2007, 8(1), 46–50 CrossRef CAS.
- M. Fernandez, Y. Cuadrado, J. F. Aparicio and J. F. Martin, Role of homoserine and threonine pathway intermediates as precursors for the biosynthesis of aminoethoxyvinylglycine in Streptomyces sp. NRRL 5331, Microbiology, 2004, 150(Pt 5), 1467–1474 CrossRef CAS.
- K. Watanabe, K. Hotta, A. P. Praseuth, K. Koketsu, A. Migita, C. N. Boddy, C. C. Wang, H. Oguri and H. Oikawa, Total biosynthesis of antitumor nonribosomal peptides in Escherichia coli, Nat. Chem. Biol., 2006, 2(8), 423–428 CrossRef CAS.
- J. Ju, S. G. Ozanick, B. Shen and M. G. Thomas, Conversion of (2S)-arginine to (2S,3R)-capreomycidine by VioC and VioD from the viomycin biosynthetic pathway of Streptomyces sp. strain ATCC11861, Chembiochem, 2004, 5(9), 1281–1285 CrossRef CAS.
- H. Chen, M. G. Thomas, S. E. O'Connor, B. K. Hubbard, M. D. Burkart and C. T. Walsh, Aminoacyl-S-enzyme intermediates in beta-hydroxylations and alpha,beta-desaturations of amino acids in peptide antibiotics, Biochemistry, 2001, 40(39), 11651–11659 CrossRef CAS.
- E. R. Strieter, F. H. Vaillancourt and C. T. Walsh, CmaE: A transferase shuttling aminoacyl groups between carrier protein domains in the coronamic acid biosynthetic pathway, Biochemistry, 2007, 46(25), 7549–7557 CrossRef CAS.
- Z. Chang, P. Flatt, W. H. Gerwick, V. A. Nguyen, C. L. Willis and D. H. Sherman, The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit, Gene, 2002, 296(1–2), 235–247 CrossRef CAS.
- S. Garneau-Tsodikova, A. Stapon, D. Kahne and C. T. Walsh, Installation of the pyrrolyl-2-carboxyl pharmacophore by CouN1 and CouN7 in the late biosynthetic steps of the aminocoumarin antibiotics clorobiocin and coumermycin A1, Biochemistry, 2006, 45(28), 8568–8578 CrossRef CAS.
- J. Sambrook and D. W. Russell, Molecular cloning: a laboratory manual, Cold Spring Harbor Laboratory Press, New York, 2001 Search PubMed.
- B. Gust, G. L. Challis, K. Fowler, T. Kieser and K. F. Chater, PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(4), 1541–1546 CrossRef CAS.
- V. B. Rajgarhia and W. R. Strohl, Minimal Streptomyces sp. strain C5 daunorubicin polyketide biosynthesis genes required for aklanonic acid biosynthesis, J. Bacteriol., 1997, 179(8), 2690–2696 CAS.
- M. Bierman, R. Logan, K. O’Brien, E. T. Seno, R. N. Rao and B. E. Schoner, Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp, Gene, 1992, 116(1), 43–49 CrossRef CAS.
- N. Lomovskaya, S. L. Otten, Y. Doi-Katayama, L. Fonstein, X. C. Liu, T. Takatsu, A. Inventi-Solari, S. Filippini, F. Torti, A. L. Colombo and C. R. Hutchinson, Doxorubicin overproduction in Streptomyces peucetius: cloning and characterization of the dnrU ketoreductase and dnrV genes and the doxA cytochrome P-450 hydroxylase gene, J. Bacteriol., 1999, 181(1), 305–318 CAS.
- A. Fujii, Biosynthetic aspects of bleomycin-phleomycin group antibiotics, in Bleomycin: Chemical Biochemical and Biological Aspects, ed. S. M. Hecht, Springer-Verlag, New York, 1979, pp. 75–91 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: A schematic representation of the arrangement of Cy and Ox domains within the thiazole forming NRPS modules (Fig. S1), schematic representations of gene inactivation for cluster boundary determination and the corresponding Southern blots (Fig. S2), and proposed mechanisms for the biosynthesis and incorporation of L-hydroxyvaline (Fig. S3). See DOI: 10.1039/b814075h |
| This journal is © The Royal Society of Chemistry 2009 |
