Infusion of dye molecules into Red clover necrotic mosaic virus†
LiNa
Loo
a,
Richard H.
Guenther
b,
Steven A.
Lommel
b and
Stefan
Franzen
*a
aDepartment of Chemistry, North Carolina State University, Raleigh, NC 27695, USA. E-mail: Stefan_Franzen@ncsu.edu; Fax: +1 (919)515-8920; Tel: +1 (919)515-8915
bDepartment of Plant Pathology, North Carolina State University, Raleigh, NC 27695, USA
First published on 23rd November 2007
Abstract
The Red clover necrotic mosaic viruscapsid is utilized to package and release molecules through reversible depletion and re-addition of divalent cations.
Virus capsids consist of multiple copies of subunits that self-assemble to form a protein cage. These protein cages offer monodisperse and rigid structures with an interior cavity that can function as an ideal container for cargo encapsidation. Several approaches employing virus capsids as a container to package molecules1–3 and nanoparticles4–6 have been reported. We recently demonstrated that self-assembly of the Red clover necrotic mosaic virus (RCNMV) capsidprotein (CP) around nanoparticles required the attachment of oligonucleotides to mimic the viral RNA origin of assembly. Using this method, different types of nanoparticles can be encapsidated in a manner that is robust enough to withstand ultracentrifugation as required for purification.7 To further explore the versatility of the RCNMV capsid for nano-cargo packaging and delivery we describe a small cargo/molecule infusion process.
RCNMV is a soil-transmitted 36 nm icosahedral plant infecting virus with a capsid composed of 180 copies of a 37 kDa CP packaging either 1 copy each of a 4 kb single-stranded RNA-1 and a 1.5 kb RNA-2 or 4 copies of RNA-2.8 Divalent ions are an integral part of the RCNMV structure with 390 ± 30 Ca2+ and 420 ± 25 Mg2+ per virus.9 Selective removal of Ca2+ results in minor reorientation of the CP shell (S) and protruding (P) domains. However, depletion of both divalent ions (Ca2+ and Mg2+) induces significant conformational changes to the S and P domain that leads to formation of pores (11–13 Å) within the capsid. The pores observed at the pseudo-3-fold axes expose the capsid interior to the outer environment. Here, we take advantage of the reversible opening and closure of pores within the capsid to incorporate dye and doxorubicin molecules into the interior cavity of RCNMV. Based on the presence of anionic genome in the capsid, our hypothesis is that the molecules with appropriate compensating charge will enter the inner cavity through the opened pores.
The protocol to package dye or doxorubicin molecules within the RCNMV capsid is illustrated in Scheme 1. The divalent ions associated with RCNMV capsid were removed by dialyzing against 200 mM ethylenediaminetetraacetic acid (EDTA) at pH 8, followed by incubation with molecules at a mole ratio of 2000 for dye and 5000 for doxorubicin per capsid. The formation of pores within the capsid facilitates the infusion of molecules into the interior cavity of the capsid. The encapsidation of molecules within an RCNMV capsid is completed by dialyzing against 200 mM Ca2+, pH 6 to close the pores. In the final step, filtration was performed to remove the excess molecules until fluorescence intensities of the RCNMV-infused dye (RCNMVdye) and RCNMV-infused doxorubicin (RCNMVdox) were as low as the background signal (untreated capsids). To determine the number of molecules infused, RCNMVdye and RCNMVdox were liberated by treatment with 200 mM EDTA at either pH 8 or pH 10 to re-open pores or disrupt the capsid, respectively. The released dye molecules were collected and their concentration was determined by fluorescence.
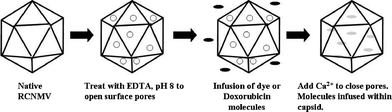 | ||
| Scheme 1 Steps involved to infuse and encapsidate molecules within the RCNMV capsid. Native RCNMV treated with EDTA, pH 8 induced opening of surface pores, which allowed molecules to infuse into the interior cavity. Encapsidation of molecules within the protein shell is completed by dialyzing against Ca2+. | ||
Three differently charged dye molecules were tested for capsid infusion: rhodamine (positive), luminarosine (neutral) and fluorescein (negative). Doxorubicin, a fluorescent, positively-charged chemotherapeutic agent, was also studied for infusion within the RCNMV capsid. The positively-charged rhodamine exhibited an infusion yield of 83 ± 10 molecules per virus. The internalization was due at least in part to the electrostatic interaction with the anionic viral genome. The neutral charged dye, luminarosine, loaded to 76 ± 5 molecules per virus. Luminarosine is a fluorescent nucleobase with the ability to interact with nucleotides , which may explain its relatively high loading. In contrast, fluorescein, with only 2 ± 1 molecules per virus, had the lowest infusion rate, presumably due to electrostatic repulsion between its negative charge and the anionic RNA viral genome. Moreover, the amino acid residues on the surface of the capsid surrounding the site of pore formation possess a net negative charge,9 which may prevent the fluorescein from entering the capsid. These measurements support the hypothesis that dye infusion is facilitated by an electrostatic interaction with the viral genome. The RCNMV capsid exhibits a high infusion rate for doxorubicin, with 4300 ± 1300 molecules infused per capsid based on the fluorescence assay .
In order to determine the capability of the infused molecules to diffuse through the pores, RCNMVdye and RCNMVdox were exposed to 200 mM EDTA at pH 8 to re-open the surface pores. A low but detectable amount of dye was observed to diffuse from the surface pores. This observation suggested an alternative mechanism of dye release by diffusion through re-opened pores without complete capsid disruption. However, the fluorescence intensity of doxorubicin from RCNMVdox was as low as the background signal, which implies doxorubicin does not diffuse through the re-opened pores. This observation is different from rhodamine and luminarosine, where approximately 10% of the total infused dyes diffused out the re-opened pores after EDTA, pH 8 treatment (Table 1). Doxorubicin is an anthracycline drug that is known to intercalate DNA. We hypothesize that the interaction between doxorubicin and the viral genome inhibits the diffusion of doxorubicin through the pores.
| Dye | Charge | EDTA pH 10a | EDTA pH 8b |
|---|---|---|---|
| a Virus disassembled to release infused molecules. b Surface pores re-opened (without virus disruption) to diffuse molecules from the capsid. | |||
| Rhodamine | Positive | 83 ± 10 | 6 ± 4 |
| Luminarosine | Neutral | 76 ± 5 | 7 ± 3 |
| Fluorescein | Negative | 2 ± 1 | 1 ± 1 |
| Doxorubicin | Positive | 4300 ± 1300 | <1 |
To determine the loading capacity of RCNMV for rhodamine, different molar ratios of rhodamine per RCNMV were incubated (150 : 1, 500 : 1, and 4000 : 1) during the infusion protocol depicted in Scheme 1. The results indicated that a maximum of ∼90 rhodamine molecules can be infused per RCNMV (see ESI† ). TEM analysis revealed that infusion of rhodamine did not result in a detectable change in the capsid diameter or morphology when compared to native RCNMV (Fig. 1). Unlike rhodamine, incubation of luminarosine at a mole ratio of 150 : 1 reported almost no uptake of the dye in the virus. This observation suggested that the electrostatic interaction with the viral genome plays a major role in the infusion mechanism, even when low numbers of excess dye molecules are available, which explains the successful infusion for rhodamine but not for luminarosine.
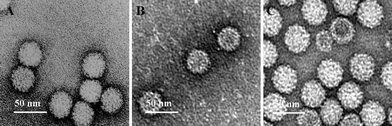 | ||
| Fig. 1 TEM images of (A) native RCNMV (B) RCNMV-infused rhodamine (RCNMVrho) and (C) RCNMV-infused doxorubicin (RCNMVdox). | ||
Selective depletion of Ca2+ by treatment with ethylene glycol tetraacetic acid (EGTA) resulted in changes to the capsid organization but did not induce the opening of pores.9 A control experiment was performed where rhodamine was incubated with RCNMV pre-treated with EGTA (instead of EDTA), at a mole ratio of 500 : 1 (rhodamine-to-RCNMV), followed by dialyzing against Ca2+. Identical protocols to remove the excess rhodamine and release of infused rhodamine were subsequently performed. Based on fluorescence measurements, 6.2 ± 3.4 rhodamine molecules infused per virus (see ESI† ). In a second control experiment, rhodamine was incubated with untreated RCNMV (not treated with EGTA or EDTA). The emission spectrum indicated the released rhodamine signal was as low as the background signal, suggesting no rhodamine infusion. Collectively these results indicated that rhodamine infusion is strictly dependent upon total divalent ion extraction and pore formation, since the failure to open the pores leads to almost no infusion of rhodamine.
The fluorescence assay indicated that an extremely high level of doxorubicin was released when RCNMVdox was disassembled by EDTA pH 10 treatment (Table 1). For further characterization, the RCNMVdox was banded by ultracentrifugation in a 20–50% sucrose gradient. The infusion of doxorubicin into the RCNMV capsid uniformly increased the density of the entire virus population, and hence, resulted in a shift of density and a band at 40% sucrose, as compared to native RCNMV which yielded a band at 30% sucrose (see ESI† ). The TEM image (Fig. 1C) indicated that the integrity of RCNMVdox remained intact and the diameter (34.5 ± 3.7 nm) is similar to the native RCNMV.
RCNMVdox was further subjected to a 20–60% iodixanol (OptiPrep) density gradient centrifugation (see ESI† ). The centrifugation of native RCNMV resulted in 2 visible bands, where the lower and upper band corresponded to native and expanded (swollen) form of the virus, respectively. On the other hand, RCNMVdox formed a distinct band that reached equilibrium at a higher density (lower in the gradient) than native RCNMV (Fig. 2A). Absorbance of the fractionated iodixanol gradients indicated a major peak of RCNMVdox at fraction 2, corresponding to 60% iodixanol, while native RCNMV was found at fraction 8, equivalent to 50% iodixanol. To eliminate the possibility that the increased density of RCNMVdox as observed was due to attachment of doxorubicin on the virus outer surface, a control experiment was performed where doxorubicin was incubated with untreated RCNMV and centrifuged under the same conditions. The distribution profile of this control sample was the same as native RCNMV (see ESI† ). The control experiment, together with the notable difference in density observed for native RCNMV and RCNMVdox further confirmed that the doxorubicin was indeed infused into the capsid.
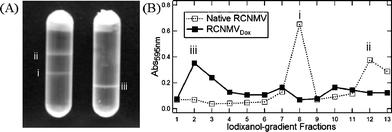 | ||
| Fig. 2 (A) Distribution profile of native RCNMV (left) and RCNMVdox (right) through a 20–60% iodixanol gradient after centrifugation. (B) Absorbance of each iodixanol gradient fraction. Data are presented with the densest fractions on the left. Peaks labelled with (i), (ii) and (iii) corresponded to bands observed on the tubes. | ||
The value from the fluorescence measurement may be artificially high due to binding of doxorubicin to the exterior of the capsid or artifacts due to ground state complexation of doxorubicin. The equilibrium ultracentrifugation measurements provide a more accurate estimate of the infused number of doxorubicin molecules per virus. Based on the refractive index measurements, the density of RCNMVdox was 1.324 g ml−1, compared to native RCNMV which was 1.247 g ml−1. Calculations based on the difference in the densities indicated that a minimum of 1000 doxorubicin molecules are infused per RCNMV. Determination of an exact value would require knowledge of the packing density of doxorubicin in the virus interior. The increase in density that arises from displacement of water by packed doxorubicin cannot be measured directly.
The loading density of doxorubicin (number of doxorubicin/particle volume) in RCNMV is comparable to the clinically approved liposomal doxorubicin formulation (Doxil),10 with 0.0053 nm−3 and 0.0051 nm−3 respectively, using the value of 1000 doxorubicin molecules per virus particle for calculation. Since RCNMV is stable in soil where the divalent ion concentrations are high (millimolar range), we tested the stability under similar conditions in solution. Results indicated that doxorubicin packaged in the RCNMVdoxcapsid showed no diffusion through the pores in the presence of millimolar divalent ion concentration, such as found in blood. This feature eliminates drug leakage during transit as observed in liposomal formulations.11 The loading efficiency, together with the tight non-leaky packaging of the RCNMV capsid, offers a superior candidate for development as a drug carrier. The key to intracellular drug delivery is that RCNMV is triggered to open surface pores and releases its genome inside a cell where the divalent ion concentrations are low (micromolar level). The packaging method described here is advantageous as the RCNMV capsid can perform as a container to protect and sequester a cargo until it reaches the targeted cell to be released. With the isoelectric point of 6.8, RCNMV maintains capsid integrity at high pH as compared to other plant viruses such as Tomato bushy stunt virus (TBSV) (isoelectric point is 4.8)12 which also utilizes a similar opening and closing of surface pores upon changes of pH and divalent ion concentrations to release its genome .
The idea of using plant viruses as a biomedical tool has been demonstrated with Cowpea mosaic virus (CPMV) and Cowpea chlorotic mottle virus (CCMV) for vaccine development13 or as bio-imaging agents.14,15 The usage of protein containers as delivery vectors has been proposed with ferritin,16 where the packaged cargo was simultaneously packaged and delivered into cells. Inspired by that, experiments are currently in progress to study the ability of RCNMV capsid as a multifunctional tool to package, deliver and target the packaged molecules into cells.
In summary, this study demonstrates a method that exploits the natural RCNMV pH and divalent ion-dependent genome release encoded into its polymeric protein capsid to encapsidate and release dye molecules. This study revealed that infusion of positive and neutral charged dye molecules was feasible, but that negatively-charged dye molecules were not readily infused into the virus. Additionally, the loading density of the neutral luminarosine and doxorubicin may be increased because of interactions with the viral genome. The results demonstrated an extremely high loading density of ∼1000 dye molecules per virus for the chemotherapeutic doxorubicin. The method reported here offers an approach that enables the use of virus capsid as a vector for the controlled loading and unloading of cargos.
SF acknowledges support through NIH grant NCI CA098194. This work was partially funded by an NSF grant MCB-0651263 to SAL.
Notes and references
- M. Knez, M. Sumser, A. M. Bittner, C. Wege, H. Jeske, T. P. Martin and K. Kern, Adv. Funct. Mater., 2004, 14, 116–124 CrossRef CAS.
- T. Douglas and M. Young, Nature, 1998, 393, 152–155 CrossRef CAS.
- Y. Ren, S. M. Wong and L. Y. Lim, Bioconjugate Chem., 2007, 18, 836–843 CrossRef CAS.
- J. Sun, C. DuFort, M. C. Daniel, A. Murali, C. Chen, K. Gopinath, B. Stein, M. De, V. M. Rotello, A. Holzenburg, C. C. Kao and B. Dragnea, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 1354–1359 CrossRef CAS.
- S. K. Dixit, N. L. Goicochea, M. C. Daniel, A. Murali, L. Bronstein, M. De, B. Stein, V. M. Rotello, C. C. Kao and B. Dragnea, Nano Lett., 2006, 6, 1993–1999 CrossRef CAS.
- L. Loo, R. H. Guenther, V. R. Basnayake, S. A. Lommel and S. Franzen, J. Am. Chem. Soc., 2006, 128, 4502–4503 CrossRef CAS.
- L. Loo, R. H. Guenther, S. A. Lommel and S. Franzen, J. Am. Chem. Soc., 2007, 129, 11111–11117 CrossRef CAS.
- V. R. Basnayake, T. L. Sit and S. A. Lommel, Virology, 2006, 345, 532–539 CrossRef CAS.
- M. B. Sherman, R. H. Guenther, F. Tama, T. L. Sit, C. L. Brooks, A. M. Mikhailov, E. V. Orlova, T. S. Baker and S. A. Lommel, J. Virol., 2006, 80, 10395–10406 CrossRef CAS.
- A. Gabizon, H. Shmeeda, A. T. Horowitz and S. Zalipsky, Adv. Drug Delivery Rev., 2004, 56, 1177–1192 CrossRef CAS.
- G. J. R. Charrois and T. M. Allen, Biochim. Biophys. Acta, 2004, 1663, 167–177 CAS.
- B. Lorber, C. Sauter, J. D. Ng, D. W. Zhu, R. Giege, O. Vidal, M. C. Robert and B. Capelle, J. Cryst. Growth, 1999, 204, 357–368 CrossRef CAS.
- F. R. Brennan, T. Bellaby, S. M. Helliwell, T. D. Jones, S. Kamstrup, K. Dalsgaard, J. I. Flock and W. D. O. Hamilton, J. Virol., 1999, 73, 930–938 CAS.
- M. Allen, J. W. M. Bulte, L. Liepold, G. Basu, H. A. Zywicke, J. A. Frank, M. Young and T. Douglas, Magn. Reson. Med., 2005, 54, 807–812 CrossRef CAS.
- K. J. Koudelka, C. S. Rae, M. J. Gonzalez and M. Manchester, J. Virol., 2007, 81, 1632–1640 CAS.
- M. Uchida, M. L. Flenniken, M. Allen, D. A. Willits, B. E. Crowley, S. Brumfield, A. F. Willis, L. Jackiw, M. Jutila, M. J. Young and T. Douglas, J. Am. Chem. Soc., 2006, 128, 16626–16633 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Protocols and supplementary figures. See DOI: 10.1039/b714748a |
| This journal is © The Royal Society of Chemistry 2008 |
