Open Access Article
Open Access ArticleA highly selective and sensitive chemiluminescent probe for leucine aminopeptidase detection in vitro, in vivo and in human liver cancer tissue†
Baoqu
Wang‡
a,
Zhenzhou
Chen‡
a,
Xiaohong
Cen‡
a,
Yuqing
Liang
a,
Liyi
Tan
a,
En
Liang
a,
Lu
Zheng
a,
Yanjun
Zheng
a,
Zhikun
Zhan
a and
Kui
Cheng
 *ab
*ab
aGuangdong Provincial Key Laboratory of New Drug Screening, Guangzhou Key Laboratory of Drug Research for Emerging Virus Prevention and Treatment, School of Pharmaceutical Sciences, Southern Medical University, Guangzhou, 510515, Guangdong Province, People's Republic of China
bDepartment of Musculoskeletal Oncology, The Third Affiliated Hospital of Southern Medical University, Guangzhou 510642, Guangdong Province, People's Republic of China. E-mail: Chengk@smu.edu.cn
First published on 27th January 2022
Abstract
Leucine aminopeptidase (LAP) is involved in tumor cell proliferation, invasion, and angiogenesis, and is a well-known tumor marker. In recent years, chemiluminescence has been widely used in the field of biological imaging, due to it resulting in a high sensitivity and excellent signal-to-noise ratio. Here, we report the design, synthesis, and evaluation of the first LAP-activated chemiluminescent probe for LAP detection and imaging. The probe initially had no chemiluminescence but produced an extremely strong chemiluminescence after the release of the dioxetane intermediate in the presence of LAP. The probe had high selectivity over other proteases and higher signal-to-noise ratios than commercial fluorophores. Real-time imaging results indicated that the chemiluminescence was remarkably enhanced at the mice tumor site after the probe was injected. Furthermore, the chemiluminescence of this probe in the cancerous tissues of patients was obviously improved compared to that of normal tissues. Taken together, this study has developed the first LAP-activable chemiluminescent probe, which could be potentially used in protein detection, disease diagnosis, and drug development.
Introduction
Cancer is a major disease that seriously endangers human health and life and causes great suffering and burden to families and society.1 Therefore, early cancer detection, diagnosis, and treatment can remarkably reduce mortality and increase cure rates for patients with cancer.2 Enzymes are an important class of biomarkers for tumor diagnosis and prognosis.3 Certain aminopeptidases are highly expressed in many malignant tumors compared to that in normal tissue.4 Leucine aminopeptidase (LAP; EC 3.4.11.1) is one of these enzymes and belongs to the M1 and M17 peptidase families as an important protein that catalyzes the hydrolysis of the N-terminal leucine residue of a protein or peptide.5 LAP is overexpressed in malignant tumor cells and is involved in tumor cell proliferation, invasion, and angiogenesis (e.g., HepG2 cells are LAP-overexpressed).6 In medical diagnosis, LAP can be used as a cancer-related biomarker for tumor tracking. Therefore, the development of highly sensitive and selective in situ detection methods that target LAP is of great importance for medical diagnosis and pathophysiology.LAP levels can be detected via several methods, but the efficient tracking of LAP activity in vitro and in vivo remains a challenge. Among the methods for tracking LAP in vivo, L-leucine-p-nitroaniline ultraviolet detection is not suitable for real-time tracking because of its low sensitivity and poor stability.7 In addition, fluorescent probes for monitoring LAP activity have greatly limited biological applications, because they present disadvantages, such as spectral crossover, autofluorescence background interference, photobleaching, and epidermal scattering under excitation by an external light source.8 Therefore, chemiluminescence detection methods that can overcome these disadvantages should be developed.9
Chemiluminescence has developed into a promising sensing and imaging tool, because it does not require excitation by an external light source, is free of light scattering and autofluorescence interference, rapidly achieves extremely high signal-to-background ratios, and improves imaging sensitivity.10 The phenoxy 1,2-dioxetane luminophore (Schaap's) is an excellent backbone for the construction of highly sensitive chemiluminescent probes.11 Some intriguing studies have aimed to improve and extend the use of Schaap's probes for the monitoring of various chemical and biological processes. Several activable chemiluminescent probes have been developed for the detection of small molecules12 (e.g., H2S, H2O2, and GSH), various biomarker enzymes13 (e.g., alkaline phosphatase, β-galactosidase, cathepsin B), and pathogens (e.g., Salmonella and Listeria monocytogenes,14Mycobacterium tuberculosis15). Here, we developed a chemiluminescent probe (probe 1) for the detection of LAP using modified Schaap's. Probe 1 was highly sensitive and specific, which allows the rapid detection of LAP activity in vitro and in vivo.
Results and discussion
Design and synthesis of probes 1 and 2
The general design of the LAP-activable chemiluminescent probe is shown in Fig. 1. Probe 1 was designed by caging a LAP recognition substrate, an acryl-substituted phenoxy 1,2-dioxetane luminophore (Int 4-1), and a self-immolative group linker p-aminobenzyl alcohol (PABA). According to relevant literature, L-leucine-containing substrates are highly specific for LAP and have been used in the design of a variety of fluorescent probes.6 The luminophore, Int 4-1, emits intense chemiluminescence for in vivo imaging under physiological conditions. The use of PABA to link the L-leucine substrate and the Int 4-1 luminophore might help reduce the spatial site resistance of the probe and facilitate interaction with the narrower and deeper LAP active site.16 After the amide bond was hydrolyzed by LAP, probe 1 was converted into Int 3-1 accompanied by a spontaneous 1,6-elimination within PABA, which departs at physiological pH (pH 7.4), to generate Int 4-1. Subsequently, the peroxide bond in Int 4-1 was cleaved via a chemical excitation process, which generates a fluorescent product 6-1 and a chemiluminescence at about 550 nm. Thus, LAP can selectively switch on the yellow green chemiluminescence of probe 1, which allows the sensitive detection of LAP activity in vitro and in vivo. The near-infrared (NIR) region is considered superior for in vivo animal imaging. NIR emission chemiluminescence is preferred for in vivo imaging because of its deeper penetration and less light scattering than other methods.17 We constructed a NIR emission chemiluminescent probe (probe 2) by applying a similar design approach. The dicyano methyl chromone was introduced at the ortho position of the phenol as an acceptor to extend the conjugated π-electron system of the phenolic luminophore, which results in the red-shift of its emission wavelength.18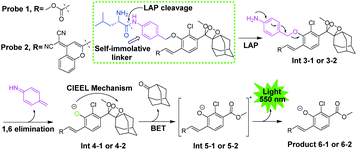 | ||
| Fig. 1 General structure and disassembly of chemiluminescent probes 1 and 2 for LAP detection. Probe 1 responds to LAP and produces intense chemiluminescence emission at 550 nm. | ||
Probe 1 was successfully synthesized and characterized (Fig. 2). Compound 5 was synthesized according to the reported method.6 Compound 6 with a chlorine at the C2 position and an electron-withdrawing acrylic substitute at the C6 position was synthesized according to the method reported by Shabat's group.13 The direct reaction between compounds 5 and 6 affords compound 7. The protecting groups of compound 7 were subsequently removed using ZnBr2 to prepare compound 8. Compound 8 was oxidized by singlet oxygen (1O2) to prepare probe 1. Then, probe 2 was successfully synthesized and characterized (see ESI, Scheme S1†).
Spectroscopic responses of probes 1 and 2 to LAP in vitro
The responses of probes 1 and 2 (10 μM) to LAP (100 U L−1) were first investigated. After probe 1 was incubated with LAP, a new UV absorption peak was observed at approximately 400 nm, accompanied by strong fluorescence emission at about 560 nm. These UV and fluorescence spectra were identical to that of the fluorescent product 6-1 (Fig. 3A). The subsequent high-performance liquid chromatography (HPLC) analysis (1 h incubation of probe with LAP, Fig. 3B) showed that probe 1 (TR = 5.13 min, Fig. S1†) was mostly converted into the fluorescent product 6-1 (TR = 4.3 min, Fig. S2 and S3†). The synthetic route to product 6-1 is illustrated in Scheme S2.† Probe 1 did not initially show chemiluminescence but displayed a clear chemiluminescence at 550 nm after incubation with LAP at 37 °C for 10 min (Fig. 3C, inset). The chemiluminescence kinetic curve of probe 1 after incubation with LAP shows a rapid increase in the signal, which reached the maximum at approximately 20 min (Fig. 3D). However, the absorption, fluorescence, and chemiluminescence spectra of probe 2 had no change before and after incubation with LAP (Fig. S4† and 3C).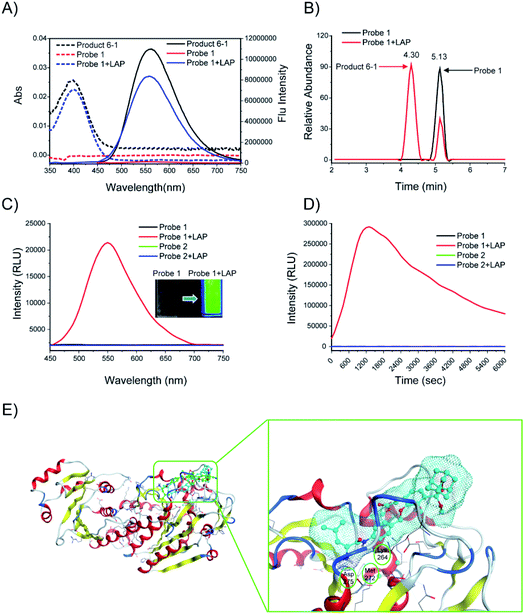 | ||
| Fig. 3 (A) Absorption (dashed line) and fluorescence (solid line) emission spectra of probe 1. Probe 1 (10 μM) was incubated with or without LAP (100 U L−1) in enzyme assay buffer at 37 °C for 6 h. (B) HPLC analysis of probe 1 (10 μM) before (black) and after (red) incubation with LAP (100 U L−1) in enzyme assay buffer (pH = 7.4) at 37 °C for 1 h. (C) Chemiluminescence spectra and images (inset) of probes 1 and 2 (10 μM). The probes were incubated with or without LAP (100 U L−1) at 37 °C for 10 min. (D) Chemiluminescence kinetic profiles of probes 1 and 2 (10 μM) after incubation with or without LAP (100 U L−1) at 37 °C for 6000 s. (E) Low-energy binding models of probes 1 (cyan) and 2 (green) bound to the LAP (PDB: 2hc9) interface. | ||
The computer docking simulation results showed that the probe 1 molecule easily reached the Zn ion coordination center through the open hydrophobic cavity of LAP, whereas probe 2 cannot fit in the catalytic active center because of steric hindrance (Fig. 3E). In addition, probe 1 has three hydrogen bonds with LAP amino acid residues (Lys264, Met272, Asp275), and the amide bonds are potentially hydrolyzed under the catalysis of these amino acids and the Zn ion, which finally releases a bright chemiluminescence signal.19 Therefore, LAP could effectively activate probe 1 to produce significant chemiluminescence, which was investigated subsequently.
The analytical conditions, including pH and temperature for the LAP assay, were studied. The hydrolysis effect of LAP on probe 1 at different temperatures was investigated. As shown in Fig. S5,† the chemiluminescence at 550 nm was remarkably enhanced as the temperature increased from 25 °C to 37 °C in the presence of LAP. The result indicated that within the experimental temperature range, probe 1 molecules have greater enzyme activity and stronger catalytic hydrolysis ability at a higher temperature. The effect of pH on the LAP activation of probe 1 was also investigated. Results showed that the maximum chemiluminescence was observed when probe 1 was incubated with LAP at pH of 6.0 to 8.0, which is close to the pH of the extracellular environment (6.5 to 7.4, Fig. S6†). Therefore, probe 1 is suitable for detecting endogenous LAP under biologically relevant conditions.
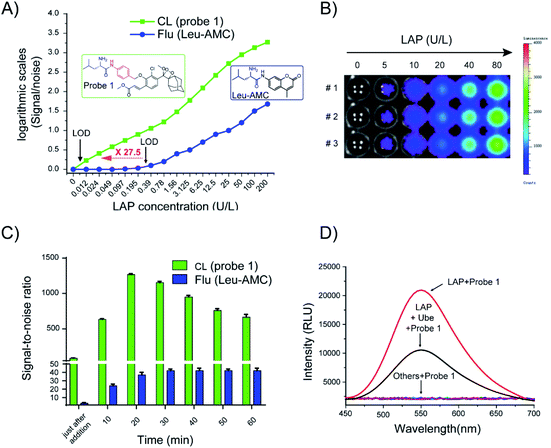
After the chemiluminescence response of probe 1 toward LAP was confirmed, the sensitivity of the probe to LAP was further determined. We compared the sensitivity of probe 1 to that of a commercially available fluorogenic probe, Leu-AMC (the synthetic route is illustrated in Scheme S3†), which fluoresces at 450 nm after cleavage by LAP. Different LAP concentrations were used to plot the signal-to-noise (S/N) ratio against the enzyme concentration in logarithmic scale for the direct comparison of the sensitivity of probe 1 and Leu-AMC (Fig. 4A). Remarkably, probe 1 exhibited a limit of detection (LOD) of 0.008 U L−1, whereas Leu-AMC detected LAP with a LOD of 0.22 U L−1 (Fig. S7†). The enhanced sensitivity of probe 1 (27.5-fold) clearly demonstrates the advantage of our chemiluminescent substrates versus currently existing fluorescent substrates for LAP detection assays. Additionally, the LOD of probe 1 was much lower than those of other reported fluorescent probes (Table S1†). The chemiluminescence images of probe 1 incubated with different LAP concentrations were acquired using the IVIS Lumia XR III system (Fig. 4B). The chemiluminescence images became bright as the LAP concentration increased. The S/N ratios of probe 1 and Leu-AMC after reaction with LAP for 1 h were measured. Probe 1 had higher S/N ratios than Leu-AMC and achieved S/N values of approximately 1260 in less than 20 min (Fig. 4C), whereas Leu-AMC had S/N ratios consistently under 43. The superior sensitivity and high S/N ratios (more than 30-fold) of probe 1 clearly prove the advantage of the chemiluminescence modality over fluorescence modality for diagnostic assays.
The specific identification of target analyzers is one of the important indicators to evaluate the usability of probe 1. Next, the specific response of probe 1 to LAP was tested. When probe 1 (10 μM) was incubated with amino acids (e.g., Hcy, L-Cys, Glu), metal ions (e.g., KCl, Na2S, Al2(SO4)3) and enzymes (β-galactosidase, cathepsin B, alkaline phosphatase pyroglutamyl aminopeptidase I, LAP), only incubation with LAP resulted in chemiluminescence (Fig. 4D, S8†). Ubenimex (Ube) is a LAP inhibitor that inhibits enzyme activity by specifically binding to the Zn ion coordination catalytic center of LAP.5 Ube treatment dramatically inhibited the chemiluminescence intensity, which indicates that probe 1 has specificity for LAP.
Detection of LAP activity in living cells
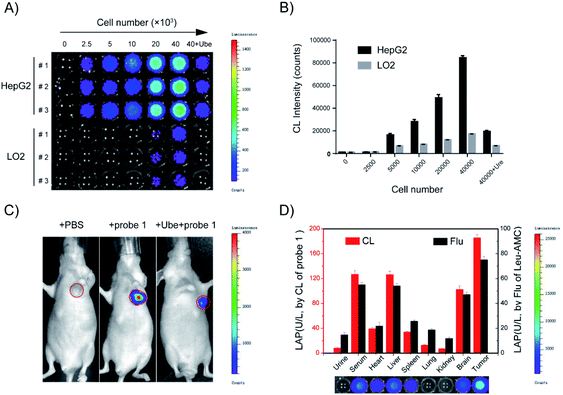
Considering that probe 1 shows an ideal optical response for LAP detection in vitro and low toxicity to HepG2 and LO2 liver cells (Fig. S9†), probe 1 was applied to distinguish normal cells from liver cancer cells by cell imaging. HepG-2 cancer cells incubated with probe 1 showed gradually enhanced chemiluminescence and the maximum signals were achieved at around 30 min. By contrast, the chemiluminescence was very weak over the course of incubation when cell lines were pretreated with Ube (Fig. S10A†). Probe 1 was incubated with different amounts of LO2 and HepG2 cells for 30 min, and the chemiluminescence images in these wells were acquired (Fig. 5A). The chemiluminescence images became brighter with increasing cell number; the chemiluminescence images of HepG-2 tumor cells were much brighter than those of LO2 cells (Fig. 5B). Notably, a good linear relationship was observed between the chemiluminescence intensity and cell number from 2500 to 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in the wells (Fig. S10B†). These results show that probe 1 can detect LAP activity in living cells in real time and distinguish tumor cells and normal cells through sensitive chemiluminescence imaging in different cells.
000 in the wells (Fig. S10B†). These results show that probe 1 can detect LAP activity in living cells in real time and distinguish tumor cells and normal cells through sensitive chemiluminescence imaging in different cells.
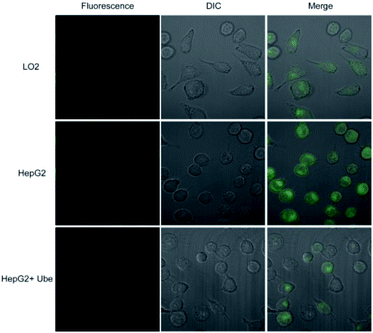
Probe 1 was incubated with LO2 cells, HepG2 cells, and HepG2 cells pretreated with Ube (40 μM) for 1 h. Results showed that the fluorescence intensity in HepG2 cells was remarkably enhanced compared to those in LO2 and Ube-pretreated HepG2 cells (Fig. 6 and S11†). Therefore, the change in fluorescence intensity is caused by endogenous LAP in cells, which indicates that LAP expression in HepG2 cells is higher than that in LO2 cells. Moreover, Ube can inhibit LAP activity in HepG2 cells and confirm the LAP-specific activation of probe 1 at the cell level.
The cell subcellular localization experiment was performed to study the location where probe 1 enters the main agglomeration of cells after enzyme activation. The experimental results showed that in HepG2 cells, the fluorescence signal activated by probe 1 overlaps with the lysosome dye signal (Fig. S12†). This phenomenon is similar to the fluorescence distribution of fluorescence product 6-1 directly incubated with HepG2 cells. This finding was obtained possibly because the fluorescent product 6-1 activated by LAP has exposed hydroxyl groups and interacts with the weak acidity of lysosomes, which results in the gathering of a large amount of hydrolyzed product 6-1 in the lysosomes.
LAP detection in liver cancer model
In vivo real-time imaging offers a powerful tool for accurately diagnosing disease and suspicious lesions with valuable spatiotemporal precision. Current fluorescent probes for imaging LAP activity are not suitable for in vivo experiments, because they are disturbed by intrinsic auto-fluorescence background signals. Accordingly, considering the prominent performance of the LAP probe in cellular chemiluminescence imaging, we examined the applicability of probe 1 for the in vivo real-time visualization of endogenous LAP activity in HepG2 tumor-bearing mice. Chemiluminescence images were collected at different times after the in situ injection of PBS, probe 1, or probe 1 with Ube pre-injection. The experimental results showed that the chemiluminescence signal rapidly responded to LAP activity and reached the maximum at 10 min after probe 1 was injected into the tumor region (Fig. 5C and S13†). The region treated with Ube exhibited a low chemiluminescence signal under the same conditions. A very small amount of chemiluminescence signal was observed in the PBS control group. Therefore, probe 1 can be used for LAP imaging in living tumors and the real-time monitoring of LAP activity in situ.Tissue samples from tumor-bearing mice were prepared and analyzed using probe 1 to further confirm the usability of probe 1 in complex biological systems. The results in Fig. 5D are consistent with the measurement trend of Leu-AMC. A higher chemiluminescence enhancement, which indicates a higher LAP level, was obtained in the serum, liver, and tumor tissues than in other tissues. The results showed that probe 1 has excellent chemiluminescence performance, accuracy, and sensitivity for LAP analysis.
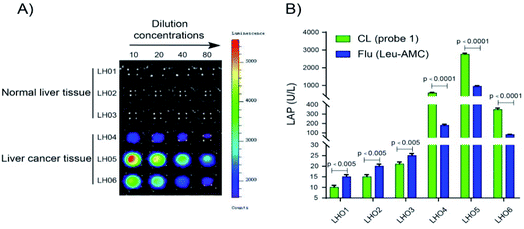
LAP detection in human tissue samples
Probe 1 was used to detect LAP in human tissue samples to determine whether it can distinguish normal tissue and liver cancer tissue. Chemiluminescence images were acquired after probe 1 was incubated with different concentrations of the supernatant (10% tissue homogenate) for 10 min at 37 °C in enzyme reaction buffer (Fig. 7). As shown in Fig. 7A, a super strong chemiluminescence signal was observed after probe 1 was incubated with the supernatant of 10% liver cancer tissue homogenate, whereas a weak chemiluminescence intensity was detected in normal tissues. LAP activity in human tissue samples (normal and liver cancer tissues) was detected using probe 1. The results are consistent with the measurement trend of Leu-AMC, which indicates that probe 1 is a reliable detection method that can be used to distinguish normal tissue and liver cancer tissue and detect LAP activity (Fig. 7B).Conclusions
In summary, we designed and synthesized a LAP-activable chemiluminescent probe and proved its mechanism and specificity for LAP detection. The sensitivity for LAP detection of the chemiluminescent probe was greatly improved compared with traditional fluorescent probes. Furthermore, our study shows that probe 1 can detect LAP-related cells in vitro and can be used to image LAP in living tumors and monitor its activity in situ in real time. In addition, the chemiluminescence of the probe in liver cancer tissues was remarkably enhanced compared to that in normal tissues, which enables the differentiation between liver cancer tissues and normal tissue through endogenous LAP detection. Therefore, the successful application of probe 1 makes it a valuable imaging tool for LAP-related physiological and pathological processes and medicine. We anticipate that the design strategy proposed in this study could be broadly used for the development of other protease probes for protein detection, disease diagnosis, and drug discovery.Data availability
All experimental supporting data and procedures are available in the ESI.†Author contributions
K. C. supervised and analyzed the experiments. B.-Q. W. designed and performed the majority experiments. Z.-Z. C. and X.-H. C. contributed to the initial implementation and data analysis. Y.-Q. L., E. L., L. Z., and Y.-J. Z. guided and revised the paper. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
All animal studies were conducted according to institutional animal care and use regulations approved by the Animal Research Center of Southern Medical University (Permit number SCKK 2016-0041). This work was supported by grants from the National Natural Science Foundation of China (no. 81773558 and 82073689), the National Natural Science Foundation of Guangdong Province (no. 2020A151501518 and 2018B030312010), and the Science and Technology Program of Guangzhou (201904010380).Notes and references
- H. Sung, J. Ferlay and R. L. Siegel, et al. , Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- F. Bray, J. Ferlay and I. Soerjomataram, et al. , Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- J. Zhang, X. Chai and X. P. He, et al. , Chem. Soc. Rev., 2019, 48, 683–722 RSC.
- H. Li, Q. Yao and W. Sun, et al. , J. Am. Chem. Soc., 2020, 142, 6381–6389 CrossRef CAS PubMed.
- (a) D. Cheng, J. Peng and Y. Lv, et al. , J. Am. Chem. Soc., 2019, 141, 6354–6361 Search PubMed; (b) S. Mizukami, K. Tonai and M. Kaneko, et al. , J. Am. Chem. Soc., 2008, 130, 14376–14377 CrossRef CAS PubMed.
- (a) Y. Chen, Mater. Today Chem., 2020, 15, 100216 CrossRef CAS; (b) L. Liu, Y. You and K. Zhou, et al. , Angew. Chem., Int. Ed., 2019, 58, 14929–14934 CrossRef CAS PubMed; (c) Q. Gong, W. Shi and L. Li, et al. , Chem. Sci., 2016, 7, 788–792 RSC; (d) X. He, L. Li and Y. Fang, et al. , Chem. Sci., 2017, 8, 3479–3483 RSC; (e) W. Zhang, F. Liu and C. Zhang, et al. , Anal. Chem., 2017, 89, 12319–12326 CrossRef CAS PubMed.
- T. Wang, Q. Sun and H. Xiong, et al. , Sens. Actuators, B, 2020, 321, 128631 CrossRef CAS.
- (a) J. Zhang, X. Chai and X. P. He, et al. , Chem. Soc. Rev., 2019, 48, 683–722 RSC; (b) A. C. Sedgwick, L. Wu and H. H. Han, et al. , Chem. Soc. Rev., 2018, 47, 8842–8880 RSC.
- N. Hananya and D. Shabat, et al. , Angew. Chem., Int. Ed., 2017, 56, 16454–17463 CrossRef CAS PubMed.
- S. Gnaim, O. Green and D. Shabat, et al. , Chem. Commun., 2018, 54, 2073–2085 RSC.
- U. Haris, H. N. Kagalwala and Y. L. Kim, et al. , Acc. Chem. Res., 2021, 54, 2844–2857 CrossRef CAS PubMed.
- (a) N. Hananya, O. Green and R. Blau, et al. , Angew. Chem., Int. Ed., 2017, 56, 11793–11796 CrossRef CAS PubMed; (b) P. Cheng, Q. Miao and J. Li, et al. , J. Am. Chem. Soc., 2019, 141, 10581–10584 CrossRef CAS PubMed; (c) J. Huang, J. Li and Y. Lyu, et al. , Nat. Mater., 2019, 18, 1133–1143 CrossRef CAS PubMed; (d) K. J. Bruemmer, O. Green and T. A. Su, et al. , Angew. Chem., Int. Ed., 2018, 57, 7630–7634 CrossRef; (e) W. An, L. S. Ryan and A. G. Reeves, et al. , Angew. Chem., Int. Ed., 2019, 58, 1361–1365 CrossRef CAS PubMed; (f) J. Sun, Z. Hu and S. Zhang, et al. , ACS Sens., 2019, 4, 87–92 CrossRef CAS PubMed; (g) L. S. Ryan, J. Gerberich and J. Cao, et al. , ACS Sens., 2019, 4, 1391–1398 CrossRef CAS PubMed; (h) N. Hananya and D. Shabat, et al. , ACS Cent. Sci., 2019, 5, 949–959 CrossRef CAS PubMed; (i) S. Ye, N. Hananya and O. Green, et al. , Angew. Chem., Int. Ed., 2020, 59, 14326–14330 CrossRef CAS PubMed; (j) T. Eilon-Shaffer, M. Roth-Konforti and A. Eldar-Boock, et al. , Org. Biomol. Chem., 2018, 16, 1708–1712 RSC; (k) W. Adam, I. Bronstein and B. Edwards, et al. , J. Am. Chem. Soc., 1996, 118, 10400–10407 CrossRef CAS; (l) T. Eilon-Shaffer, M. Roth-Konforti and A. Eldar-Boock, et al. , Org. Biomol. Chem., 2018, 16, 1708–1712 RSC.
- (a) M. Yang, J. N. Huang and J. Fan, et al. , Chem. Soc. Rev., 2020, 49, 6800–6815 RSC; (b) R. An, S. Wei and Z. Huang, et al. , Anal. Chem., 2019, 91, 13639–13646 CrossRef CAS PubMed; (c) S. Gutkin, O. Green and G. Raviv, et al. , Bioconjugate Chem., 2020, 31, 2488–2493 CrossRef CAS PubMed; (d) A. Fu, H. Wang and T. Huo, et al. , Anal. Chem., 2021, 93, 6501–6507 CrossRef CAS PubMed; (e) J. I. Scott, S. Gutkin and O. Green, et al. , Angew. Chem., Int. Ed., 2021, 60, 5573–6184 CrossRef PubMed; (f) O. Shelef, A. C. Sedgwick and S. Pozzi, et al. , Chem. Commun., 2021, 57, 11386–11389 RSC; (g) O. Shelef, A. C. Sedgwick and S. Pozzi, et al. , Chem. Commun., 2021, 57, 11386–11389 RSC.
- M. Roth-Konforti, O. Green and M. Hupfeld, et al. , Angew. Chem., Int. Ed., 2019, 58, 10361–10367 CrossRef CAS PubMed.
- B. M. Babin, G. Fernandez-Cuervo and J. Sheng, et al. , ACS Cent. Sci., 2021, 7, 803–814 CrossRef CAS PubMed.
- (a) Y. Zhang, X. Chen and Q. Yuan, et al. , Chem. Sci., 2021, 12, 14855–14862 RSC; (b) Y. Liu, L. Teng and C. Xu, et al. , Chem. Sci., 2019, 10, 10931–10936 RSC; (c) X. Huang, Q. Lei and S. Huang, et al. , Chem. Commun., 2021, 57, 6608–6611 RSC.
- (a) O. Green, S. Gnaim and R. Blau, et al. , J. Am. Chem. Soc., 2017, 139, 13243–13248 CrossRef CAS PubMed; (b) Y. Zhang, C. Yan and C. Wang, et al. , Angew. Chem., Int. Ed., 2020, 59, 9059–9066 CrossRef CAS PubMed.
- (a) M. Yang, J. Zhang and D. Shabat, et al. , ACS Sens., 2020, 5, 3158–3164 CrossRef CAS PubMed; (b) J. Huang, Y. Jiang and J. Li, et al. , Angew. Chem., Int. Ed., 2021, 60, 3999–4003 CrossRef CAS PubMed.
- (a) M. Zhang, Z. Tian and J. Wang, et al. , ACS Sens., 2021, 6, 3604–3610 CrossRef CAS PubMed; (b) M. Sakabe, D. Asanuma and M. Kamiya, et al. , J. Am. Chem. Soc., 2013, 135, 409–414 CrossRef CAS PubMed; (c) H. Fujioka, J. Shou and R. Kojima, et al. , J. Am. Chem. Soc., 2020, 142, 20701–20707 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, synthesis and characterization data, and additional spectra and imaging data. See DOI: 10.1039/d1sc06528a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |