Structural similarity of hydrogen-bonded and metal-coordinated abiotic base pairs allows oligonucleotide-like mutual stacking†,‡
Marc-Oliver M.
Piepenbrock
,
Kirsty M.
Anderson
,
Benedict C. R.
Sansam
,
Nigel
Clarke
and
Jonathan W.
Steed
*
Department of Chemistry, Durham University, South Road, Durham, UK DH1 3LE. Fax: +44 (0)191 384 4737; Tel: +44 (0)191 334 2085E-mail: jon.steed@durham.ac.uk
First published on 8th October 2008
Abstract
A hydrogen bonded artificial base-pair can replace a metal-bridged ligand pair in an almost isosteric fashion to give a hybrid π-stacked cage complex containing one base-bair and one metal-bridged linkage suggesting the components have potential as artificial nucleobases.
Introduction
The development of artificial analogues of the characteristic DNA Watson–Crick nucleobase pairs is a topic of considerable current interest.1–5 Such analogues hold the key to a potential new world of ‘artificial biology’ and are of interest as molecular probes and in chemotherapy.4,6 Recent reports have focussed on the replacement of hydrogen bonded artificial nucleobase pairs with other kinds of interaction such as halogen bonding5 and particularly metal-linked pairs of bis(chelate) ligands.7–11 It has been shown that a copper(II)-linked hydroxypyridone base pair can be incorporated into DNA implying that the metal-linked artificial system must be able to engage in the characteristic DNA π–π stacking interactions.9 Indeed key to any artificial diverse DNA-type genetic code is the existence of more than one kind of essentially isosteric base pairs that can be mutually exchanged without distortion of the overall assembly.1 Metal-linked base pairs have been incorporated into the G-quartet motif to give both canonical and unconventional hybrid artificial G-quartets comprising both metal links and hydogen bonding.10 Metal-linked and hydrogen bonded artificial base pairs should be approximately isosteric and can thus play a wide role in self-assembly. For example, it has been shown in giant self-assembled resorcarene capsules, that 24 Cu(II) ions can replace 48 hydrogen bonds to give the same capsule structure, and a mixed hydogen-bonded and Ga(III) coordinated analogue is also known.12,13 We now report a remarkable complex that contains isosteric hydrogen bonded and metal-coordinated 2-pyridylurea base pairs within the same molecule as heteromeric π-stacked duplexes.Results and discussion
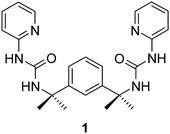
Ligand 1 is readily prepared by reaction of 2-aminopyridine with 1,3-bis-(1-isocyanato-1-methyl-ethyl)-benzene. The steric hindrance of the two quaternary carbon atoms has been shown to give a characteristic C-shaped conformation in the related 3-pyridyl isomer14 and hence the two ureidopyridine functionalities are predisposed to stack one above another without chelating. Reaction of 1 with an equimolar amount of either copper(II) chloride or bromide gives the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes [(CuX)2(μ–κ–N,N′,O,O′-1)2](X)2·3H2O (X
1 complexes [(CuX)2(μ–κ–N,N′,O,O′-1)2](X)2·3H2O (X![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cl, 2; Br, 3). Both compounds were characterised by X-ray crystallography and are isomorphous. Each complex features two square pyramidal copper(II) centres with the basal plane being occupied by two transpyridyl groups from two different ligands, and two urea oxygen atoms resulting in the formation of a double six-membered chelate ring motif. This N,O-chelate mode for 2-pyridylureas is unusual but has been previously observed for a series of octahedral compounds of type [M(N,N′-di-2-pyridylurea)2(NO3)2] (M = Mn, Fe, Co, Ni, Cu and Zn).15,16 The axial site is occupied by long bonds to the coordinated halide ions in each case. The complex appears to contract in the middle compared to the 5.06 Å length of the o-xylylene spacers such that free space in the central void is minimised—the stacking distance between the basal plane centroids of the square pyramidal metal centres is 3.80 Å, Fig 1a. The crystal packing involving the halide anions and water may be divided into two discrete segments. One halide and two water molecules form an infinite chain of 4-membered rings comprising two halide anions and two water molecules, {(H2O)2Cl−}n, running in between the offset stacks of binuclear complexes. The other halide anion and water molecule, along with the coordinated halide anions take part in a complex and unusual hydrogen bonding arrangement with the urea groups of the ligands. While the uncoordinated halide anion is bound on one side by a conventional R12(6) motif17,18 on the other side only one urea NH proton interacts with the anion. The anion also accepts a fourth hydogen bond from a water molecule. The result is an unusual R23(12) hydrogen bonded ring comprising two urea groups of the metal complex, a water molecular and halide anion. The metal-coordinated halide anion is also bound on one side by a urea group exhibiting an R12(6) motif but on the other side again a urea group donates one hydrogen atom to the coordinated halide and one to water, Fig. 1b.
Cl, 2; Br, 3). Both compounds were characterised by X-ray crystallography and are isomorphous. Each complex features two square pyramidal copper(II) centres with the basal plane being occupied by two transpyridyl groups from two different ligands, and two urea oxygen atoms resulting in the formation of a double six-membered chelate ring motif. This N,O-chelate mode for 2-pyridylureas is unusual but has been previously observed for a series of octahedral compounds of type [M(N,N′-di-2-pyridylurea)2(NO3)2] (M = Mn, Fe, Co, Ni, Cu and Zn).15,16 The axial site is occupied by long bonds to the coordinated halide ions in each case. The complex appears to contract in the middle compared to the 5.06 Å length of the o-xylylene spacers such that free space in the central void is minimised—the stacking distance between the basal plane centroids of the square pyramidal metal centres is 3.80 Å, Fig 1a. The crystal packing involving the halide anions and water may be divided into two discrete segments. One halide and two water molecules form an infinite chain of 4-membered rings comprising two halide anions and two water molecules, {(H2O)2Cl−}n, running in between the offset stacks of binuclear complexes. The other halide anion and water molecule, along with the coordinated halide anions take part in a complex and unusual hydrogen bonding arrangement with the urea groups of the ligands. While the uncoordinated halide anion is bound on one side by a conventional R12(6) motif17,18 on the other side only one urea NH proton interacts with the anion. The anion also accepts a fourth hydogen bond from a water molecule. The result is an unusual R23(12) hydrogen bonded ring comprising two urea groups of the metal complex, a water molecular and halide anion. The metal-coordinated halide anion is also bound on one side by a urea group exhibiting an R12(6) motif but on the other side again a urea group donates one hydrogen atom to the coordinated halide and one to water, Fig. 1b.
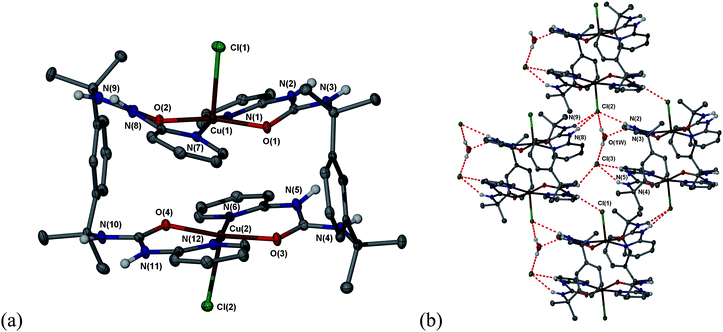 | ||
| Fig. 1 (a) Molecular structure of the binuclear copper(II) complex 2; (b) crystal packing in 2 showing the complex interactions from urea to both coordinated and uncoordinated Cl−. Selected bond distances: Cu1–O2 1.9229(16), Cu1–O1 1.9313(16), Cu1–N1 2.030(2), Cu1–N7 2.0336(19), Cu1–Cl1 2.5095(7) Å. Selected hydrogen bond distances: N3⋯O1W 2.866(3), Cl2⋯N9 3.219(2), Cl2⋯N8 3.354(2), Cl2⋯N2 3.365(2), Cl2⋯O1W 3.179(3), O1W⋯Cl3 3.183(3), Cl3⋯N5 3.163(2), Cl3⋯N4 3.262(2) Å. The bromide complex is isomorphous (30% ellipsoids). | ||
Reaction of 1 with copper(II) bromide in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio forms an interesting, related ‘supercomplex’ [{Cu(κ–N,O–1)2}2(μ–Br)](Br)3 (4). Complex 4 may be regarded as comprising two units resembling 2 and 3 linked by a long, linear bridging Br− ligand.19 However, while 2 and 3 are bimetallic complexes with a 1
1 ratio forms an interesting, related ‘supercomplex’ [{Cu(κ–N,O–1)2}2(μ–Br)](Br)3 (4). Complex 4 may be regarded as comprising two units resembling 2 and 3 linked by a long, linear bridging Br− ligand.19 However, while 2 and 3 are bimetallic complexes with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio between 1 and Cu, the units within 4 are monometallic with one of the copper(II) ions being replaced by a hydrogen bonded bridge (Fig. 2). This base pairing between one set of the pyridyl urea functionalities in 1 results in a hydrogen bonded dimer based on an eight-membered inter-ligand hydrogen bonded ring, graph set R22(8) that is essentially isosteric with the copper(II) bridged base pair that links the other ends of the same two ligands. The 8-membered ring motif differs from the classic R12(6) urea α-tape motif20–22 because the urea moiety adopts an anti conformation as a result of intramolecular hydrogen bonding from the γ-NH unit to the uncoordinated pyridyl N-atom, a phenomenon seen in other 2-pyridyl urea derivatives.23 The diagonal non-bonded O⋯O and N⋯N distances across the 2-pyridyl urea functionalities are very similar in the copper-bridged and hydrogen bonded pairs (O⋯O 3.58 vs. 3.83 and N⋯N 3.72 vs. 3.98 Å) and hence overall the mixed copper/hydrogen-bonded unit in 4 is essentially the same size and shape as the di-copper complexes 2 and 3. The stacking distance between the metal-coordinated basal plane atoms and the hydrogen bonded ring is 3.29 Å, rather smaller than the distance in 2 and 3 (in which the metal centre is more pyramidalised) but comparable with the 3.3 Å typical interplanar π−π stacking distances in oligonucleotides.24 The bromide-bridged supercomplex is supported by a total of eight NH⋯Br−hydrogen bonds of the usual R12(6) type from the urea groups near the metal to the lattice anions.
1 ratio between 1 and Cu, the units within 4 are monometallic with one of the copper(II) ions being replaced by a hydrogen bonded bridge (Fig. 2). This base pairing between one set of the pyridyl urea functionalities in 1 results in a hydrogen bonded dimer based on an eight-membered inter-ligand hydrogen bonded ring, graph set R22(8) that is essentially isosteric with the copper(II) bridged base pair that links the other ends of the same two ligands. The 8-membered ring motif differs from the classic R12(6) urea α-tape motif20–22 because the urea moiety adopts an anti conformation as a result of intramolecular hydrogen bonding from the γ-NH unit to the uncoordinated pyridyl N-atom, a phenomenon seen in other 2-pyridyl urea derivatives.23 The diagonal non-bonded O⋯O and N⋯N distances across the 2-pyridyl urea functionalities are very similar in the copper-bridged and hydrogen bonded pairs (O⋯O 3.58 vs. 3.83 and N⋯N 3.72 vs. 3.98 Å) and hence overall the mixed copper/hydrogen-bonded unit in 4 is essentially the same size and shape as the di-copper complexes 2 and 3. The stacking distance between the metal-coordinated basal plane atoms and the hydrogen bonded ring is 3.29 Å, rather smaller than the distance in 2 and 3 (in which the metal centre is more pyramidalised) but comparable with the 3.3 Å typical interplanar π−π stacking distances in oligonucleotides.24 The bromide-bridged supercomplex is supported by a total of eight NH⋯Br−hydrogen bonds of the usual R12(6) type from the urea groups near the metal to the lattice anions.
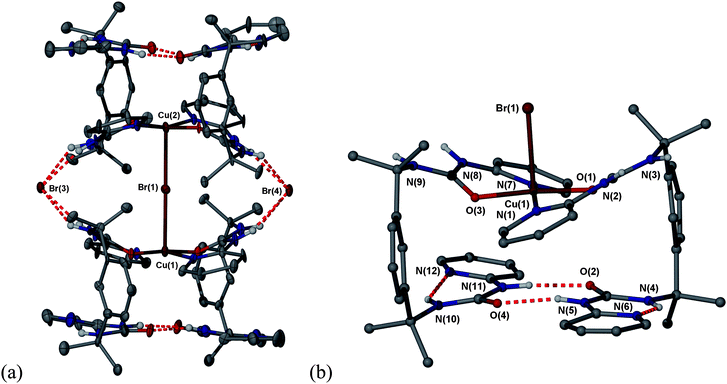 | ||
| Fig. 2 (a) The mixed hydrogen bonded/copper(II) ‘supercomplex’ 4 showing the bridging Br− anion and supporting urea⋯Br−hydrogen bonds (30% ellipsoids);(b) one half of the ‘supercomplex’ showing the similarity of the metal-linked and hydrogen bonded base pairs. Selected bond distances: Br1–Cu1 2.767(4), Br1–Cu2 2.769(4), Cu1–O3 1.911(8), Cu1–O1 1.925(9), Cu1–N1 1.987(11), Cu1–N7 2.015(11) Å. Selected hydrogen bond distances: N4⋯N6 2.634(19), N5⋯O4 2.863(18), N10⋯N12 2.70(2), N11⋯O2 2.874(16), N⋯Br− 3.270(11) to 3.469(11) Å. | ||
In conculsion, we have shown that a preorganised C-shaped binucleating ligand forms discrete 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 cage-like binuclear complexes with copper(II) salts. The analogous 2
1 cage-like binuclear complexes with copper(II) salts. The analogous 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex replaces the missing copper(II) centre with an 8-membered hydrogen bonded ring motif to give a hydrogen bonded base pair that is almost isosteric with the metal-bridged analogue and exhibits π-stacking comparable to that found in naturally occurring nucleobases. These versatile artificial base pairs thus show promise as part of a diverse system of artificial biology.
1 complex replaces the missing copper(II) centre with an 8-membered hydrogen bonded ring motif to give a hydrogen bonded base pair that is almost isosteric with the metal-bridged analogue and exhibits π-stacking comparable to that found in naturally occurring nucleobases. These versatile artificial base pairs thus show promise as part of a diverse system of artificial biology.
Experimental
Synthesis
1,3-bis-(1-[2-Pyridylureido]-1-methyl-ethyl)-benzene (1) A solution of 2-aminopyridine (3.80 g, 40.4 mmol) in dry CHCl3 (70 mL) was added dropwise to a stirred solution of 1,3-bis-(1-isocyanato-1-methyl-ethyl) benzene (4.88 g 20.0 mmol) in dry CHCl3 (50 mL) under reflux. After complete addition the reaction was allowed to reflux under an inert N2 atmosphere for 72 h. The resulting precipitate formed was collected by filtration and washed with CHCl3 (3 × 15 mL). The resulting white powder was obtained in a good yield (7.20 g, 83%). 1H NMR (400 MHz, DMSO-d6, J/Hz): 1.45 (12H, s, C–(CH3)2), 6.70 (2H, dd, pyrH, J = 7.2, 5.3), 7.08 (1H, t, ArH), 7.07 (2H, s, NH–C(CH3)2), 7.10 (2H, d, pyrH, J = 8.2), 7.29 (1H, s, ArH), 7.44 (2H, dd, pyrH, J = 8.2, 7.2), 7.95 (2H, d, ArH, J = 5.0), 8.53 (2H, broad m, pyrH), 8.90 (2H, s, NH-pyr). νmax(cm−1): 3332,3215 (NH), 1680,1656 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). m/z (ES+-MS): 433.1 ([M + H]+, 100%), 865.3 ([2M + H]+, 15%). Anal. calcd. for C24H28N6O2: C 66.65% H 6.53% N 19.43% Found C 66.59% H 6.47% N 19.30%
O). m/z (ES+-MS): 433.1 ([M + H]+, 100%), 865.3 ([2M + H]+, 15%). Anal. calcd. for C24H28N6O2: C 66.65% H 6.53% N 19.43% Found C 66.59% H 6.47% N 19.30%
Crystal preparation
[(CuCl)2(μ–κ–N,N′,O,O′−1)2](Cl)2·3H2O (2) Ligand 1 (0.0300 g, 6.94 × 10−5 mol) was dissolved in hot THF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 3
H2O = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (10 mL) in a closed screw-cap vial. When the solution had cooled down to room temperature 100 μL of a CuCl2 solution (0.1182 g, 6.93 × 10−4 mol in 1 mL MeOH) was injected into the vial. Crystal growth was at room temperature in the sealed vial. The crystals obtained took the form of green needles. νmax(cm−1): 3470,3414 (OH), 3269 (NH) 1670,1650 (C
1 (10 mL) in a closed screw-cap vial. When the solution had cooled down to room temperature 100 μL of a CuCl2 solution (0.1182 g, 6.93 × 10−4 mol in 1 mL MeOH) was injected into the vial. Crystal growth was at room temperature in the sealed vial. The crystals obtained took the form of green needles. νmax(cm−1): 3470,3414 (OH), 3269 (NH) 1670,1650 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). Anal. calcd. for C48H62N12O7Cu2Cl4: C 48.53% H 5.26% N 14.15% Found C 48.43% H 5.12% N 13.95%
O). Anal. calcd. for C48H62N12O7Cu2Cl4: C 48.53% H 5.26% N 14.15% Found C 48.43% H 5.12% N 13.95%
[(CuBr)2(μ–κ–N,N′,O,O′−1)2](Br)2·3H2O (3) Ligand 1 (0.0300 g, 6.94 × 10−5 mol) was dissolved in hot THF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 3
H2O = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (10 mL) in a closed screw-cap vial. When the solution had cooled down to room temperature 100 μL of a CuBr2 solution (0.1549 g, 6.93 × 10−4 mol in 1 mL MeOH) was injected into the vial. Crystal growth was at room temperature in the sealed vial. The crystals obtained took the form of green prisms. νmax(cm−1): 3486,3432 (OH) 3267 (NH), 1666,1649 (C
1 (10 mL) in a closed screw-cap vial. When the solution had cooled down to room temperature 100 μL of a CuBr2 solution (0.1549 g, 6.93 × 10−4 mol in 1 mL MeOH) was injected into the vial. Crystal growth was at room temperature in the sealed vial. The crystals obtained took the form of green prisms. νmax(cm−1): 3486,3432 (OH) 3267 (NH), 1666,1649 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). Anal. calcd. for C48H62N12O7Cu2Br4: C 42.21% H 4.53% N 12.31% Found C 41.72% 4.37% N 12.07%
O). Anal. calcd. for C48H62N12O7Cu2Br4: C 42.21% H 4.53% N 12.31% Found C 41.72% 4.37% N 12.07%
[{Cu(κ–N,O–1)2}2(μ–Br)](Br)3 (4) Ligand 1 (0.1000 g, 2.31 × 10−4 mol) was dissolved in hot MeOH (10 mL) in a closed screw-cap vial. When the solution had cooled down to room temperature 100 μL of a CuBr2 solution (0.2581 g, 1.15 × 10−3 mol, in 1 mL MeOH) was injected into the vial. Crystal growth was at room temperature in the sealed vial. The obtained crystals took the form of square blue plates. νmax(cm−1): 3265,3217 (NH), 1683,1650 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). Anal. calcd. for C48H56N12O4Cu1Br2: C 52.97% H 5.19% N 15.44% Found C 52.13% H 5.21% N 15.06%.
O). Anal. calcd. for C48H56N12O4Cu1Br2: C 52.97% H 5.19% N 15.44% Found C 52.13% H 5.21% N 15.06%.
Crystals were also grown in the same way using 100 μL of a CuBr2 solution (0.516 g, 2.31 × 10−3 mol in 1 mL MeOH). In this way crystals were obtained in the form of green hexagonal plates. Although too small for crystal structure determination, elemental analysis revealed a composition similar to that expected for [(CuX)2(μ–κ–N,N′,O,O′−1)2](Br)2·3H2O (3). νmax(cm−1): 3432 (OH) 3268, 3226 (NH), 1659 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). Found C 42.40% H 4.36% N 12.25%.
O). Found C 42.40% H 4.36% N 12.25%.
Acknowledgements
We thank the EPSRC for funding and for use of the SRS component of the National Crystallographic Service. We are especially gratefull to Prof. W. Clegg and the EPSRC sponsored national X-ray service for synchrotron data collection on compound 4.References
- N. Paul, V. C. Nashine, G. Hoops, P. M. Zhang, J. Zhou, D. E. Bergstrom and V. J. Davisson, Chem. Biol., 2003, 10, 815 CrossRef CAS.
- D. V. Jarikote, O. Kohler, E. Socher and O. Seitz, Eur. J. Org. Chem., 2005, 3187 CrossRef CAS.
- T. Hirano, K. Kuroda, H. Kodama, M. Kataoka and Y. Hayakawa, Lett. Org. Chem., 2007, 4, 530 Search PubMed.
- L. Bethge, D. V. Jarikote and O. Seitz, Bioorg. Med. Chem., 2008, 16, 114 CrossRef CAS.
- R. Tawarada, K. Seio and M. Sekine, J. Org. Chem., 2008, 73, 383 CrossRef CAS.
- M. Shionoya and K. Tanaka, Curr. Opin. Chem. Biol., 2004, 8, 592 CrossRef CAS.
- K. Tanaka, M. Tasaka, H. H. Cao and M. Shionoya, Eur. J. Pharm. Sci., 2001, 13, 77 CrossRef CAS.
- M. Tasaka, K. Tanaka, M. Shiro and M. Shionoya, Supramol. Chem., 2001, 13, 671 CrossRef CAS.
- K. Tanaka, A. Tengeiji, T. Kato, N. Toyama, M. Shiro and M. Shionoya, J. Am. Chem. Soc., 2002, 124, 12494 CrossRef CAS.
- E. Freisinger, I. B. Rother, M. S. Luth and B. Lippert, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 3748 CrossRef CAS.
- A. Kusel, J. H. Zhang, M. A. Gil, A. C. Stuckl, W. Meyer-Klaucke, F. Meyer and U. Diederichsen, Eur. J. Inorg. Chem., 2005, 4317 CrossRef.
- R. M. McKinlay, G. W. V. Cave and J. L. Atwood, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 5944 CrossRef CAS.
- R. M. McKinlay, P. K. Thallapally and J. L. Atwood, Chem. Commun., 2006, 2956 RSC.
- A. M. Todd, K. M. Anderson, P. Byrne, A. E. Goeta and J. W. Steed, Cryst. Growth Des., 2006, 6, 1750 CrossRef CAS.
- M. Tiliakos, P. Cordopatis, A. Terzis, C. P. Raptopoulou, S. P. Perlepes and E. Manessi-Zoupa, Polyhedron, 2001, 20, 2203 CrossRef CAS.
- F. Kyriakidou, A. Panagiotopoulos, S. P. Perlepes, E. ManessiZoupa, C. P. Raptopoulou and A. Terzis, Polyhedron, 1996, 15, 1031 CrossRef.
- J. Bernstein, R. E. Davis, L. Shimoni and N.-L. Chang, Angew. Chem., Int. Ed. Engl., 1995, 34, 1555 CrossRef CAS.
- D. R. Turner, B. Smith, A. E. Goeta, I. R. Evans, D. A. Tocher, J. A. K. Howard and J. W. Steed, CrystEngComm, 2004, 6, 633 RSC.
- P. Baran, D. Valigura, I. Svoboda and H. Fuess, Z. Kristallogr., 1992, 202, 142 CAS.
- L. S. Reddy, S. Basavoju, V. R. Vangala and A. Nangia, Cryst. Growth Des., 2006, 6, 161 CrossRef CAS.
- P. Byrne, D. R. Turner, G. O. Lloyd, N. Clarke and J. W. Steed, Cryst. Growth Des., 2008, 8, 3335 CrossRef CAS.
- M. C. Etter, Acc. Chem. Res., 1990, 23, 120 CrossRef CAS.
- J. T. Lenthall, K. M. Anderson, S. J. Smith and J. W. Steed, Cryst. Growth Des., 2007, 7, 1858 CrossRef CAS.
- C. R. Calladine and H. R. Drew, Understanding DNA: The Molecule and How it Works, 2nd edn, Academic Press, San Diego, 1997 Search PubMed.
Footnotes |
| † CCDC reference number 696257. For crystallographic data in CIF or other electronic format see DOI: 10.1039/b815252g |
| ‡ Crystal data for 2: C48H62Cl4Cu2N12O7, M = 1187.98, green plate, 0.30 × 0.10 × 0.05 mm3, monoclinic, space groupP21/n (No. 14), a = 10.6915(3), b = 26.3108(7), c = 18.9615(5) Å, β = 99.035(1)°, V = 5267.7(2) Å3, Z = 4, Dc = 1.498 g cm−3, F000 = 2464, SMART 6k, Mo Kα radiation, λ = 0.71073 Å, T = 120(2) K, 2θmax = 58.4°, 88778 reflections collected, 14225 unique (Rint = 0.0772). Final GooF = 1.008, R1 = 0.0398, wR2 = 0.0864, R indices based on 9875 reflections with I > 2σ(I) (refinement on F2), 690 parameters, 0 restraints. Lp and absorption corrections applied, μ = 1.073 mm−1. |
| This journal is © The Royal Society of Chemistry 2009 |