Observation of the formation of supported bilayers by amphiphilic peptidyl-RNA†
Adina N.
Lazar
a,
Anthony W.
Coleman
*a,
Silvia
Terenzi
b and
Peter
Strazewski
c
aAssemblages Moléculaires d'Intérêt Biologique (UMR CNRS 5086), Institut de Biologie et Chimie des Protéines, 7 passage du Vercors, F-69367 Lyon, France. E-mail: aw.coleman@ibcp.fr; Fax: +334 72 72 26 90; Tel: +334 72 72 26 40
bLaboratoire de Chimie Physique des Polymères et Membranes, École Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland. E-mail: silvia.terenzi@epfl.ch
cLaboratoire de Synthèse de Biomolécules (UMR 5181), Université Claude Bernard Lyon 1, 43 bvd du 11 novembre 1918, F-69622 Villeurbanne Cedex, France. E-mail: strazewski@univ-lyon1.fr
First published on 11th November 2005
Abstract
Amphiphilic peptidyl-RNA conjugates, molecules that mimic natural peptidyl-transfer RNA, are capable of self-assembling on glass substrates as vesicles and supported bilayers.
Amphiphilic peptidyl-RNA conjugates are likely to have played an important role at the origin of RNA-controlled peptide synthesis that may have taken place in lipid bilayer vesicles. Largely lipophilic peptides could have served as primordial molecular anchoring devices that would enable their RNA carriers to be transiently immobilized, compartmentalized and thus highly concentrated on or in liposomes.
We have recently reported on the synthesis, properties and the preliminary observation of the formation of vesicular structures derived from amphiphilic peptidyl-RNA molecules in the absence of lipids.1 In these molecules, the oligomeric RNA chains form the polar ‘head group’, while suitable helix forming peptides form the hydrophobic group. The spreading of such vesicular structures at surfaces may allow well defined anchored structures to be obtained. Such structures will resemble the Supported Lipid Bilayers (SLBs) formed by natural lipids. SLBs are formed by the deposition and opening of liposomes at surfaces.2 They represent a highly significant model of biological membranes,3 particularly as the presence of a hydration layer of about 1 nm height between the bilayer and the substrate allows biomolecules inserted into SLBs to retain their biological activity.4
Atomic Force Microscopy (AFM) has proved to be the key tool for the investigation of SLBs and other supported lipid structures at surfaces. AFM has allowed nanometric resolution imaging of the in situ formation of SLBs, the formation of rafts in SLBs and especially the interactions between biological macromolecules and SLBs.5–7
In the present communication we report on the observation of the formation of thick (about 21 nm) supported bilayers by peptidyl-RNA molecules on optical glass surfaces.8 The structure and a model of the peptidyl-RNA molecule, Ala21RNA22, used in this study is presented in Fig. 1.
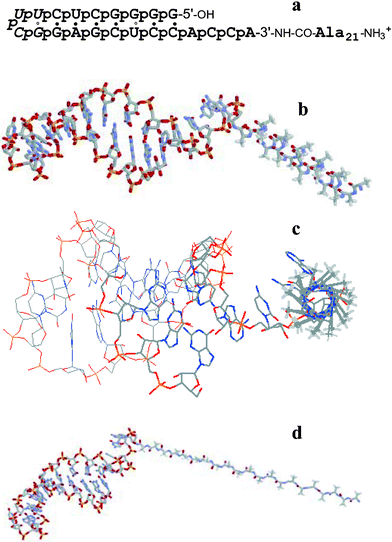 | ||
| Fig. 1 The peptidyl-RNA molecule analysed here: (a) nucleotide and amino acid sequence: 7 base pair stem closed by UUCG loop (italic). The RNA part mimics the aminoacyl acceptor stem of E. coli transfer RNA specific for alanine and contains 21 negative charges (‘p’ for phosphodiester), the peptide N-terminus bears one positive charge in neutral water. The connection between RNA and peptide is stabilised through an amide bond (3′-NH–CO) instead of the natural, somewhat labile ester bond in peptidylated transfer RNA molecules; (b,c) two views of the shape of the conjugate bearing a right-handed α-helical peptide conformation; (d) shape of the conjugate where Ala21 adopts an elongated β-strand conformation, as in an antiparallel β-sheet.‡ Color code: N blue, O red, P yellow, C grey, methyl H off-white (all non-methyl H atoms are not shown). | ||
Combining molecular modelling of the peptide sequence and structural studies allowed the estimation of the molecular dimensions of the conjugate molecule. The length of the polyanionic RNA hairpin, as taken from NMR data,9 is between 4.2 and 4.9 nm, its maximal diameter is 2.23 nm (based on the A-RNA double helix structure). Two main conformations may be taken into account for the peptide fragment. For a five-turn α-helical conformation of Ala21 the length of about 3 nm and a diameter of 0.86 nm were calculated. For an extended β-strand conformation lengths of 6.7 nm (parallel β-strand) or 7.2 nm (antiparallel β-strand) are possible, the β-strand diameter is 0.65 nm. If the peptide populated 310-helix conformations,10 the average peptide length would be up to 31% longer than an α-helix. If, less likely, the peptide adopted a π-helix conformation,11 it would be somewhat shorter. Partial folds or β-strands/sheets12 would make the peptide even longer than 7.2 nm. Thus the possible lengths of the conjugate are 7–8 nm bearing an α-helical peptide or 10–12 nm bearing a β-strand. These measurements do not take into account contributions from counter cations (ammonium or sodium) and hydration layers.
Circular dichroism spectra of the conjugate in neutral water showed the predominance of α-helical peptide conformations (double-minimum at 210–212 nm and ∼225 nm, reinforced through the addition of 33% (v/v) 2,2,2-trifluoroethanol).† However, even a highly diluted analyte solution produced significant amounts of precipitated conjugate that escaped CD measurement. It is known that α-helical peptides can slowly rearrange into β-sheets upon ageing and/or interacting with a lipophilic environment.13 Therefore it may well be that, upon ageing of the solution, the conjugate is thermodynamically driven into self-aggregation where β-sheet formation becomes favoured.
Dynamic Light Scattering (DLS) of the conjugate molecules in water showed a highly polydisperse vesicle size distribution with the major populations having diameters of approximately 430 nm and 150 nm.†
A 10 µl droplet of the vesicular dispersion was deposited onto an optical glass slide and dried at 40 °C during 24 h prior to Atomic Force Microscopy (AFM) imaging in the non-contact mode.§ The film of peptidyl-RNA proved to be inhomogeneous. Three types of phases are distinguished on the surface: a central vesicular area, surrounded by planar fractal type structures of supported bilayers and an intermediate phase between the two.
A 25 × 25 µm image shown in Fig. 2 presents the vesicular portion of the sample. These vesicles have diameters in the range 400–650 nm with 100–200 nm range in height. The sizes observed can be compared to the 430 nm population observed by dynamic light scattering, reasonable similarity being present in the volumes, taking into account the vertical flattening of the vesicles. The observed height implies these vesicles to be multilamellar in structure. In concentrated zones there is a relatively strong tendency of vesicles to form clumps on the surface, although no apparent fusion is observed.
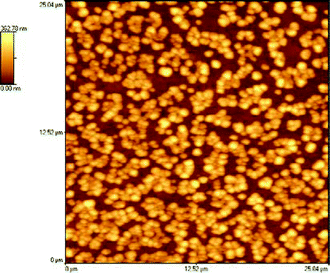 | ||
| Fig. 2 The vesicular phase of the peptidyl-RNA sample. | ||
The intermediate phase† represents post-vesicle fusion and spreading, with formation of separated blocks of supported bilayers (SBs), of random-geometry phase.
A topographic analysis on a 25 × 25 µm image of this surface shows bilayers of approximatively 21 ± 1 nm in height in coexistence with dispersed particles of a mean diameter of 200 nm—all values were calculated from 50 measurements (Fig. 3). The observed height is in agreement with that expected from the molecular models, taking into account the presence of a hydration layer.
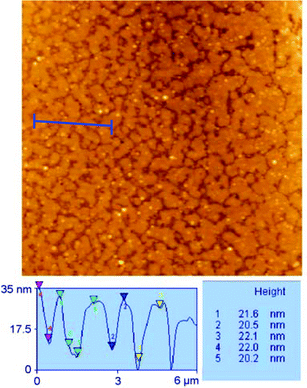 | ||
| Fig. 3 SB formation phase by vesicle fusion; topographic analysis of the bilayers—the sample can be seen to be slightly tilted with regard to the measuring line. | ||
In the area of supported bilayers, the topography is regular (Fig. 4), the network is very dense and flat (RMS value measured on the bilayer ∼2 nm). Holes and canyons are present in the overall bilayer coverage; these tend to be oriented along a single direction (approximately 40% alignment along the X scanning axis). The depth of the holes is, as expected, about 21 nm.
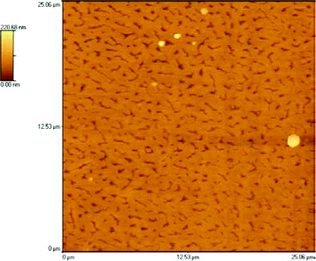 | ||
| Fig. 4 Fractal type SB structures of synthetic peptidyl-RNA molecules. | ||
The present results constitute a proof of the flexibility and the capacity of synthetic peptidyl-RNAs to self-assemble into highly stable and organised structures.
Notes and references
- S. Terenzi, E. Biala, N.-Q. Nguyen-Trung and P. Strazewski, Angew. Chem., Int. Ed., 2003, 42, 2909–2912 CrossRef CAS.
- R. P. Richter, A. Mukhopadhyay and A. Brisson, Biophys. J., 2003, 85, 3035–3047 CrossRef CAS.
- Introduction to Biological Membranes, ed. M. K. Jain and R. C. Wagner, John Wiley, New York, 1988 Search PubMed.
- M. Ross, S. Krol, A. Janshoff and H. J. Galla, Eur. Biophys. J., 2002, 31, 52–61 Search PubMed.
- J. T. Groves and G. B. Steven, Acc. Chem. Res., 2002, 35, 149–157 CrossRef CAS.
- D. Fotiadis, S. Scheuring, S. A. Muller, A. Engel and D. J. Muller, Micron, 2002, 33, 385–397 CrossRef CAS.
- P. E. Milhiet, V. Vie, M.-C. Giocondi and C. Le Grimellec, Single Molecules, 2001, 2, 109–112 Search PubMed.
- (a) C. A. Keller and B. Kasemo, Biophys. J., 1998, 75, 1397–1402 CrossRef CAS; (b) P. S. Cremer and S. G. Boxer, J. Phys. Chem. B, 1999, 103, 2554–2559 CrossRef CAS; (c) J. F. Nagle and S. Tristram-Nagle, Biochim. Biophys. Acta, 2000, 1469, 159–195 CrossRef CAS.
- A. Ramos and G. Varani, Nucleic Acids Res., 1997, 25, 2083–2090 CrossRef CAS.
- F. B. Sheinerman and C. L. Brooks III, J. Am. Chem. Soc., 1995, 117, 10098–10103 CrossRef CAS.
- R. Sudha, M. Kohtani, G. A. Breaux and M. F. Jarrold, J. Am. Chem. Soc., 2004, 126, 2777–2784 CrossRef CAS.
- C. M. Bishop, W. F. Walkenhorst and W. C. Wimley, J. Mol. Biol., 2001, 301, 975–988 CrossRef CAS.
- (a) W. C. Wimley, K. Hristina, A. S. Ladokhin, L. Silvestro, P. H. Axelsen and S. H. White, J. Mol. Biol., 1998, 277, 1091–1110 CrossRef CAS; (b) N. Sinha, C. J. Tsai and R. Nussinov, Protein Eng., 2001, 14, 93–103 CrossRef CAS; (c) C. Paul, J. Wang, W. C. Wimley, R. M. Hochstrasser and P. H. Axelsen, J. Am. Chem. Soc., 2004, 126, 5843–5858 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: CD spectra, DLS and AFM of the intermediate phase. See DOI: 10.1039/b509971b |
| ‡ The Ala21 α-helix was calculated with PyMOL® from http://pymol.sourceforge.net. The RNA hairpin was taken from an NMR study on this compound,9i.e., model 4 in 1IKD (Brookhaven database). Both fragments were joined to one another, the new amide bond was set to the usual length (1.47 Å) and the amide torsional angle to trans (180°), resulting in a tilt between the helical axis of the A-RNA and the peptide backbone axis of about 37° in the α-helical and 52° in the β-stranded conjugate. However, the spacial arrangement between RNA and peptide is by no means the only one, since the single-stranded part of the RNA (ACCA-3′, here fully stacked) is quite flexible and the chosen Cα–CO torsional angle ψ in the C-terminal, RNA-bound alanine moiety is only one of many possible angles (here ψC-terminal = 165° in the α-helical and 132° in the β-stranded conjugate). Therefore all kinds of RNA–peptide connection angles are imaginable in the unaggregated molecule, including perpendicular or coaxial arrangements. Note that the negative end of the macro dipole of the Ala21 α-helix points, owing to the unidirectional arrangement of the intrahelical CO⋯HN hydrogen bonds, towards the RNA hairpin, and thus, reinforces the amphiphilicity of the all-helical conjugate molecule. |
| § The measurements by Atomic Force Microscopy were carried out using an Explorer Thermomicroscope equipped with a scanner of 100 µm, in non-contact mode. The scanning speed is of 1–2 Hz and the resolution of 500 × 500. Cantilevers are made of silicon, the resonance frequency is f0 = 260 kHz and the force constant is 45 N. Topographic and internal sensor images were collected sequentially from 80 × 80 µm, 50 × 50 µm, 25 × 25 µm, 15 × 15 µm, 10 × 10 µm, 7.5 × 7.5 µm to 5 × 5 µm. The images were processed with the SPMLab 5.01 software and are unfiltered. |
| This journal is © The Royal Society of Chemistry 2006 |
