Efficient generation of dendritic arrays of cross-linked hemoglobin: symmetry and redundancy†
Dongxin
Hu
and
Ronald
Kluger
*
Davenport Laboratories, Department of Chemistry, University of Toronto, 80 St. George Street, Toronto, Ontario, Canada M5S 3H. E-mail: rkluger@chem.utoronto.ca
First published on 16th November 2007
Abstract
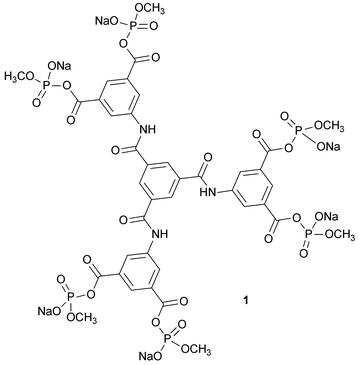
Chemically connected protein arrays have significant diverse applications including the production of red cell substitutes, bioconjugate drug delivery, and protein therapies. In order to make materials of defined structure, there is a need for efficient and accessible reagents. While chemical cross-linking with a multi-subunit protein can be achieved in high yield, connecting proteins to one another in a dendritic assembly along with concurrent cross-linking has met with limited success. This has now been overcome through the design and implementation of a readily prepared reagent with added reaction sites that compensate for competing hydrolysis. N,N′,N″-Tris[bis(sodium methyl phosphate)isophthalyl]-1,3,5-benzenetricarboxamide (1), a hexakis(methyl phosphate) isophthalyl trimesoyl tris-amide, was designed and synthesized in high yield in three stages from a reactive trimesoyl core. This material has three pairs of coplanar cross-linking reaction sites in a symmetrical array. The presence of three sets of sites greatly increases the probability that at least two sets will produce cross-links within hemoglobin tetramers (in competition with hydrolysis) and thereby connect two cross-linked tetramers at the same time. Reaction of 1 with deoxyhemoglobin at pH 8.5 gives a material that contains two cross-linked hemoglobin tetramers connected to one another and to a constituent αβ dimer. Products were characterized by SDS-PAGE, MS, enzyme digestion and HPLC. The isolated dendritic-hemoglobin with 2.5 tetrameric components has the same oxygen affinity as native hemoglobin (P50 = 5.0 torr) and retains cooperativity (n50 = 2.0). Analysis of circular dichroism spectra indicates that the assembly retains proper folding of the globin chains while the hemes are in an altered environment.
Introduction
Chemical cross-linking of proteins with site-directed reagents1–3 can convert unstable proteins into stable, homogeneous functional entities.4 Extension of the concept to include connection of proteins to one another to create defined dendritic assemblies5,6 is a further challenge with a number of important applications as illustrated by red cell substitutes7,8 and extends to diverse fields including drug conjugation and protein therapeutics.9 We note that successful clinical trials have been reported for materials made from heterogeneous arrays generated in the reaction of glutaraldehyde and hemoglobin,10 while trials with smaller species were not successful. By creating similarly-sized assemblies of proteins but with defined structures, information on structure and efficacy can be accessed. Thus, we have sought to design reagents that produce cross-links within a multimeric protein along with covalent linkages between proteins .11–13 With these reagents, selective inter-protein linking occurs in low yield. The major reason for the low yields is the competitive hydrolysis of the reactive functional group on the reagent, which results in the loss of a reactive essential site. In considering ways to overcome this, we reasoned that the hydrolytic loss of a single reactive group would be compensated by additional symmetrical reaction sites available within the reagent. These built-in redundancies increase the probability of the proper reaction and also provide a statistical advantage in competing with hydrolysis.14,15We now report the successful implementation of such a strategy based on the convenient synthesis of a multifunctional reagent N,N′,N″-tris[bis(sodium methyl phosphate)isophthalyl]-1,3,5-benzenetricarboxamide (1). Our approach to the synthesis of 1 is based on using symmetrical multi-sited core materials.11–13 In a practical test, reaction of 1 with hemoglobin efficiently introduces both cross-links and inter-protein links. The isolated dendritic protein assemblies are fully functional in cooperative oxygen binding and release.
Experimental
Solvents
THF was dried by distillation from metallic sodium. Acetone was dried by distilling from dry gypsum.Hemoglobin
The protein was purified from human red cells by the method described by Winslow et al.16 Purified hemoglobin was stored in doubly distilled water at 0–5 °C. Concentrations of hemoglobin solutions were determined using the cyanomethemoglobin assay described by Salvati and Tentori.17 The purity of hemoglobin was determined using the reverse-phase HPLC (RP HPLC) analysis described by Jones.18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. The solid product was washed five times with water to remove the organic solvent. The (wet) solid was lyophilized overnight to produce a pale yellow powder (3.35 g, 95.8% yield). 1H NMR (DMSO-d6): δ ppm 13.38 (broad peak, COOH), 10.96 (s, 3H, CONH), 8.84 (s, 3H, ArH), 8.73 (d, 6H, J = 1.2 Hz, ArH), 8.26 (t, 3H, J = 1.2 Hz, ArH); MS (ESI, methanol): C33H21N3O15, 698.1([M − H+]−). Mp: >300 °C.
000 rpm. The solid product was washed five times with water to remove the organic solvent. The (wet) solid was lyophilized overnight to produce a pale yellow powder (3.35 g, 95.8% yield). 1H NMR (DMSO-d6): δ ppm 13.38 (broad peak, COOH), 10.96 (s, 3H, CONH), 8.84 (s, 3H, ArH), 8.73 (d, 6H, J = 1.2 Hz, ArH), 8.26 (t, 3H, J = 1.2 Hz, ArH); MS (ESI, methanol): C33H21N3O15, 698.1([M − H+]−). Mp: >300 °C.
Reaction with deoxyhemoglobin
Carbonmonoxyhemoglobin (HbCO, 5 mL, 1.0 mM) was equilibrated through a Sephadex G-25 column (250 × 35 mm) into sodium borate buffer (0.05 M, pH 8.5) at 4 °C. The solution (∼0.5 mM) was oxygenated and deoxygenated to give deoxyhemoglobin (deoxyHb).12 Different amounts of 1 were added as a solid into the deoxyHb solution. The reaction was carried out for 18 h at 37 °C under nitrogen. Carbon monoxide was then passed over the modified hemoglobin mixture for 15 min to protect the hemes. The modified HbCO was transferred to MOPS buffer (0.1 M, pH 7.2) through a Sephadex G-25 column (250 × 35 mm) at 4 °C. The mixture was concentrated and stored at 4 °C.SDS-PAGE analysis
The molecular weights of constituent proteins were estimated using polyacrylamide gel (12%, Tris-HCl) electrophoresis. Two dimensional Tris-HCl polyacrylamide gels comprised of 12% separating gel (pH 8.8) and 5% stacking gel (pH 6.8) both with 10% sodium dodecyl sulfate. Protein samples were treated with 2-mercaptoethanol and sodium dodecyl sulfate.20 Globin chains were further denatured by heating at 100 °C for 15 min before loading on the gel. Finished gels were stained with Coomassie Brilliant Blue. Details are described in earlier papers.11Analytical reverse-phase HPLC
A 330 Å pore-size C-4 Vydac column (4.6 × 250 mm) was used to separate modified globin chains. Gradient elution was initiated with 20% acetonitrile and ran to 60% acetonitrile with 0.1% (v/v) TFA as developer.18 The effluent was monitored at 220 nm.Size exclusion HPLC
Superdex G-75 HR (10 × 300 mm) column was used to investigate the size distribution of the modified protein mixtures. Samples were eluted under conditions that dissociate the hemoglobin tetramers into dimers (25 mM Tris-HCl, pH 7.4, 0.5 M magnesium chloride).21 The effluent was monitored at 280 nm and 414 nm.Isolation of modified hemoglobins
The modified hemoglobins were separated on gel-filtration chromatography (Sephadex G-100, 1000 × 35 mm) eluting with 25 mM Tris-HCl, pH = 7.4 containing 0.5 M magnesium chloride.21 Fractions were concentrated and analyzed using Superdex G-75 size exclusion HPLC, C4 reverse-phase analytical HPLC and SDS-PAGE.Tryptic and Glu-C digest
Dehemed native β globin chains and cross-linked β globin chains were collected from preparative C4 reverse-phase HPLC and lyophilized. The globin chains were then dissolved with 8 M urea in 80 mM ammonium bicarbonate (pH 8.5) and kept for 4 h at room temperature. The protein samples were hydrolyzed by TPCK-treated trypsin (2% w/w) at 37 °C for 24 h followed by endoproteinase Glu-C Staphylococcus aureus V8 (1% w/w) at 37 °C for 72 h in 80 mM ammonium bicarbonate (pH 8.5). The hydrolytes were analyzed by HPLC reverse-phase on a Vydac C18 column (250 × 4.5 mm). Peptide fragments were separated using gradients of acetonitrile from 0% to 100% with 0.1% THF as the developer.19 The peaks for peptide fragments were labeled based upon MALDI-TOF mass analysis results of hydrolytes.22 The effluents were monitored at 220 nm for all peptide fragments, 280 nm for peptide fragments containing tryptophan and tyrosine and 258 nm for phthalates.22Oxygen-binding analysis
Purified cross-linked hemoglobin (∼1 g L−1) and native hemoglobin (∼1 g L−1) were transferred to sodium phosphate buffer (I = 0.1 M, pH = 7.4) and oxygenated. Oxygen-binding curves were obtained using a Hemox-Analyzer at 25 °C. The data were fitted to the Adair equation to obtain P50 and n50.Circular dichroism spectroscopy conformational study
Native hemoglobin and cross-linked dendritic-hemoglobin in CO-bound form were studied by circular dichroism spectrometry in both far (200–260 nm) and near (260–470 nm) ultra-violet regions at room temperature.23 Hemoglobin samples were prepared as 10 mL solutions of the same globin chain (heme) concentration in the sodium phosphate buffer (I = 0.02 M, pH = 7.4). The globin chain (heme) concentration of the hemoglobin samples was 0.5 µM for the far-UV study and 5.0 µM for the near-UV study.Results and discussion
The tris[bis(sodium methyl phosphate)isophthalyl]-1,3,5-benzenetricarboxamide (1) contains six anionic electrophilic sites that are expected to acylate amino groups with high efficiency in cationic regions of proteins . The six sites are arranged in three pairs with a span in each pair that is expected to connect different β subunits of hemoglobin through reaction at the ε-amino group of β-Lys-82 or the N-terminal amino group of β-Val-1.11–13 Once a cross-link within a hemoglobin tetramer has formed between a pair of sites, the remaining sites can combine with additional tetramers and various cross-linked species are connected to one another. While tetrafunctional reagents can cross-link within two tetramers,11–13 they are inefficient due to the competing hydrolysis of the reagent: if hydrolysis inactivates only one site of four, the reagent becomes incapable of producing species with two cross-linked hemoglobins.15 With an additional set of reaction sites, as in 1, the chances of forming connected tetramers is increased by 50%, and hydrolysis of a site leaves the reagent still able to produce cross-linked bis-tetramers. Furthermore, where there are single modifications (no cross-link forms) due to one site being hydrolyzed, the resulting modified subunit will be connected to the bis-tetramer. The collection of altered protein materials all have defined structures.Multifunctional reagent 1 was synthesized through a series of convergent coupling reactions (Scheme 1). Trimesoyl chloride reacts readily with three molar equivalents of 5-aminoisophthalic acid to form the hexa-carboxylic acid precursor. This was quantitatively converted to the corresponding acid chloride, which combines with sodium dimethyl phosphate to give the dimethyl phosphoryl derivative of each carboxylate. The six highly reactive phosphodiesters of the anhydrides in the reagent were converted to the monoesters (1) by six-fold mono-demethylation with sodium iodide. The structure of 1 was confirmed by a combination of analytical procedures.
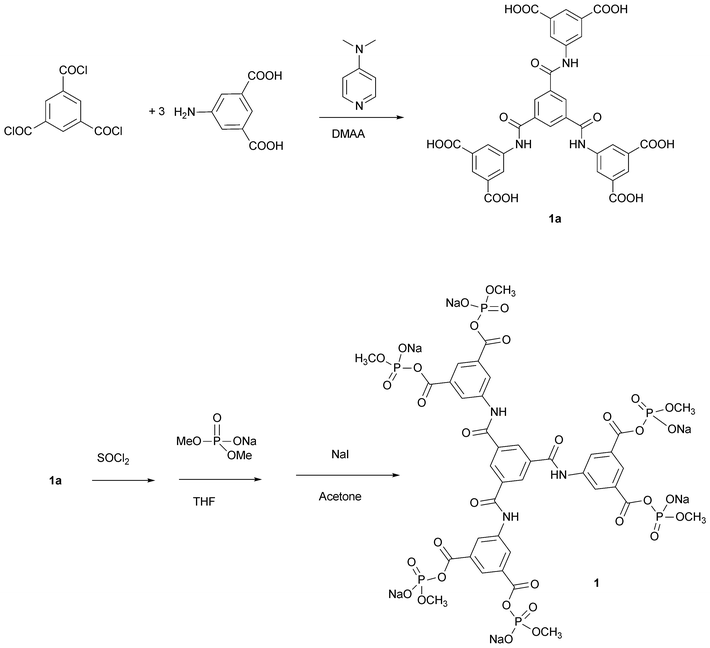 | ||
| Scheme 1 Synthesis of reagent 1. | ||
We used AM-1 semi-empirical methods to calculate the lowest energy conformation of 1. The structure resembles a three-bladed propeller with the trimesoyl core as its hub and each isophthalyl group twisted out of plane (Fig. 1). The carbonyl carbons in the acyl phosphate moieties of the meta-substituted rings are separated by about 5 Å, which is ideal for cross-linking, while the functional groups on adjacent rings are separated by about 12 Å, which gives enough separation to connect different tetramers.11
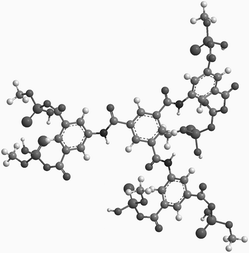 | ||
| Fig. 1 Molecular model of 1 from semi-empirical molecular orbital calculations. | ||
Reactions of 1 with deoxyhemoglobin were most effectively achieved in pH 8.5 solution, where unprotonated amino groups are present in substantial proportions. The site of reaction was determined by the established combination of reverse-phase HPLC analysis and enzyme-digest peptide mapping.22 This revealed that β chains are exclusively modified, as expected. (Modified β chains elute after the native α chains, while native β chains elute ahead of the α chains, Fig. 2).
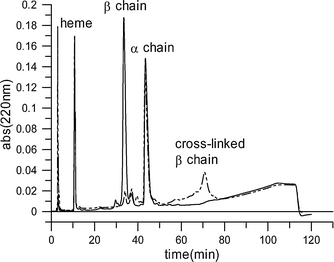 | ||
| Fig. 2 C-4 Reverse-phase HPLC analysis of modified hemoglobin globin chains (dashed line) compared with globin chains in the native protein (solid line). | ||
SDS-PAGE analysis employs conditions that denature the globin chains and separate all units into monomers.20 Unmodified hemoglobin produces a single band for the free α and β chains at a position corresponding to their similar molecular weight of ∼16 kDa. Chemical cross-linking of globin subunits causes new peaks to appear that correspond to a mass that is the sum of the (n) constituent subunits at n × 16 kDa.
When deoxyhemoglobin was combined with one equivalent of 1 (six reacting sites in one equivalent of 1), the highest molecular weight component gives a band at ∼80 kDa on SDS-PAGE (ESI† ), which corresponds to five covalently linked β subunits (from connections of two cross-linked tetramers and one αβ dimer). For comparison, a band at ∼32 kDa is the expected result from two covalently linked β subunits arising from ββ′-cross-linked hemoglobin tetramers and has been observed previously.22 Reactions of deoxyhemoglobin with 1.5 equivalents of 1 results in a product at ∼48 kDa, indicating the presence of three covalently linked β chains (a cross-linked tetramer and a dimer connected to one another). Reactions of deoxyhemoglobin with less than one equivalent or with more than two equivalents of 1 gives bands no higher in weight than 32 kDa, indicating that dendritic cross-linking is not taking place under these conditions.
Size exclusion HPLC can be conducted under conditions that cause hemoglobin tetramers without cross-links to dissociate into αβ dimers (pH 7.4 Tris-HCl, 0.5 M magnesium chloride21). Native hemoglobin was added to an aliquot of the modified hemoglobin mixture and eluted under the same conditions as the modified material was eluted on its own in order to verify the assignment of each peak based on coincident elution. Under these conditions, modified hemoglobin produces two peaks, which accounts for the two main cross-linked hemoglobin products. They appear ahead of the peak for the free αβ dimers of native hemoglobin. Integration of the peak area indicates that the highest weight component (∼160 kDa, five αβ dimers) accounts for ∼47% of the modified protein , a very high yield in comparison with previous efforts.11–13 A schematic representation of this structure is shown in Fig. 3.
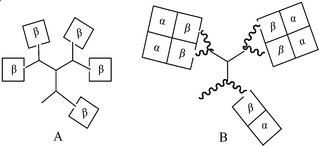 | ||
| Fig. 3 Schematic structures of cross-linked dendritic-hemoglobins: A is obtained in SDS-PAGE and RP HPLC; B is from gel-filtration chromatography . | ||
This analysis of the cross-linked dendritic-hemoglobin that was isolated by gel-filtration chromatography confirmed that the purified sample contains a single entity with uniform molecular size (Fig. 2 and Fig. 4). SDS-PAGE analysis (ESI† ) showed the expected band for the cross-linked β subunits at ∼80 kDa and the band at ∼16 kDa for the free α subunits.
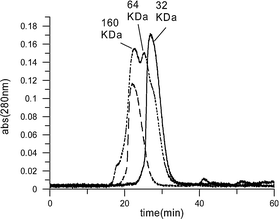 | ||
| Fig. 4 Size-exclusion (G-75) gel filtration HPLC analysis of modified hemoglobins run under conditions that cause dissociation of tetramers into dimers:21 native hemoglobin (solid line); modified hemoglobin mixture (dotted line); cross-linked dendritic-hemoglobin (dashed line). | ||
Peptide mapping of the product through tryptic and Glu-C digestion combined with C-18 reverse-phase HPLC fragment analysis (ESI† ) and MALDI-TOF mass analysis showed that ε-amino groups of β lysine 82 were converted to amides and were thus not susceptible to digestion at that peptide as in the native protein (the N-terminal amino group was also modified. This changes the elution pattern of the first tryptic peptide). This result is consistent with the specificity of previously reported isophthalyl-derived regents.11–13
Measurements of oxygen-binding properties of the product showed that the 160 kDa entity binds and releases oxygen cooperatively (n50 = 2.0) with the same overall oxygen-affinity (P50 = 5.0 torr) as native hemoglobin (Fig. 5 and 6). This indicates that the modification does not affect the potential for the material to function an oxygen transport agent.
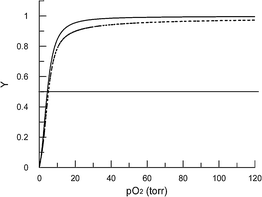 | ||
| Fig. 5 Oxygen-binding curves for cross-linked dendritic-hemoglobin (solid line, P50 5.0 torr) and native hemoglobin (dashed line, P50 5.0 torr). | ||
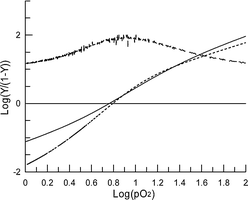 | ||
| Fig. 6 Hill plot of data from Fig. 5. The solid curve is for cross-linked dendritic-hemoglobin (n50 = 2.0) and the dotted line is for the native protein (n50 = 2.7). The slope of the Hill plot of cross-linked dendritic-hemoglobin is plotted as the upper dashed line. | ||
CD spectra of native hemoglobin and cross-linked hemoglobins are nearly identical in the far UV region (Fig. 7: 220 nm to 260 nm), where the signals arise from the globin chains. However, there are significant differences in the near-UV–VIS region (240 nm to 470 nm), where the heme environment is primarily responsible for the absorbance.23 This altered heme environment after modification accounts for the different oxygen-binding properties of chemically modified hemoglobin species.
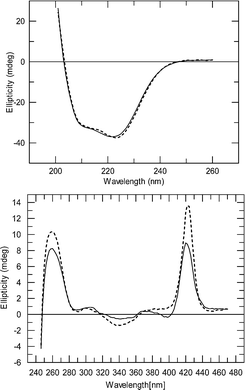 | ||
| Fig. 7 CD spectra of native hemoglobin (dashed line) and cross-linked dendritic-hemoglobin (solid line) in the CO-bound state. The far UV region (upper) was recorded at low concentration and shows the protein spectrum. The UV–visible region (lower) was recorded at high concentration and shows the heme spectrum. | ||
Conclusions
The reaction of one molar-equivalent of 1 with deoxyhemoglobin provides for the first time a practical route to a dendritic species with cross-linked hemoglobin tetramers connected to one another. A major operational advantage of 1 is the presence of additional reaction sites that can compensate for competing hydrolysis and which may also assist in orientation of the reacting groups. The symmetry of 1 permits its efficient preparation. The reagent has six acylation sites and it is only required that four react to produce bis-tetramers. The relatively rigid core structure resulting from the extended conjugation among the aromatic rings and the amide linkages prevents the compound from folding onto itself, keeping the reacting sites maximally separated. Although the core structure of the linker is probably hydrophobic, the charges of the monomethyl phosphates make the molecule very water-soluble. It reacts rapidly with dissolved hemoglobin in aqueous buffer. The oxygen-binding properties of the product give an indication of the effects of protein –protein interactions within the cross-linked conjoined hemoglobin (five covalently linked αβ dimers). The somewhat reduced cooperativity (compared to the native protein ) is typical of cross-linked bis-tetramers (n50 = 2.0) and is only somewhat lower than that of native hemoglobin (n50 = 2.7). The overall oxygen affinity is close to that of the native protein (P50 = 5.0 torr). The covalent links stabilize the inherent tetrameric association, which is essential for the cooperativity and moderate oxygen affinity. It is likely that the relative rigidity of the cross-link and the large hydrophobic core of the product affect the environment of the hemes and reduce cooperativity.Acknowledgements
We thank the Natural Sciences and Engineering Research Council of Canada for support through a Strategic Project.References
- R. Kluger, Synlett, 2000, 12, 1708–1720.
- R. Kluger, A. S. Grant, S. L. Bearne and M. R. Trachsel, J. Org. Chem., 1990, 55, 2864–2868 CrossRef CAS.
- S. R. Snyder, E. V. Welty, R. Y. Walder, L. A. Williams and J. A. Walder, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 7280–7284 CrossRef CAS.
- M. F. Perutz, M. G. Rossmann, A. F. Cullis, H. Muirhead, G. Will and A. C. T. North, Nature, 1960, 185, 416–422 CAS.
- J. Simoni, G. Simoni, C. D. Lox, S. D. Prien and G. T. Shires, Artif. Cells, Blood Substitutes, Immobilization Biotechnol., 1997, 25, 193–210 CAS.
- A. Bellelli, P. L. Benedetti, M. Coletta, R. Ippoliti and M. Brunori, Biochem. Biophys. Res. Commun., 1988, 156, 970–977 CrossRef CAS.
- J. G. Riess, Chem. Rev., 2001, 101, 2797–2919 CrossRef CAS.
- R. M. Winslow, J. Intern. Med., 2003, 253, 508–517 CrossRef CAS.
- T. M. S. Chang, Ann. N. Y. Acad. Sci., 1999, 875, 71–83 CrossRef CAS.
- S. A. Gould, E. E. Moore, D. B. Hoyt, P. M. Ness, E. J. Norris, J. L. Carson, G. A. Hides, I. H. G. Freeman, R. DeWoskin and G. S. Moss, J. Am. Coll. Surg., 2002, 195, 445–452 CrossRef.
- R. Kluger, J. Lock-O'Brien and A. Teytelboym, J. Am. Chem. Soc., 1999, 121, 6780–6785 CrossRef CAS.
- N. Gourianov and R. Kluger, J. Am. Chem. Soc., 2003, 125, 10885–10892 CrossRef CAS.
- N. Gourianov and R. Kluger, Biochemistry, 2005, 44, 14989–14999 CrossRef CAS.
- R. Kluger, Y. Song, J. Wodzinska, C. Head, T. S. Fujita and R. T. Jones, J. Am. Chem. Soc., 1992, 114, 9275–9279 CrossRef CAS.
- K. Paal, R. T. Jones and R. Kluger, J. Am. Chem. Soc., 1996, 118, 10380–10383 CrossRef CAS.
- K. D. Vandegriff, A. Malavalli, J. Wooldridge, J. Lohman and R. M. Winslow, Transfusion (Malden, MA, US), 2003, 43, 509–516 Search PubMed.
- A. M. Salvati, L. Tentori and G. Vivaldi, Clin. Chim. Acta, 1965, 11, 477–479 CrossRef CAS.
- R. T. Jones, Methods Enzymol., 1994, 231, 322–343 CAS.
- J. B. Shelton, J. R. Shelton and W. A. Schroeder, J. Liq. Chromatogr., 1984, 7, 1969–1977 CrossRef CAS.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CAS.
- G. Guidotti, J. Biol. Chem., 1967, 242, 3685–3693 CAS.
- R. T. Jones, C. G. Head, T. S. Fujita, D. T. B. Shih, J. Wodzinska and R. Kluger, Biochemistry, 1993, 32, 215–223 CrossRef CAS.
- J. J. M. De Llano and J. M. Manning, Protein Sci., 1994, 3, 1206–1212 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: SDS-PAGE analysis and reverse-phase HPLC fragment analysis. See DOI: 10.1039/b714684a |
| This journal is © The Royal Society of Chemistry 2008 |