On the use of cyclic voltammetry for the study of anodic electron transfer in microbial fuel cells
Katja
Fricke
,
Falk
Harnisch
and
Uwe
Schröder
*
University of Greifswald, Institute of Biochemistry, Felix-Hausdorff-Strasse 4, Greifswald, 17487, Germany
First published on 17th June 2008
Abstract
In this communication we discuss, by means of the metal reducing bacterium Geobacter sulfurreducens, a strategy to use cyclic voltammetry for the study of anodic bioelectrocatalytic electron transfer in microbial fuel cells.
Although the mechanisms of bioelectrocatalytic substrate oxidation processes in microbial fuel cells and, especially, of anodic electron transfer are of utmost importance for the performance of microbial fuel cells,1 little is known, so far, of the nature of the underlying mechanisms. For this reason, research activities in this field have considerably intensified over the past years. Different concepts and mechanisms for electron transfer from the biocatalyst to the fuel cell anode have been proposed.2 Thus, it can be distinguished between direct electron transfer (DET) and mediated electron transfer (MET) mechanisms. Examples of DET are, the electron transferviamembrane bound cytochromes (e.g., from Geobacter sp.,3Rhodoferax ferrireducens)4 or via conductive bacterial pili (“nanowires”), as recently proposed for Shewanella oneidensis MR-1.5MET, on the other hand, has been reported to occur either by primary metabolites (e.g., hydrogen, formate)6–8 or by secondary metabolites, such as phenazine derivates9 or quinones10 or flavines, as has just been discovered for Shewanella oneidensis MR-1.11
Many of these proposed transfer mechanisms are of a putative nature and are controversially discussed. Often, the involved redox species are barely identified or understood. Some of the greatest challenges in their study are (i) the complexity of the microbial metabolism, (ii) the often extremely low concentrations of the involved redox species and (iii) the complex (electro)chemical nature of bacterial cultures and even of microbial cell membranes, which may contain several redox active species that do not necessarily contribute to the bioelectrocatalytic current flow.
Cyclic voltammetry is a standard tool in electrochemistry12 and has regularly been exploited to study and to characterize the electron transfer interactions between microorganisms or microbial biofilms and microbial fuel cell anodes.13–15 In most of these publications the microorganisms have been studied either in their bioelectrocatalytically active state or in the inactive state. In this communication, we demonstrate that by applying cyclic voltammetry at different stages of microbial growth and metabolic activity, valuable information on the anodic electron transfer processes in microbial fuel cells can be gained. We also show that due to the complexity of the underlying processes, the simplified assumptions and models of anodic electron transfer are difficult to prove.
The experiments described in this communication are based on Geobacter sulfurreducens biofilm modified graphite electrodes, obtained when graphite electrodes are immersed in the inoculated medium and are poised at a constant anodic potential (here +0.3 V, vs.Ag/AgCl, sat. KCl), in order to form an electrochemically active bacterial biofilm.16Fig. 1 illustrates such biofilm formation and bioelectrocatalytic current generation by means of a chronoamperometric experiment. The figure shows that 168 h after inoculation with G. sulfurreducens, the electrode reached a first activity maximum, followed by a decrease in the current, caused by substrate exhaustion. After replenishment of the bacterial medium at 210 h the current generation commenced again, reaching a maximum current density of about 75 μA cm−2, 265 h after inoculation.
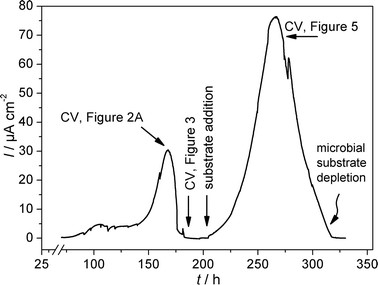 | ||
| Fig. 1 Chronoamperometric plot of the formation and the bioelectrocatalytic activity of a G. sulfurreducens biofilm at a graphite electrode; semi-batch experiment, substrate: 10 mM L−1 acetate. | ||
By recording cyclic voltammograms at different stages of biofilm formation and substrate availability (and thus different stages of current generation), valuable information on the electron transfer mechanism can be gained. Fig. 2A shows a typical cyclic voltammogram of a G. sulfurreducens biofilm modified electrode recorded at the first maximum of the bioelectrocatalytic activity (see Fig. 1). The figure shows a typical sigmoidal shape based on, at first glance, one single underlying redox centre. Yet, the first derivative of this voltammetric curve (Fig. 2B) reveals that the oxidative as well as reductive potential sweep possess two inflection points, reflected by two maxima in the derivative curve. This behaviour has also recently been reported for artificially immobilized G. sulfurreducenscells.17
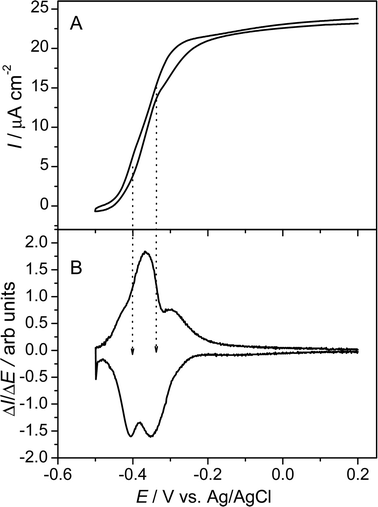 | ||
| Fig. 2 (A) Cyclic voltammogram of a metabolizing G. sulfurreducens biofilm. The voltammogram was recorded at maximum biofilm activity 168 h after the start of the chronoamperometric experiment (see Fig. 1). The scan rate was 5 mV s−1. (B) First derivatives of the voltammetric curve over the potential. | ||
A more detailed view on the electrochemical features of the biofilm are obtained at substrate depletion (in this experiment 190 h after inoculation; see Fig. 1). Under these non-catalytic conditions, the voltammetric behaviour strongly depends on the scan rate of the voltammetric experiment. As illustrated in Fig. 3A, for scan rates above 20 mV s−1 the voltammogram shows one major redox system, at a formal potential of −0.331 V. The shape of the voltammogram appears typical for bacteria, whose electrochemical activity is ascribed to outer membrane cytochromes.18,19 At lower scan rates (see Fig. 3B), the simplicity of the voltammogram gives way to more complex behaviour. Now, four major redox systems can be distinguished, indicated as systems 1–4. Besides the two less evolved redox systems, at −0.515 V (Ef,1) and +0.059 V (Ef,4), the major redox system in Fig. 3A is split into two systems, with formal potentials of −0.376 V and −0.295 V (Ef,2 and Ef,3). Both systems possess a fine structure, i.e., they may consist of further redox processes. None of the redox peaks are obtained in the sterile culture medium or at a blank (biofilm-less) graphite electrode in the inoculated medium. Additionally, all peaks remain present when the medium is exchanged to a fresh, sterile medium, which demonstrates that all redox signals are caused by biofilm based redox compounds. A comparison of Fig. 2B and 3B reveals that both redox processes associated with the formal potentials Ef,2 and Ef,3 contribute to the bioelectrocatalytic anodic electron transfer (see Fig. 2), whereas, systems 1 and 4 appear electrocatalytically inactive. From the literature, it can be derived that electron transfer from G. sulfurreducens to a solid electron acceptor is accomplished by outer membrane cytochromes, like OmcB, OmcE and OmcS.20–22 For OmcB, a formal potential of −190 mV vs.SHE (corresponding to −387 mV vs.Ag/AgCl) has been reported.23 This potential most likely corresponds to redox system 2 (−376 mV, vs.Ag/AgCl), found in our experiments. The presence of a second involved species at more positive potential now indicates a parallel, competing path, possibly via a similar membrane associated species.
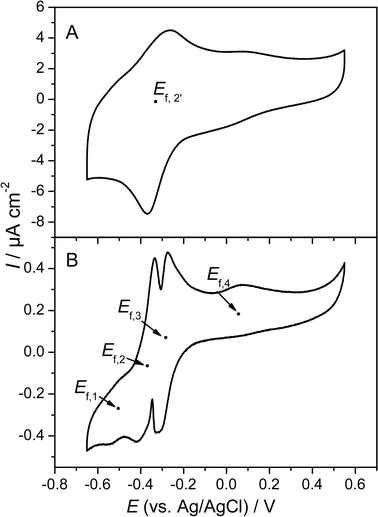 | ||
| Fig. 3 Cyclic voltammograms of a G. sulfurreducens biofilm. The voltammograms were recorded in a substrate depleted culture medium (see Fig. 1). Scan rate: (A) 50 mV s−1, (B) 1 mV s−1. | ||
The anodic electron transfer of G. sulfurreducens is considered to be an archetype for direct electron transfer.24 From an electrochemical point of view, one would (for redox systems 2 and 3) expect typical thin film behaviour, manifested as e.g., a linear relationship of the peak current with the scan rate (i∼v) in cyclic voltammetry. Yet, how much the electrochemistry of living bacterial cells (and bacterial biofilms) may differ from that of isolated and immobilized redox enzymes or other “simple” redox compounds is illustrated in Fig. 4. These experimental results show a scan rate dependence of the peak currents of i∼v0.50 (reduction peak) i∼v0.70 (oxidation peak) as derived from the slope of the double logarithmic plot of the peak currents versus scan rate (not shown). This dependence strongly indicates diffusion control, a result that cannot be explained by the current knowledge of DET. It can be speculated that the electron hopping between the heme centres of the bacterial outer membrane cytochromes, or electron transfer between electrochemically linked redox enzymes show diffusion characteristics in cyclic voltammetry.
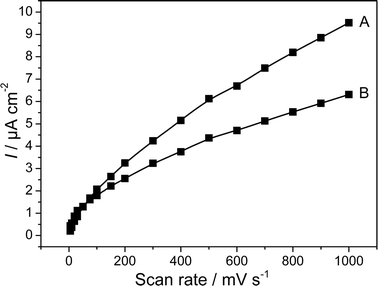 | ||
| Fig. 4 Absolute values of the peak currents of a G. sulfurreducens biofilm electrode as a function of the scan rate. The voltammograms were recorded in a substrate depleted culture. (A) Oxidation peak, (B) reduction peak. | ||
At increasing bioelectrocatalytic activity, as is found after replenishment of the medium (see Fig. 1), the electrocatalytic curve (Fig. 5) did not reveal any fine structure. Here, the electrocatalytic wave possessed only a single inflexion point (and thus, only one single maximum in first the derivative ΔI/ΔE), at a potential of −0.335 V, corresponding to the arithmetic mean of systems 2 and 3 (Fig. 2 and Fig. 3). Yet, after subsequent substrate exhaustion, again a cyclic voltammogram equivalent to that shown in Fig. 3B was obtained, which showed that both redox systems, 2 and 3, were still present as separate features. It can be assumed, that at high catalytic activity these features may be resolved only at extremely small scan rates.
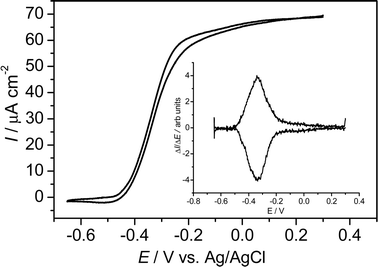 | ||
| Fig. 5 Cyclic voltammogram of a metabolizing G. sulfurreducens biofilm. The voltammogram was recorded at a scan rate of 5 mV s−1 268 h after the start of the chronoamperometric experiment (see Fig. 1). Inset: first derivatives of the voltammetric curve over the potential. | ||
The height of the bioelectrocatalytic current strongly correlates with the peak currents measured under substrate depletion. For example, the increase of the bioelectrocatalytic performance from 30 μA cm−2 at 168 h by a factor of 2.5 to 76 μA cm−2 at 265 h (see chronoamperometric curve, Fig. 1) was accompanied by a proportional increase (factor 2.5) of the peak currents measured by cyclic voltammetry at the subsequent periods of substrate depletion (at 190 h and 320 h, respectively). This finding indicates that the increase of the electrocatalytic biofilm activity is either caused by a growing cell density at the electrode surface, or by an increase in the number of membrane bound electron transfer proteins in the individual cells.
In summary, we demonstrate the principle suitability of cyclic voltammetry to study biofilm based anodic electron transfer processes in microbial fuel cells. The experiments underline the advantages of performing voltammetric experiments at different stages of biofilm growth and activity. We also demonstrate that the complexity of microbial and bioelectrochemical processes make evaluation and interpretation of the voltammetric results a challenging endeavour. The results show that data gained by a purely electrochemical study needs to be combined with data from, e.g., molecular biology and metabolomics, in order to gain a better understanding of the mechanisms of cell–electrode interactions at the molecular level.
Experimental
G. sulfurreducens strain ATCC 51573 was purchased from DSMZ. All incubations were performed at 30 °C. Growth medium contained (per L): 0.1 g of KCl, 0.15 g of NH4Cl, 0.6 g of Na2HPO4, 0.82 g of Na acetate, 2.5 g of NaHCO3, 10 mL of trace element solution, 10 mL of vitamin solution and 1 mL of selenite–tungstate solution. The medium was adjusted to pH 6.8 and flushed with N2–CO2 (80 : 20).All electrochemical experiments were carried out under potentiostatic control utilizing a three electrode arrangement, consisting of the working electrode, a Ag/AgCl reference electrode (sat. KCl, Sensortechnik Meinsberg, Germany, 0.195 V vs.SHE) and a counter electrode (platinum wire or a carbon rod electrode). The experiments were conducted with PGSTAT 30 Autolab systems (Ecochemie, Netherlands). Sealed and thermostated vessels (100 mL) served as electrochemical cells which hosted the fermentation medium and the electrodes. All experiments were performed under strictly anoxic conditions.
Acknowledgements
The authors gratefully acknowledge support by the DFG. U.S. acknowledges the grant of a Heisenberg Fellowship by the DFG, F.H. thanks the Deutsche Bundesstifung Umwelt and the Studienstiftung des Deutschen Volkes for PhD Scholarships.References
- U. Schröder, Phys. Chem. Chem. Phys., 2007, 9, 2619–2629 RSC.
- B. E. Logan, B. Hamelers, R. Rozendal, U. Schröder, J. Keller, S. Freguia, P. Aelterman, W. Verstraete and K. Rabaey, Environ. Sci. Technol., 2006, 40, 5181–5191 CrossRef CAS.
- D. R. Bond, D. E. Holmes, L. M. Tender and D. R. Lovley, Science, 2002, 295, 483–485 CrossRef CAS.
- S. K. Chaudhuri and D. R. Lovley, Nat. Biotechnol., 2003, 21, 1229–1232 CrossRef.
- Y. A. Gorby, S. Yanina, J. S. McLean, K. M. Rosso, D. Moyles, A. Dohnalkova, T. J. Beveridge, I. S. Chang, B. H. Kim, K. S. Kim, D. E. Culley, S. B. Reed, M. F. Romine, D. A. Saffarini, E. A. Hill, L. Shi, D. A. Elias, D. W. Kennedy, G. Pinchuk, K. Watanabe, S. i. Ishii, B. Logan, K. H. Nealson and J. K. Fredrickson, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11358–11363 CrossRef CAS.
- J. Nießen, U. Schröder, M. Rosenbaum and F. Scholz, Electrochem. Commun., 2004, 6, 571–575 CrossRef CAS.
- M. Rosenbaum, U. Schröder and F. Scholz, Appl. Microbiol. Biotechnol., 2005, 68, 753–756 CrossRef CAS.
- M. Rosenbaum, F. Zhao, U. Schröder and F. Scholz, Angew. Chem., Int. Ed., 2006, 45, 6658–6661 CrossRef CAS.
- K. Rabaey, J. Rodríguez, L. Blackall, J. Keller, D. Batstone, W. Verstraete and K. H. Nealson, ISME J., 2007, 0, 1–10 Search PubMed.
- M. E. Hernandez and D. K. Newman, Cell. Mol. Life Sci., 2001, 58, 1562–1571 CrossRef CAS.
- E. Marsili, D. B. Baron, I. D. Shikhare, D. Coursolle, J. A. Gralnick and D. R. Bond, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3968–3973 CrossRef CAS.
- J. Heinze, Angew. Chem., 1984, 96, 823–840 CAS.
- K. Rabaey, N. Boon, S. D. Siciliano, M. Verhaege and W. Verstraete, Appl. Environ. Microbiol., 2004, 70, 5373–5382 CrossRef CAS.
- J. P. Busalmen, A. Esteve-Nunez and J. M. Feliu, Environ. Sci. Technol., 2008, 42, 2445–2450 CrossRef CAS.
- T. Zhang, C. Cui, S. Chen, X. Ai, H. Yang, P. Shen and Z. Peng, Chem. Commun., 2006, 2257–2259 RSC.
- D. R. Bond and D. R. Lovley, Appl. Environ. Microbiol., 2003, 69, 1548–1555 CrossRef CAS.
- S. Srikanth, E. Marsili, M. Flickinger and D. R. Bond, Biotechnol. Bioeng., 2008, 99, 1065–1068 CrossRef CAS.
- H. S. Park, B. H. Kim, H. S. Kim, H. J. Kim, G. T. Kim, I. S. Chang, Y. K. Park and H. I. Chang, Anaerobe, 2001, 7, 297–306 Search PubMed.
- H. J. Kim, H. S. Park, M. S. Hyun, I. S. Chang, M. Kim and B. H. Kim, Enzyme Microb. Technol., 2002, 30, 145–152 CrossRef CAS.
- X. Qian, G. Reguera, T. Mester and D. R. Lovley, FEMS Microbiol. Lett., 2007, 277, 21–27 CrossRef CAS.
- L. Shi, T. C. Squier, J. M. Zachara and J. K. Frederickson, Mol. Microbiol., 2007, 65, 12–20 CrossRef CAS.
- K. A. Weber, L. A. Achenbach and J. D. Coates, Nature, 2006, 4, 752–764 CAS.
- T. S. Magnuson, N. Isoyoma, A. L. Hodges-Myerson, G. Davidson, M. J. Maroney, G. G. Geesey and D. R. Lovley, Biochem. J., 2001, 359, 147–152 CrossRef CAS.
- D. R. Lovley, Nat. Rev. Microbiol., 2006, 4, 497–508 Search PubMed.
| This journal is © The Royal Society of Chemistry 2008 |
