Probing the mechanism of a fungal glycosyltransferase essential for cell wall biosynthesis. UDP-Chitobiose is not a substrate for chitin synthase†
Robert Chang‡, Adam R. Yeager‡ and Nathaniel S. Finney*
Department of Chemistry and Biochemistry - 0358 University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92093-0358, USA
First published on 5th December 2002
Abstract
Chitin synthase is responsible for the biosynthesis of chitin, an essential component of the fungal cell wall. There is a long-standing question as to whether “processive” transferases such as chitin synthase operate in the same manner as non-processive transferases. The question arises from analysis of the polysaccharide structure – in chitin, for instance, each sugar residue is rotated ≈180° relative to the preceding sugar in the chain. This requires that the enzyme account for the alternating “up/down” configuration during biosynthesis. An enzyme with a single active site, analogous to the non-processive transferases – would have to accommodate a distorted glycosidic linkage at every other synthetic step. An alternative proposal is that the enzyme might assemble the disaccharide donor, addressing the “up/down” conformational problem prior to polymer synthesis. We present compelling evidence that this latter hypothesis is incorrect.
We wish to report our recent efforts to clarify an important aspect of the mechanism of chitin synthase, the processive glycosyltransferase responsible for the polymerization of uridinediphosphoryl-N-acetylglucosamine (UDP-GlcNAc) to form chitin (poly-β-1,4-GlcNAc).1,2 Chitin is a rigid rod-like oligosaccharide, and an essential component of the fungal cell wall. As it is absent in mammals, chitin synthase (CS) has long been regarded as a promising target for antifungal therapeutics. Among the challenges in developing inhibitors of CS are the dearth of inhibitory natural products and the ubiquity of UDP-GlcNAc-dependent transferases.3 With regard to the latter issue, there would be clear value in identifying differences between CS and the numerous other N-acetylglucosaminyl transferases, as this would allow for the selective targeting of this important enzyme.
There is a long-standing question as to whether “processive” transferases such as chitin synthase, which synthesize polymeric oligosaccharides without releasing synthetic intermediates, operate in the same manner as the comparatively well understood non-processive transferases, which transfer a single activated sugar to an acceptor substrate and then release the product.4–6 This question arises from the conformation of the extended structure of polysaccharides. In chitin, a representative case, each residue is ≈180° out of phase with the preceding sugar in the polymer chain (Fig. 1), and the enzyme must somehow account for this alternating “up/down” configuration during biosynthesis. If processive and non-processive transferases share a common mechanism – addition of a monosaccharide donor (UDP-N-acetylglucosamine, UDP-GlcNAc) to the non-reducing end of the growing polymer – then every other synthetic step must lead to a distorted intermediate (Fig. 2, path a).7,8 While this mechanism cannot be excluded, there is clear incentive for the consideration of other mechanisms. One alternative proposal is that the enzyme or an associated protein preassembles the disaccharide donor (UDP-chitobiose), allowing extension by two residues at a time and resolving the “up/down” conformational issue prior to polymer synthesis (Fig. 2, path b).6b–d We describe here the synthesis of UDP-chitobiose, the intermediate required by the disaccharide preassembly hypothesis, and show that it is not a kinetically competent substrate for chitin synthase.
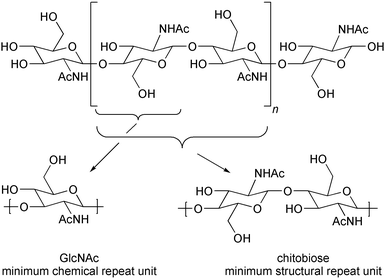 | ||
| Fig. 1 | ||
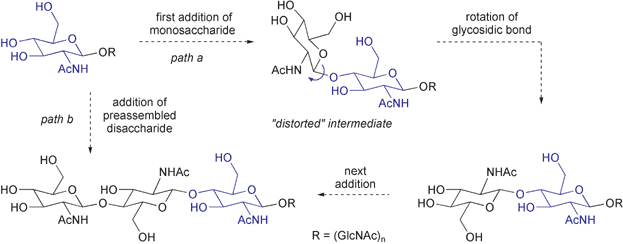 | ||
| Fig. 2 | ||
Synthesis of UDP-chitobiose (UDP-Chi) began with a modified procedure for the degradation of chitin in hot AcOH–H2SO4, which consistently provided a ca. 10% yield of peracylated chitobiose without chromatography (Scheme 1).9,10 Similar to peracylated GlcNAc, the anomeric acetate of peracetylchitobiose could be selectively removed by treatment with hydrazine acetate.9,11 Subsequent phosphorylation with tetrabenzyl pyrophosphate and removal of the benzyl groups afforded the corresponding anomeric phosphate, isolated as the ammonium salt.12 Coupling of this phosphate with the morpholidate of UMP,13 followed by deacylation, afforded UDP-Chi in 30% yield from peracetyl chitobiose.
![a) H2SO4, Ac2O, △, ≈10%; b) N2H4·HOAc, DMF, 83%; c) i. LDA, THF ii. [(BnO)2PO]2O, 77%; d) H2, Pd/C, CH3OH–NEt3, 99%; e) UMP morpholidate, tetrazole, py, 55%; f) CH3OH–NH3, 85%.](/image/article/2003/OB/b208953j/b208953j-s1.gif) | ||
| Scheme 1 a) H2SO4, Ac2O, △, ≈10%; b) N2H4·HOAc, DMF, 83%; c) i. LDA, THF ii. [(BnO)2PO]2O, 77%; d) H2, Pd/C, CH3OH–NEt3, 99%; e) UMP morpholidate, tetrazole, py, 55%; f) CH3OH–NH3, 85%. | ||
Chitin synthase assays rely on the use of radiolabelled nucleotide sugar substrates, which necessitated the synthesis of 3H-UDP-Chi (Scheme 2). The acetamide of the reducing sugar was selectively deacylated by conversion to the oxazoline, via the anomeric chloride, followed by selective hydrolysis of the oxazoline to afford the anomeric acetate with an ammonium group at C2.14 Acylation with Ac2O–3H-Ac2O provided tritiated peracetyl chitobiose in 37% overall yield, which was converted to the UDP derivative by the preceding method.
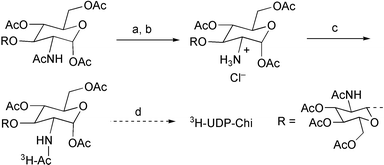 | ||
| Scheme 2 a) HCl, AcOH–Ac2O, 61%; b) 1 eq. H2O, acetone, 72%; c)AcO–3H-Ac2O, py, 83%; d) as Scheme 1. | ||
Chitin synthase activity was assayed with membrane preparations from yeast (S. cerevisiae) by the procedure of Orlean.15 Reactions were run for 1 hour, at which time the protein was denatured and the chitin precipitated by the addition of cold trichloroacetic acid. Assay mixtures were then filtered and rinsed to isolate precipitated chitin, and total incorporated radioactivity was measured by scintillation counting. “100% activity” was typically 12–13,000 cpm h−1, with a time-independent background of 250–300 cpm; the background radioactivity for high-concentration 3H-UDP-Chi assays was slightly higher due to higher initial radioactivity (Table 1). All data are the average of 2 or more experimental trials.
Initial evaluation revealed that UDP-Chi is not a viable substrate for chitin synthase: even at elevated [UDP-Chi], no radioactivity incorporation above background was observed (Table 1, entries 1–4). Control experiments ruled out the possibility that UDP-Chi was a viable substrate but required the presence of UDP-GlcNAc as a co-substrate or polymerization initiator (Table 1, entries 5–7).16 Further, measurement chitin formation at fixed [UDP-GlcNAc] with variable [UDP-Chi] indicated that UDP-Chi is an inhibitor of chitin synthase, with Ki = 3.3 mM (IC50 = 10.0 mM).17,18 This is similar to the KM for UDP-GlcNAc (0.5 mM).15 Measurement of chitin formation with [UDP-Chi] = 7.5 mM and variable [UDP-GlcNAc] demonstrated that addition of sufficient UDP-GlcNAc (≥15 mM) leads to full recovery of enzymatic activity, which indicated that the inhibition is competitive.19 These data support the idea that UDP-GlcNAc and UDP-Chi bind to a common site in chitin synthase, and that the failure of the enzyme to convert UDP-Chi to chitin truly reflects lack of reactivity rather than lack of binding affinity.
These experiments thus reasonably exclude the involvement of UDP-Chi as a transient intermediate in chitin biosynthesis. Given the homology among processive glycosyltransferases, it is likely that this conclusion can be extended to other enzymes, such as cellulose synthase.20 With this mechanism excluded, there remain two limiting cases to consider: first, as noted, that chitin synthase has a single active site and somehow compensates for the formation of a high-energy intermediate required by every other synthetic step; second, that chitin synthase in fact has two active sites, one for each orientation of the GlcNAc electrophile. Efforts are underway to test this latter hypothesis.
Acknowledgements
We thank the NIH (GM60875), the Hellman Foundation and the UC Systemwide Biotechnology Program for direct support and the NIH and NSF for infrastructure support (GM62116, GM61894, CHE-9709183). We thank Professor Peter Orlean (University of Illinois) and Dr Enrico Cabib (NIH) for assay advice, Professor Orlean for donation of yeast strains, Dr Herman van Halbeek, Mr Steven Adams and Mr Dennis Koehler (UCSD) for technical assistance, and Professors Scott Singleton (Rice University) and Jack Kyte (UCSD) for helpful discussions.References and Notes
- Representative reviews of fungal chitin synthase (EC 2.4.1.16). (a) C. A. Munro and N. A. R. Gow, Med. Mycol., 2001, 39, 41–53 Search PubMed; (b) M. H. Valdivieso, A. Duran and C. Roncero, EXS, 1999, 87, 55–69 Search PubMed; (c) R. A. Merz, M. Horsch, L. E. Nyhlen and D. M. Rast, EXS, 1999, 87, 9–37 Search PubMed; (d) C. E. Bulawa, Ann. Rev. Microbiol., 1993, 47, 505–534 Search PubMed; (e) E. Cabib, Advances Enzymol., 1987, 59, 59–101 Search PubMed.
- The significance of glycosyltransferases derives from the fact that mono- or polysaccharides are involved in virtually all biological processes. For an excellent overview of the field, see (a) Essentials of Glycobiology, eds. A. Varki, R., Cummings, J. Esko, H. Freeze, G. Hart and J. Marth, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1999. Search PubMedFor recent reviews; (b) R. A. Dwek and T. D. Butters, Chem. Rev., 2002, 102, 283–284 CrossRef CAS; (c) C. R. Bertozzi and L. L. Kiessling, Science, 2001, 291, 2357–2364 CrossRef CAS; (d) A. Dove, Nature Biotechnol., 2001, 19, 913–917 Search PubMed; (e) A. Kobata, Glycoconjugate J., 2001, 17, 443–464 Search PubMed; (f) K. M. Koeller and C.-H. Wong, Nature Biotechnol., 2000, 18, 835–841 Search PubMed.
- While natural product inhibitors are known, their effectiveness has been limited by low affinity and selectivity. The closely-related polyoxins and nikkomycins are among the most potent inhibitors, with Ki values in the low mM range. None of these have sufficient in vivo, activity for therapeutic use, although polyoxin D is used as an agricultural fungicide. See: C. A. Kauffman and P. L. Carver, Drugs, 1997, 53, 539–49 Search PubMed.
- For recent reviews of mechanism-based inhibitors of glycosidases and non-processive glycosyltransferase, see (a) P. Compain and O. R. Martin, Bioorg. Med. Chem., 2001, 99, 3077–3092 Search PubMed; (b) P. Sears and C.-H. Wong, Angew. Chem., Int. Ed., 1999, 38, 2300–2324 CrossRef.
- Several crystal structures of non-processive transferases have been determined. For discussion of common features, see (a) G. J. Davies, Nature Struct. Biol., 2001, 8, 98–100 Search PubMed; (b) K. Persson, H. D. Ly, M. Dieckelmann, W. W. Wakarchuk, S. G. Withers and N. C. J. Strynadka, Nature Struct. Biol., 2001, 8, 166–175 Search PubMed; (c) S. J. Charnok, B. Henrissat and G. J. Davies, Plant Physiol., 2001, 125, 527–531 CrossRef.
- Representative mechanistic discussion (a) refs 5a,c; (b) I. M. Saxena, R. M. Brown, Jr. and T. Dandekar, Phytochemistry, 2001, 57, 1135–1148 CrossRef CAS; (c) I. M. Saxena and R. M. Brown, Jr., Curr. Opp. Plant Biol., 2000, 3, 523–531 Search PubMed; (d) I. M. Saxena, R. M. Brown, Jr., M. Fevre, R. A. Geremia and B. Henrissat, J. Bacteriol., 1995, 177, 1419–1424 CAS.
- It has been shown that the growing chitin chain is extended from the non-reducing terminus: J. Sugiyama, C. Boisset, M. Hashimoto and T. Watanabe, J. Mol. Biol., 1999, 286, 247–255 Search PubMed.
- More important than the energetic cost of the distortion is the fact that the transition states leading to the “normal” and “distorted” intermediates would have very different geometries, both of which would have to be stabilized by a common set of active-site functional groups.
- H. Kazahura, S. Nisimura, K. Shimahara and Y. Takiguchi, Carbohydr. Res., 1989, 194, 223–231 CrossRef.
- All new compounds were fully characterized by 1H- and 13C-NMR, IR and high-resolution mass spectrometry. Experimental details are available upon request.
- J. K. Coward and J. Lee, J. Org. Chem., 1992, 57, 4126–4135 CrossRef.
- H. G. Khorana and A. R. Todd, J. Chem. Soc., 1953, 2257–2260 RSC.
- (a) C.-H. Wong and V. Wittman, J. Org. Chem., 1997, 57, 2144–2147; (b) J. G. Moffatt, Methods Enzymol., 1966, 8, 136–142 Search PubMed.
- Y. Eshdat and N. Sharon, Methods Enzymol., 1977, 46, 403–414 Search PubMed.
- P. Orlean, J. Biol. Chem., 1987, 262, 5732–5739 CAS . While the original procedure employed 14C-UDP-GlcNAc, we found the less-expensive 3H-UDP-GlcNAc to be an acceptable substitute.
- Chitin synthase does not require an initial acceptor other than UDP-GlcNAc to initiate polymerization. (See refs 1,6.). The fate of the UDP fragment from the initiating sugar is unclear, and thus a unique requirement for UDP-GlcNAc as initiator could not be excluded a priori.
- Ki values were obtained from the relationship Ki = IC50/(1+[substrate]/Km) (ref. 18). Assays were performed at [UDP-GlcNAc] = 1.0 mM. KM = 0.5 mM, so this reduces to Ki = IC50/3. IC50 was determined by non-linear least-squares fitting of a plot of inhibition (%) vs. log [UDP-Chi] (measured at [UDP-GlcNAc] = 1 mM. using the Prism3 software package (Graphpad Inc., San Diego, CA); log IC50 for UDP-Chi was determined to be 1.0 ± 0.1 (95% confidence limits), providing IC50 = 10.0 +2.6/−2.1 (7.9 − 12.6).
- Y. Cheng and W. H. Prusoff, Biochem. Pharmacol., 1973, 22, 3099–3108 CrossRef CAS.
- I. H. Segel, Enzyme Kinetics, John Wiley & Sons, New York, NY, 1975 Search PubMed.
- The method of Scheme 1 has been used to prepare UDP-cellobiose. R. Chang, N. S. Finney, unpublished results.
Footnotes |
| † Electronic supplementary information (ESI) available: A. General experimental details; B. Synthesis of UDP-chitobiose (UDP-Chi); C. Procedure for chitin synthase assay; D. Data from UDP-Chi assays and E. Ki. determination. See http://www.rsc.org/suppdata/ob/b2/b208953j/ |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2003 |
