Open Access Article
Open Access ArticleMultiplex surface-enhanced Raman scattering detection of deoxynivalenol and ochratoxin A with a linear polymer affinity agent†
Rebeca S.
Rodriguez
 ,
Victoria M.
Szlag
,
Theresa M.
Reineke
,
Victoria M.
Szlag
,
Theresa M.
Reineke
 and
Christy L.
Haynes
and
Christy L.
Haynes
 *
*
Department of Chemistry, University of Minnesota, 207 Pleasant Street SE, Minneapolis, Minnesota 55455, USA. E-mail: chaynes@umn.edu
First published on 26th October 2020
Abstract
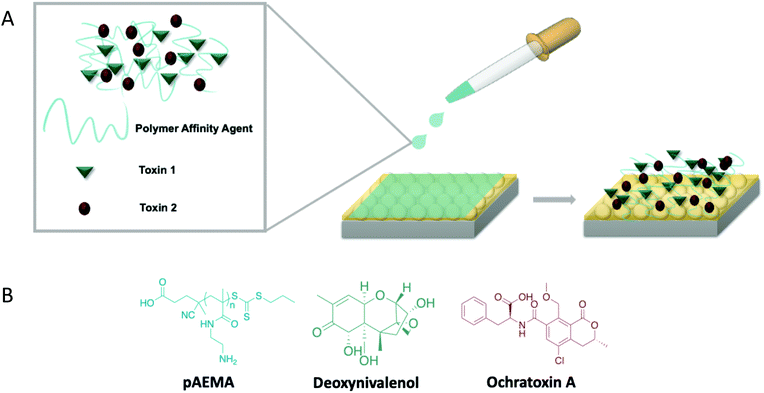
A linear, methacrylamide polymer affinity agent was explored to capture two mycotoxins, deoxynivalenol (DON) and ochratoxin A (OTA), for multiplex surface-enhanced Raman scattering (SERS) detection. These mycotoxins are naturally occurring small molecules from fungi that can be dangerous at low concentrations. SERS detection was completed for each polymer-toxin complex at concentrations relevant to current safety regulation by the FDA: 1 ppm for DON and 5 ppb for OTA. Visibly distinguishable vibrational modes were observed in the multiplex spectra that were attributed to each mycotoxin individually, thus, not requiring any additional chemometric analysis. Density functional theory (DFT) was used to model DON and OTA to accurately label the vibrational modes in the experimental spectra as well as provide insight on the binding between both targets and the affinity agent. Fully modeled vibrations of these toxins are novel contributions due to OTA never being modeled and there being only a few published vibrational modes of DON. DFT guides empirical observations regarding hydrogen bonding at multiple sites of each mycotoxin target molecule through the amine groups on the polymer, confirming the capabilities of a single polymer affinity agent to facilitate multiplex detection of a class of molecules through less-specific interactions than traditional affinity agents.
Introduction
Polymers can serve as affinity agents to capture and detect a wide variety of analytes. Synthetic affinity agents are compelling because they are relatively inexpensive and robust, and can be easily synthesized to tune and exploit certain interactions. Whether in conjunction with other affinity agents for increased specificity1 or synthesized specifically to bind to a target,2 these agents can be used to identify and detect biological toxins,3 food contaminants,4 and other small molecules.5 Polymeric molecularly imprinting (immobilizing a target as a template in a polymer matrix) has long served as the synthetic mechanism to generate polymer-based affinity agents for sensors. However, these systems are difficult to characterize and reproduce, due to their insoluble nature, and sensing measurements have not been directly made in a complex matrix.6–8 In addition, because they are cast to bind a specific target, they cannot detect multiple targets at once without adding additional synthetic steps. These relatively thick polymer templates make it difficult to use surface analytical techniques for target detection due to the fact that the majority of the sensing volume is occupied by the polymer matrix rather than the matrix with captured analytes.In contrast, linear polymer affinity agents allow one to synthetically control the chain length of the polymer and multivalent display of functional groups, where each monomer repeat unit serves as a potential binding site for the target analyte.9,10 Previous work by Szlag and coauthors11 designed a single-point-attachment polymer affinity agent with pendant saccharide moieties on the repeat unit structure to specifically bind a protein used as a bioterror agent. Leveraging simple chemistry with an attractive analytical technique like surface-enhanced Raman scattering (SERS), one can monitor binding of the target to the polymer in both purified and complex matrices. Expanding on this concept, if one allows for similar bonding interactions, the choice of the polymer repeat unit can facilitate multiplex capture of an entire class of molecules.
In light of multiplex detection, SERS is a particularly useful signal transduction mechanism because of its low limits of detection, its ability to provide a spectral “fingerprint” unique to the analyte of interest, and its compatibility with aqueous samples.9,12,13 When an analyte is immobilized near a plasmonic metal surface, one can observe an enhanced intensity of the analyte's signature “fingerprint” due to the vibrational modes of the analyte inelastically scattering light. These surfaces supporting localized plasmons are usually characterized by nanoscale roughness to generate a small-volume, but intense, electromagnetic (EM) field. This EM field extends only a few nanometers from the plasmonic surface; thus, it is imperative to contain both affinity agent and target analyte within the enhancement field to fully take advantage of SERS capabilities.14 Polymer chain length has proven to be a particularly important factor when using polymer affinity agents for SERS detection. With a long chain length, the analyte signature cannot be seen. With a short chain length, there might be insufficient repeat unit binding sites for target–analyte interaction due to dense packing of the short polymer chains at the substrate surface.15
In this work, we focus on the use of SERS and linear polymers as affinity agents for multiplex analyte detection. Recent literature on the optimization of linear polymer affinity agents concluded that the order of polymer and target attachment to the sensing substrate is pivotal to reach relevant levels of detection.16 While traditional affinity agents are often anchored to the sensing substrate first,17,18 polymer affinity agents exhibit heightened flexibility in solution, thus enabling optimal polymer-target binding that is not achievable when pre-anchored to the substrate. This increases the amount of analyte that can readily associate with the polymer. To demonstrate the full potential of linear polymer affinity agents, this work focuses on multiplex detection of two different small molecule targets of interest: deoxynivalenol and ochratoxin A. These molecules are mycotoxins, small molecule toxins that are naturally produced from fungi that contaminate various types of crops and feedstocks.19 Mycotoxins are an especially important class of small molecules to detect due to their toxicity at very low exposure levels and ability to biomagnify in the environment.20–22
Deoxynivalenol (DON), also known as vomitoxin, is a toxin naturally produced by Fusarium bacteria species (F. graminearum and F. culmorum).19 It is one of the most common mycotoxin contaminants in grains such as wheat and corn.23 When ingested, DON is an immunotoxin and can cause severe dehydration due to vomiting and diarrhea; thus, it is regulated by the FDA at 1 ppm for humans.24,25 Ochratoxin A (OTA), is a toxin naturally produced by various Aspergillus and Penicillium bacteria species (A. ochraceus, P. verrucosum, A. carbonarius, and A. nigier).23 OTA tends to contaminate various grains, pork, and alcoholic beverages such as beer and wine. OTA is a nephrotoxin, a teratogen, a potential carcinogen, and has been linked to neurodegenerative diseases.19,26 It is regulated at a much lower concentration compared to DON, 5 ppb, due to its extremely adverse effects.26 Both of these toxins are relevant targets for detection due to their prevalence in food, dangerous effects on livestock and humans, and their toxicity at very low concentrations. Multiplex detection of the two, simultaneously, is also important because they both contaminate grains and grain products.
While various analytical techniques have been used to detect each of these toxins at low limits of detection,27–33 direct multiplex detection of these toxins together with a single affinity agent has not been done. In much of the multiplex sensing literature focused on other targets, the collected data have required post-measurement chemometric analyses to distinguish between each target.34 In this work the toxins’ spectral signals do not require additional separation or filtering as seen with many other immunosensors.35–37 This sensing scheme aims to provide molecule-specific information regarding the target and the capture agent. In this work we use a linear polymer affinity agent to directly detect both DON and OTA simultaneously via surface-enhanced Raman scattering without the need for chemometric analysis. Additionally, previous literature reports of multiplex detection required multiple affinity agents, such as aptamers and antibodies, to enable such detection.34,38 Each affinity agent is incredibly specific to the target and must often be paired with SERS tags or labels to achieve low limits of detection.39–41 Thus, the use of a single non-traditional affinity agent, such as the polymer reported herein which relies on non-specific interactions, could remediate the need for multiple affinity agents. Paired with density functional theory (DFT), not only are we able to predict the vibrational stretches of each small molecule, but this information provides additional insight on association of the analyte and polymers at the molecular level. This multiplex sensing scheme is simple, relatively inexpensive, and requires minimal analysis, displaying the overall potential a linear polymer affinity agent has for detecting various classes of small molecules (Fig. 1).
Materials and methods
Materials
N-(2-Aminoethyl) methacrylamide hydrochloride (AEMA·HCl) was purchased from Polysciences, Inc., (Warrington, PA) and was purified by recrystallization in ethanol.42 Deoxynivalenol from Fusarium graminearum and Fusarium culmorum cultures and ochratoxin A from Petromyces albertensis (OTA, ≥98%) were purchased from Sigma-Aldrich. The polymerization initiator, 4,4′-azobis(4-cyanovaleric acid) (V501, ≥98.0%), was purchased from Sigma-Aldrich as well. The chain transfer agent (CTA), 4-cyano-4-(propylsulfanylthiocarbonyl)-sulfanylpentanoic acid (CPP), was synthesized as previously reported in literature.43 Silica nanospheres, with a 590 nm diameter (10% solids), were purchased from Bangs Laboratories, Inc (Fishers, IN). Pure gold (99.999%) was purchased from Kurt J. Lesker, (Clairton, PA). Purchased reagents did not undergo any further purification unless noted.Polymer synthesis and characterization
Synthesis of this 29 repeat unit polymer is fully described in a previous paper.15 Briefly, AEMA·HCl was dissolved in 90% 1 M acetate buffer, alongside 10% ethanol, the initiator, 4,4′-azobis(4-cyanovaleric acid) (V501), and the CTA 4-cyano-4-(propylsulfanylthiocarbonyl) sulfanylpentanoic acid (CPP). The reaction mixture was degassed via 3 cycles of freeze–pump–thaw and polymerized at 70–80 °C overnight (∼18 h). The polymerization was stopped by exposing the mixture to ambient air. The mixture was dialyzed in a 100–500 Da MWCO membrane against 3 L of Milli-Q water for 24 h. This was then lyophilized, resulting in a dry, light yellow solid with a yield of 78%. The polymer molecular weight was characterized via aqueous mobile phase (0.1 M Na2SO4 in 1.0 vol% acidic acid) size exclusion chromatography (SEC). The instrument used was an Agilent 1260 Infinity Quaternary LC System with Eprogen columns [CATSEC1000 (7 μm, 50 × 4.6), CATSEC100 (5 μm, 250 × 4.6), CATSEC300 (5 μm, 250 × 4.6), and CATSEC1000 (7 μm, 250 × 4.6)]. The system was equipped with a Wyatt HELEOS II light scattering detector (λ = 662 nm) and an Optilab rEX refractometer (λ = 658 nm). The SEC traces are shown in the ESI† (Fig. S1).Isothermal titration calorimetry (ITC)
ITC measurements were performed using a MicroCal PEAQ-ITC Automated (Malvern Instruments, Westborough, MA) at 25 °C as previously discussed in previous literature.15,16 The sample cell and injection syringe were cleaned with 20% Contrad 70 detergent, water, and methanol. The instrument syringe was flushed with 10% bleach twice after each experiment. A 22 vol% DMSO, 16 vol% methanol, 62 vol% acetate buffer (pH 5) mixture was used to make a 4.0 mM polymer repeat unit solution and 0.26 mM OTA and DON samples. The instrument automatedly transferred polymer into the mycotoxin samples (or into blank solvent for the background titration). The titration uses a 1.5 μL injection volume and 150 s injection intervals. Raw ITC profiles (Fig. S2a and b, ESI†) are measured as heat flow rate against time where each peak refers to the injected sample. Integration of these peaks, with respect to time, produces the final ITC plot depicting total heat absorption at each injection vs. polymer repeat unit (RU)/toxin ratio (Fig. S2c, ESI†).Film over nanosphere (FON) fabrication and characterization
FONs were fabricated as previously reported in literature.16,27,44 590 nm-diameter silica nanospheres were dropcast on 1 cm × 1 cm silicon wafers to form a nanosphere mask. A 95.3 nm pure gold film was deposited under vacuum, measured by a quartz crystal microbalance (Denton Vacuum, Moorestown, NJ). FONs with a localized surface plasmon resonance (LSPR) λmax between 750 and 850 nm, measured using a fiber optic probe (Ocean Optics, Dunedin, Florida) with a flat gold film as the reflective standard, were used for these studies.Surface-enhanced Raman scattering (SERS)
A 1 mM polymer solution (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 MeOH/water) was mixed with 50% by volume solutions of varying concentrations of DON and OTA and left to interact for 6 h. FON substrates were then incubated in 200 μL of the complexed mixture in a 24-wellplate for 18 h. Substrates were then washed with 1–2 mL of Milli-Q water and air dried. Measurements were performed using a Snowy Range Instruments SnRI ORS System with a 785 nm laser, 9 mW incident power, and a 10 s integration time. Each condition was measured on three substrates, and five spots on each substrate was measured for a total of 15 averaged spectra. The FON average spectrum was baselined in OriginLab's Origin 9.1 (using eleven anchor points created by the first and second derivative with a Savitsky–Golay smoothing and connected by B-spline interpolation, with the same number of points as the input spectrum) and normalized by the incident power and integration time.
60 MeOH/water) was mixed with 50% by volume solutions of varying concentrations of DON and OTA and left to interact for 6 h. FON substrates were then incubated in 200 μL of the complexed mixture in a 24-wellplate for 18 h. Substrates were then washed with 1–2 mL of Milli-Q water and air dried. Measurements were performed using a Snowy Range Instruments SnRI ORS System with a 785 nm laser, 9 mW incident power, and a 10 s integration time. Each condition was measured on three substrates, and five spots on each substrate was measured for a total of 15 averaged spectra. The FON average spectrum was baselined in OriginLab's Origin 9.1 (using eleven anchor points created by the first and second derivative with a Savitsky–Golay smoothing and connected by B-spline interpolation, with the same number of points as the input spectrum) and normalized by the incident power and integration time.
Density functional theory
Non-resonant Raman spectra of DON and OTA were calculated using density functional calculations in GaussView 6.0.16 with a basis set of B3LYP/6-311G++(d,p) following previous literature that calculated a small number of DON vibrational bands.45 Rotational and vibrational bands were labeled for each molecule and can be found in a comprehensive table in the ESI† (Table S1).Safety consideration
Both ochratoxin A and deoxynivalenol are dangerous mycotoxins that can be carcinogenic. For this reason, anything contaminated must be disposed of appropriately. Samples were prepared in a biosafety level II hood (BSCII). Samples should be handled with double nitrile gloves or layered nitrile and polyethylene gloves. Due to potential aerosol exposure outside of the hood, a fitted respirator is used at all times when handling all mycotoxin-exposed wafers. All instrumentation, benchtops, and hoods are sprayed with a 10% bleach solution and wiped down to avoid any harmful effects. All contaminated solid waste, including everything used in the decontamination process, is disposed of appropriately.Results and discussion
DFT modeling of deoxynivalenol and ochratoxin A
Raman detection of DON in agricultural products by Yuan et al.45 modeled the small molecule with density functional theory (DFT). However, the authors only published a few vibrational band assignments; thus, computational modeling of the molecule was completed here to fully assign all possible peaks in the experimental spectrum for DON. Previous Raman detection of OTA has relied on dye molecules,29 changes in aptamer conformation,46 and other intensity-amplifying attachments30 in order to sense the small molecule. No literature to date has vibrational band assignments for OTA itself. For this reason, DFT calculations were performed for both mycotoxins, and their vibrational band assignments were labeled accordingly. An additional advantage of DFT-based generation of Raman spectra for DON and OTA is that it allows researchers to avoid collecting normal Raman spectra from toxic solid or high concentration mycotoxin solutions. The Raman spectrum of DON was computed following the same basis set as previously reported and agreed well with the past published calculations. The spectra of the two mycotoxins (Fig. 2) have some overlap; nonetheless, there are a sufficient number of stand-alone peaks for each molecule, indicating the potential for successful multiplexing. Previous computational work has labeled the vibrational modes of the AEMA monomer and the CTA anchoring group that make up the affinity agent.15 The computed Raman spectrum for the AEMA monomer can be seen in the ESI† (Fig. S3).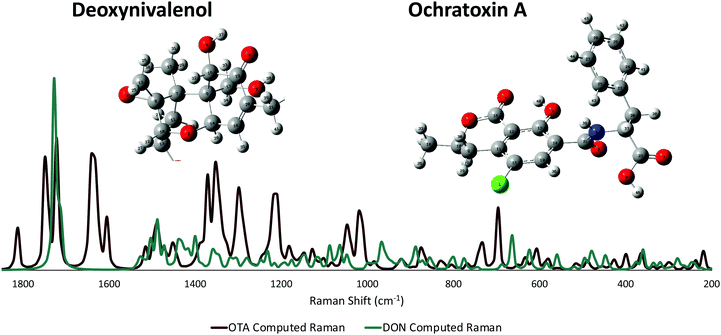 | ||
| Fig. 2 Non-resonant computed Raman spectra for both DON and OTA. The modeled atoms are numbered. An enlarged version of the numbered atoms can be seen in the ESI† (Fig. S4 and S5). | ||
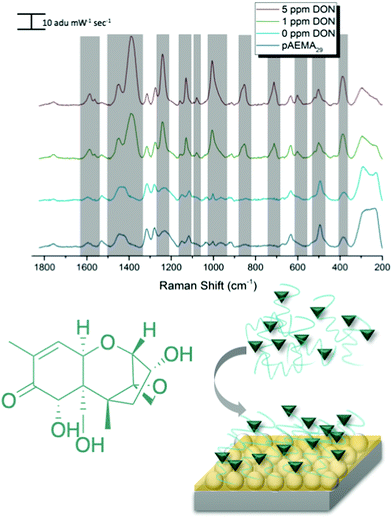
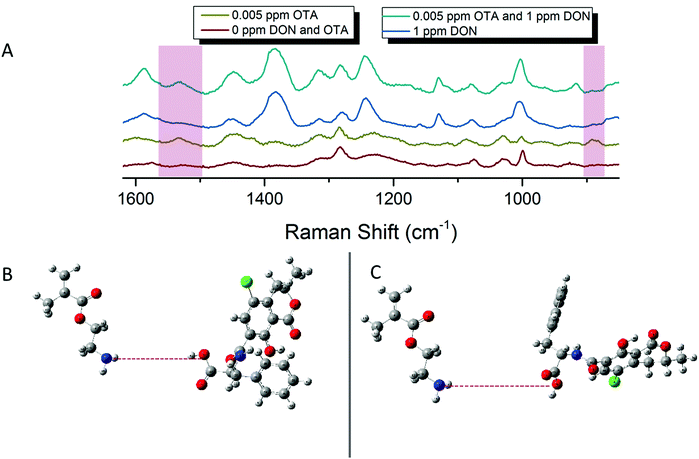
SERS detection of deoxynivalenol with pAEMA29
As mentioned previously, traditional affinity agents are anchored to the sensing substrate, and then the target is flowed or attached to the agent subsequently. In contrast, we allowed DON and pAEMAa29 to complex in solution, through what is hypothesized to be hydrogen bonding, and then anchored this complex to the FON substrate. Comparing the anchored complex at various concentrations of DON to DON-free polymer and the 0 ppm condition (just the solution conditions the toxin is diluted in) in Fig. 3 visibly reveals distinct spectral features, changes in peak intensity, and peak shifts. DON's vibrational signature is apparent at 1 ppm, the concentration at which the toxin is regulated. Additionally, FONs were incubated in varying DON solutions, without the polymer affinity agent, and no visible peaks were seen at relevant DON concentrations (Fig. S6, ESI†). By comparing vibrational modes in the DON computational spectrum, the monomer/CTA spectrum, and the experimental spectra, we can further confirm binding between small molecule and polymer. A comparison of DON and monomer/CTA DFT vibrational modes to experimental vibrational modes can be seen in Table 1. A graph displaying SERS intensity as it relates to DON concentration at a specific wavenumber shift can be seen in Fig. S7 (ESI†).| DON DFT calculated Raman shift (cm−1 shift) | DON experimental Raman shift (cm−1 shift) | Monomer/CTA calculated Raman shift (cm−1 shift) | Monomer/CTA experimental Raman shift (cm−1 shift) |
|---|---|---|---|
| 1530: scissoring of H's on C12, twisting of H's on C15 | 1590 and 1566 shoulder | 1610: monomer bending of N2 H's | 1607 |
| 1450: asymmetric out of plane rocking of H's on C17 and wagging of H38 on O5, wagging of H37 on O4, slight wagging of H's on C11 | 1455 | 1423 and 1428: O1–C4–N1 symmetric stretch, N1–C5 stretch, rocking of H on N1 and asymmetric bending of C1 H's respectively | 1433 |
| 1384: rocking of H37 on O4, symmetric angle bending of C18–C14 displacing H35 and H28, rocking of Hs on C21, ring rocking of cyclohexene | 1388 | ||
| 1245: strong stretching of H22 on C10, symmetric stretching of H27 on C13 and O3 on C13, strong angle wagging of H38 on O5 and H33 on C17 | 1241 | 1238: twisting/wagging of H's on N1, C5, C6, and N2 | 1234 |
| 853: ring breathing C10, H's on C11 in plane stretch, H32 rocking, H35 bend | 852 |
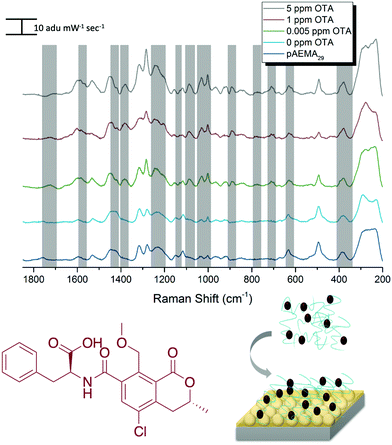
SERS detection of ochratoxin A with pAEMA29
Following the same protocol as with DON, FONs were incubated in an OTA and pAEMA29 mixture at varying concentrations, and the captured spectra can be seen in Fig. 4. Distinct and visible spectral changes are observed at concentrations as low as the OTA regulatory limit (0.005 ppm/5 ppb). A select table comparing OTA and monomer/CTA DFT vibrational modes to the experimental modes can be seen in Table 2. FONs were incubated in varying OTA concentrations, without polymer affinity agent and it was not possible to detect OTA at relevant concentrations (Fig. S8, ESI†). A graph displaying SERS intensity as it relates to OTA concentration at a specific wavenumber shift can be seen in Fig. S9 (ESI†). It is important to note that variation in FON enhancement during random sampling makes it difficult to correlate SERS intensity linearly to concentration. FONs have variation in enhancement substrate-to-substrate and spot-to-spot.47 We have randomly chosen FONs and spots on the FON when obtaining spectra which is why there is variation in SERS intensity.| OTA DFT calculated Raman shift (cm−1) | OTA experimental Raman shift (cm−1) | Monomer/CTA calculated Raman shift (cm−1) | Monomer/CTA èxperimental Raman shift (cm−1) |
|---|---|---|---|
| 1378: strong H32 wagging on C17, asymmetric wagging of Hs on C20, slight twisting of benzene ring Hs (H42, H44, etc.) | 1384 | ||
| 1082: strong C9 H's stretch causing a strong inner ring distortion on C9, C10, C11, wagging of H39, asymmetric stretch of benzene ring at C11, wagging of H40 | 1089 | ||
| 898: in plane wagging of H's on C18, angle breath of C10 and C18, C9 stretching | 891 | ||
| 714: asymmetric out of plane breathing of benzene ring, strong displacement of H41, H42, and H45 | 713 | ||
654: outward bond angle stretch of C23![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O7 and O6 and H46, twisting of H's on C20, ring distortion at C11 and twisting of H29, ring twist displacing H43 O7 and O6 and H46, twisting of H's on C20, ring distortion at C11 and twisting of H29, ring twist displacing H43 |
676 | 654: S3–C5 stretch (trithiocarbonate) | 650 |
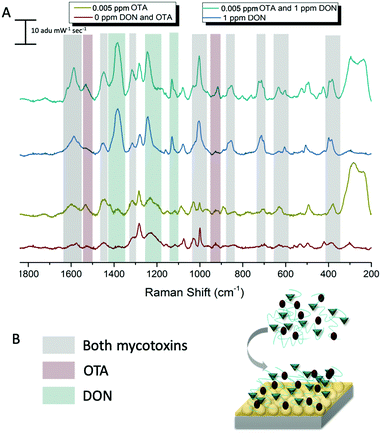
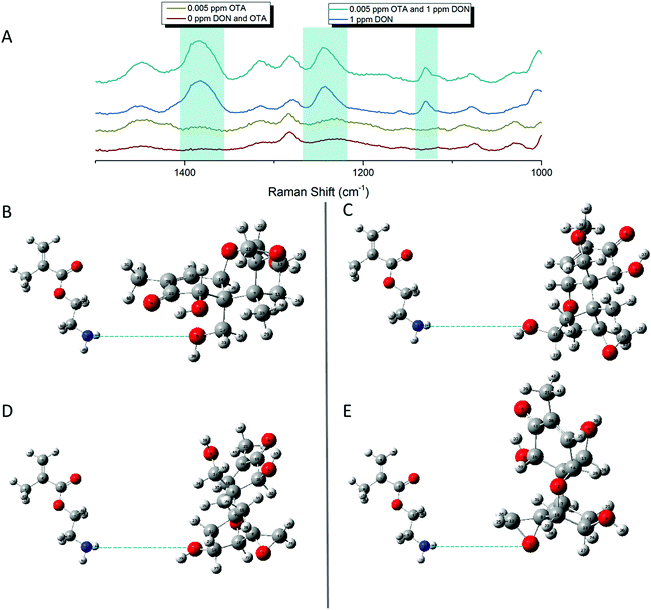
Multiplex SERS detection of deoxynivalenol and ochratoxin A with pAEMA29
In an effort to detect both toxins simultaneously, both DON and OTA were placed in solution with pAEMA29 to give both the opportunity to complex with the polymer affinity agent via the hypothesized hydrogen bonding/association. The multiplexed spectra, seen in Fig. 5, display vibrational band character from both mycotoxins. While there is significant overlap between the two spectra, as predicted by the computational spectra in Fig. 2, there are still stand-alone vibrational modes that correspond to each toxin individually, implying that each toxin can be distinguished from the other.As previously stated, our proposed association between mycotoxin and our polymer system is through hydrogen bonding interactions, while most other mycotoxin and affinity agent sensing systems rely on much more specific interactions. While multiplex mycotoxin sensing via hydrogen bonding interactions has not previously been reported, there is literature precedent that shows sensors composed of organic materials can rely on hydrogen bonding interactions to achieve low limits of detection.48,49 ITC work screening polymers against mycotoxins showed that when the hydrogen groups on the polymer are methylated, no binding is observed.15 Previous computational work done by Piletska et al.50 screened various monomers to bind to two different mycotoxins for chromatography applications. Their modeling revealed a strong binding energy (−41.94 kcal mol−1) between OTA and a monomer structure with amine groups. Unfortunately, the monomer modeled in that work is not easily amenable to controlled polymerizations, thus was not used in our studies. However, we hypothesized that the same interactions could be exploited with our sensing system due to the amine groups on our polymer and the moieties on both OTA and DON that can readily interact with amines through hydrogen bonding. Thus, by monitoring the stand-alone, unique peaks to each mycotoxin in the multiplex spectra, we can confirm the types of interactions occurring while sensing the two small molecules simultaneously.
The carboxyl group on the phenylalanine moiety of OTA can hydrogen bond and electrostatically interact with the amino group on the primary amine monomers.50 In the multiplex spectra, the 1535 cm−1 shift band, unique to OTA, can be referenced to the 1527 cm−1 shift band in the computational spectrum. When monitoring this computational mode in real time, the stretch is attributed to strong vibrations on the benzyl ring of the phenylalanine moiety of OTA. This indicates hydrogen bonding at the carboxyl of the OTA phenylalanine. The second stand-alone peak in the multiplex spectra, at 916 cm−1 shift, can be referenced to the 917 cm−1 shift in the computational spectrum due to strong asymmetric stretching of the tertiary carbons attached to the carboxyl group previously mentioned. This further confirms hydrogen bonding between OTA and the linear polymer affinity agent. An enlarged image of the multiplex spectra and hypothesized binding can be seen in Fig. 6.
Following the same hypotheses for OTA and pAEMA29, observing the unique DON peaks in the multiplex spectra, can help reveal molecular details about DON interactions with the polymer affinity agent. The peak at 1455 cm−1 shift in the multiplex spectra can be referenced to 1450 cm−1 shift from the computational spectrum, strong asymmetric rocking of the hydrogens on a tertiary carbon bound to a hydroxyl group, indicating hydrogen bonding at the hydroxyl. At 1234 cm−1 shift, the polymer and blank spectra have a broadened peak that shifts and sharpens into a peak at 1241 cm−1 shift that can be referenced to the 1245 cm−1 shift in the computational spectrum of DON. This vibrational mode is attributed to both strong stretching of the hydrogens on the bicyclic ring near the hydroxyl (O3) and symmetric stretching of the hydrogens in the out-of-plane hydroxyl group (O5) as labeled in Fig. 7. Lastly, the sharp 1130 cm−1 shift band of DON in the multiplex spectra can be referenced to strong stretches at 1142 and 1146 cm−1 shifts in the computational spectrum. These strong vibrations are due to hydrogen stretches on the bicyclic ring near the hydroxyl (O3), with movement in the epoxide ring and symmetric stretching in the epoxide ring hydrogens with symmetric stretching of the hydrogens in the out-of-plane hydroxyl group (O5), respectively as labeled in Fig. 7D and E. While full analysis of OTA and DON with pAEMA in a food matrix is out of the scope of this work, we have included a spectra of wheat beer on bare FON (Fig. S10, ESI†). We predict that we will be able to observe DON and OTA character in this complex matrix because of the position of the multiplex vibrational modes when compared to the beer signature. There are no visible peaks at 916 cm−1 shift (OTA), 1130 cm−1 shift (DON), or 1234 cm−1 shift (DON) in the beer only spectra, setting precedent for the ability to multiplex in the complex media these toxins contaminate.
Conclusions
Here we have exploited the use of basic molecular hypotheses to design a linear polymer affinity agent that can bind multiple targets. pAEMA, a simple and inexpensive linear polymer, can be used to complex at least two small molecule mycotoxins, DON and OTA, and anchor onto a sensing substrate through trithiocarbonate and gold interactions. We have shown that we can detect both toxins individually using SERS, with no additional sensing probe molecule. Sensing of both toxins simultaneously is possible and visibly distinguishable without any chemometric analysis of the data. We have computationally modeled the OTA Raman spectrum for the first time as well as added to the overall vibrational labeling of the DON Raman spectrum. Moreover, DFT can assist not only in the labeling of strong vibrational modes, but in the ability to monitor the stretches in real-time, allowing us to make clear conclusions about the fundamental interactions occurring between target and affinity agent during sensing. Through this work, we have noted that hydrogen bonding associates the two mycotoxins to the polymer through the amine groups. Additionally, there seems to be hydrogen bonding occurring at multiple sites on the small molecules based on the location of the vibrational shifts. Both toxins are able to interact and complex to the polymer at concentrations relevant to their regulation limits: 1 ppm and 5 ppb for DON and OTA, respectively. This further shows that linear polymer affinity agents can serve as unique capture agents due to their easily modifiable pendant groups, inexpensive nature, and non-specificity towards solely one target. We note that any properties, physical or chemical, that change the overall polymer will impact the polymer's ability to act as an effective affinity agent. However, these changes may promote reversibility of the sensor in future work. Future work will explore their use in complex matrices that are relevant to these targets in order to make this a commercially viable sensing system as well as continued optimization of the sensing substrates to facilitate relatively fast and efficient multiplex sensing.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Funding sources
Isothermal titration calorimetry was carried out using an ITC-200 microcalorimeter, funded by the NIH Shared Instrumentation Grant S10-OD017982. RSR would like to acknowledge both the Mistletoe Foundation and the Lester C. and Joan M. Krogh Endowed Fellowship through the Chemistry department of UMN for funding.Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
We would like to acknowledge 3M for their collaboration and funding and the Minnesota Super Computing Institute for their facilities. RSR would like to acknowledge both the Mistletoe Foundation and the Lester C. and Joan M. Krogh Endowed Fellowship through the Chemistry department of UMN for funding. RSR would also like to thank Daniel Graham from the Goodpaster Group and members of the Reineke group, specifically Derek Saxon and Rishad Dalal, for their thoughtful discussions.Notes and references
- N. Malmstadt, D. E. Hyre, Z. Ding, A. S. Hoffman and P. S. Stayton, Affinity Thermoprecipitation and Recovery of Biotinylated Biomolecules via a Mutant Streptavidin−Smart Polymer Conjugate, Bioconjugate Chem., 2003, 14(3), 575–580, DOI:10.1021/bc020055l.
- J. Xu, S. Ambrosini, E. Tamahkar, C. Rossi, K. Haupt and B. Tse Sum Bui, Toward a Universal Method for Preparing Molecularly Imprinted Polymer Nanoparticles with Antibody-like Affinity for Proteins, Biomacromolecules, 2016, 17(1), 345–353, DOI:10.1021/acs.biomac.5b01454.
- A. Ait Lahcen, F. Arduini, F. Lista and A. Amine, Label-Free Electrochemical Sensor Based on Spore-Imprinted Polymer for Bacillus Cereus Spore Detection, Sens. Actuators, B, 2018, 276, 114–120, DOI:10.1016/j.snb.2018.08.031.
- H. Munawar, K. Smolinska-Kempisty, A. Garcia Cruz, F. Canfarotta, E. Piletska, K. Karim and S. A. Piletsky, Molecularly Imprinted Polymer Nanoparticle-Based Assay (MINA): Application for Fumonisin B1 Determination, Analyst, 2018, 143(14), 3481–3488, 10.1039/C8AN00322J.
- C. S. Storer, Z. Coldrick, D. J. Tate, J. M. Donoghue and B. Grieve, Towards Phosphate Detection in Hydroponics Using Molecularly Imprinted Polymer Sensors, Sensors, 2018, 18(2), 531, DOI:10.3390/s18020531.
- J. J. BelBruno, Molecularly Imprinted Polymers, Chem. Rev., 2019, 119(1), 94–119, DOI:10.1021/acs.chemrev.8b00171.
- J. O. Mahony, K. Nolan, M. R. Smyth and B. Mizaikoff, Molecularly Imprinted Polymers—Potential and Challenges in Analytical Chemistry, Anal. Chim. Acta, 2005, 534(1), 31–39, DOI:10.1016/j.aca.2004.07.043.
- G. Vasapollo, R. D. Sole, L. Mergola, M. R. Lazzoi, A. Scardino, S. Scorrano and G. Mele, Molecularly Imprinted Polymers: Present and Future Prospective, Int. J. Mol. Sci., 2011, 12(9), 5908–5945, DOI:10.3390/ijms12095908.
- V. M. Szlag, R. S. Rodriguez, J. He, N. Hudson-Smith, H. Kang, N. Le, T. M. Reineke and C. L. Haynes, Molecular Affinity Agents for Intrinsic Surface-Enhanced Raman Scattering (SERS) Sensors, ACS Appl. Mater. Interfaces, 2018, 10(38), 31825–31844, DOI:10.1021/acsami.8b10303.
- J. Lee, J. W. Sohn, Y. Zhang, K. W. Leong, D. Pisetsky and B. A. Sullenger, Nucleic Acid-Binding Polymers as Anti-Inflammatory Agents, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(34), 14055–14060, DOI:10.1073/pnas.1105777108.
- V. M. Szlag, M. J. Styles, L. R. Madison, A. R. Campos, B. Wagh, D. Sprouse, G. C. Schatz, T. M. Reineke and C. L. Haynes, SERS Detection of Ricin B-Chain via N-Acetyl-Galactosamine Glycopolymers, ACS Sens., 2016, 1(7), 842–846, DOI:10.1021/acssensors.6b00209.
- C. L. Haynes, A. D. McFarland and R. P. Van Duyne, Surface-Enhanced Raman Spectroscopy, Anal. Chem., 2005, 77(17), 338A–346A, DOI:10.1021/ac053456d.
- K. C. Bantz, A. F. Meyer, N. J. Wittenberg, H. Im, Ö. Kurtuluş, S. H. Lee, N. C. Lindquist, S.-H. Oh and C. L. Haynes, Recent Progress in SERS Biosensing, Phys. Chem. Chem. Phys., 2011, 13(24), 11551–11567, 10.1039/C0CP01841D.
- K. M. Mayer and J. H. Hafner, Localized Surface Plasmon Resonance Sensors, Chem. Rev., 2011, 111(6), 3828–3857, DOI:10.1021/cr100313v.
- V. M. Szlag, S. Jung, R. S. Rodriguez, M. Bourgeois, S. Bryson, G. C. Schatz, T. M. Reineke and C. L. Haynes, Isothermal Titration Calorimetry for the Screening of Aflatoxin B1 Surface-Enhanced Raman Scattering Sensor Affinity Agents, Anal. Chem., 2018, 90(22), 13409–13418, DOI:10.1021/acs.analchem.8b03221.
- V. M. Szlag, R. S. Rodriguez, S. Jung, M. R. Bourgeois, S. Bryson, A. Purchel, G. C. Schatz, C. L. Haynes and T. M. Reineke, Optimizing Linear Polymer Affinity Agent Properties for Surface-Enhanced Raman Scattering Detection of Aflatoxin B1, Mol. Syst. Des. Eng., 2019, 4, 1019–1031, 10.1039/C9ME00032A.
- A. R. Campos, Z. Gao, M. G. Blaber, R. Huang, G. C. Schatz, R. P. Van Duyne and C. L. Haynes, Surface-Enhanced Raman Spectroscopy Detection of Ricin B Chain in Human Blood, J. Phys. Chem. C, 2016, 120(37), 20961–20969, DOI:10.1021/acs.jpcc.6b03027.
- J. Chen, B. Park, Y. Huang, Y. Zhao and Y. Kwon, Label-Free SERS Detection of Salmonella Typhimurium on DNA Aptamer Modified AgNR Substrates, J. Food Meas. Charact., 2017, 11(4), 1773–1779, DOI:10.1007/s11694-017-9558-6.
- J. W. Bennett and M. Klich, Mycotoxins, Clin. Microbiol. Rev., 2003, 16(3), 497–516, DOI:10.1128/CMR.16.3.497-516.2003.
- D. Milicevic, K. Nesic and S. Jaksic, Mycotoxin Contamination of the Food Supply Chain - Implications for One Health Programme, Procedia Food Sci., 2015, 5, 187–190, DOI:10.1016/j.profoo.2015.09.053.
- R. a. Gonçalves, D. Schatzmayr, U. Hofstetter and G. Santos, a. Occurrence of Mycotoxins in Aquaculture: Preliminary Overview of Asian and European Plant Ingredients and Finished Feeds, World Mycotoxin J., 2017, 10(2), 183–194, DOI:10.3920/WMJ2016.2111.
- L. Escrivá, G. Font, L. Manyes and H. Berrada, Studies on the Presence of Mycotoxins in Biological Samples: An Overview, Toxins, 2017, 9(8), 251, DOI:10.3390/toxins9080251.
- J. I. Pitt and J. D. Miller, A Concise History of Mycotoxin Research, J. Agric. Food Chem., 2017, 65(33), 7021–7033, DOI:10.1021/acs.jafc.6b04494.
- S.-H. Wang, X.-Y. Du, Y.-M. Huang, D.-S. Lin, P. L. Hart and Z.-H. Wang, Detection of Deoxynivalenol Based on a Single-Chain Fragment Variable of the Antideoxynivalenol Antibody, FEMS Microbiol. Lett., 2007, 272(2), 214–219, DOI:10.1111/j.1574-6968.2007.00765.x.
- FDA Mycotoxin Regulatory Guidance: A Guide for Grain Elevators, Feed Manufacturers, Grain Processors and Exporters, National Grain and Feed Association August 2011.
- A. Pfohl-Leszkowicz and R. A. Manderville, Ochratoxin A: An Overview on Toxicity and Carcinogenicity in Animals and Humans, Mol. Nutr. Food Res., 2007, 51(1), 61–99, DOI:10.1002/mnfr.200600137.
- D. Kim, A. R. Campos, A. Datt, Z. Gao, M. Rycenga, N. D. Burrows, N. G. Greeneltch, C. A. Mirkin, C. J. Murphy, R. P. Van Duyne and C. L. Haynes, Microfluidic-SERS Devices for One Shot Limit-of-Detection, Analyst, 2014, 139(13), 3227–3234, 10.1039/c4an00357h.
- B. C. Galarreta, M. Tabatabaei, V. Guieu, E. Peyrin and F. Lagugné-Labarthet, Microfluidic Channel with Embedded SERS 2D Platform for the Aptamer Detection of Ochratoxin A, Anal. Bioanal. Chem., 2013, 405(5), 1613–1621, DOI:10.1007/s00216-012-6557-7.
- E.-O. Ganbold, C. M. Lee, E.-M. Cho, S. J. Son, S. Kim, S.-W. Joo and S. I. Yang, Subnanomolar Detection of Ochratoxin A Using Aptamer-Attached Silver Nanoparticles and Surface-Enhanced Raman Scattering, Anal. Methods, 2014, 6(11), 3573–3577, 10.1039/C4AY00440J.
- N. H. Kim, S. J. Lee and M. Moskovits, Aptamer-Mediated Surface-Enhanced Raman Spectroscopy Intensity Amplification, Nano Lett., 2010, 10(10), 4181–4185, DOI:10.1021/nl102495j.
- Y. Li, H. Wang, B. Jia, C. Liu, K. Liu, Y. Qi and Z. Hu, Study of the Interaction of Deoxynivalenol with Human Serum Albumin by Spectroscopic Technique and Molecular Modelling, Food Addit. Contam., Part A, 2013, 30(2), 356–364, DOI:10.1080/19440049.2012.742573.
- Y. Peng, L. Geng, X. Liu, M. Liu, H. Wu and J. Li, A Label-Free Small Molecular Hydrogel-Based Electrochemical Immunosensor for Ultrasensitive Detection of Deoxynivalenol, Anal. Methods, 2019, 11, 5948–5952, 10.1039/C9AY02018G.
- D. Kong, X. Wu, Y. Li, L. Liu, S. Song, Q. Zheng, H. Kuang and C. Xu, Ultrasensitive and Eco-Friendly Immunoassays Based Monoclonal Antibody for Detection of Deoxynivalenol in Cereal and Feed Samples, Food Chem., 2019, 270, 130–137, DOI:10.1016/j.foodchem.2018.07.075.
- J. Li, H. Yan, X. Tan, Z. Lu and H. Han, Cauliflower-Inspired 3D SERS Substrate for Multiple Mycotoxins Detection, Anal. Chem., 2019, 91(6), 3885–3892, DOI:10.1021/acs.analchem.8b04622.
- G. S. Shephard, Determination of Mycotoxins in Human Foods, Chem. Soc. Rev., 2008, 37(11), 2468–2477, 10.1039/B713084H.
- E. W. Sydenham, G. S. Shephard, P. G. Thiel, C. Bird and B. M. Miller, Determination of Fumonisins in Corn:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Evaluation of Competitive Immunoassay and HPLC Techniques, J. Agric. Food Chem., 1996, 44(1), 159–164, DOI:10.1021/jf950203k.
Evaluation of Competitive Immunoassay and HPLC Techniques, J. Agric. Food Chem., 1996, 44(1), 159–164, DOI:10.1021/jf950203k. - A. Rahmani, S. Jinap and F. Soleimany, Validation of the Procedure for the Simultaneous Determination of Aflatoxins Ochratoxin A and Zearalenone in Cereals Using HPLC-FLD, Food Addit. Contam., Part A, 2010, 27(12), 1683–1693, DOI:10.1080/19440049.2010.514951.
- Y. Zhao, Y. Yang, Y. Luo, X. Yang, M. Li and Q. Song, Double Detection of Mycotoxins Based on SERS Labels Embedded Ag@Au Core–Shell Nanoparticles, ACS Appl. Mater. Interfaces, 2015, 7(39), 21780–21786, DOI:10.1021/acsami.5b07804.
- X. Wang, S.-G. Park, J. Ko, X. Xiao, V. Giannini, S. A. Maier, D.-H. Kim and J. Choo, Sensitive and Reproducible Immunoassay of Multiple Mycotoxins Using Surface-Enhanced Raman Scattering Mapping on 3D Plasmonic Nanopillar Arrays, Small, 2018, 14(39), 1801623, DOI:10.1002/smll.201801623.
- A. Foubert, N. V. Beloglazova and S. De Saeger, Comparative Study of Colloidal Gold and Quantum Dots as Labels for Multiplex Screening Tests for Multi-Mycotoxin Detection, Anal. Chim. Acta, 2017, 955, 48–57, DOI:10.1016/j.aca.2016.11.042.
- Y. Li, Q. Chen, X. Xu, Y. Jin, Y. Wang, L. Zhang, W. Yang, L. He, X. Feng and Y. Chen, Microarray Surface Enhanced Raman Scattering Based Immunosensor for Multiplexing Detection of Mycotoxin in Foodstuff, Sens. Actuators, B, 2018, 266, 115–123, DOI:10.1016/j.snb.2018.03.040.
- Z. Deng, H. Bouchékif, K. Babooram, A. Housni, N. Choytun and R. Narain, Facile Synthesis of Controlled-Structure Primary Amine-Based Methacrylamide Polymers via the Reversible Addition-Fragmentation Chain Transfer Process, J. Polym. Sci., Part A: Polym. Chem., 2008, 46(15), 4984–4996, DOI:10.1002/pola.22826.
- X. Xu, A. E. Smith, S. E. Kirkland and C. L. McCormick, Aqueous RAFT Synthesis of PH-Responsive Triblock Copolymer MPEO−PAPMA−PDPAEMA and Formation of Shell Cross-Linked Micelles, Macromolecules, 2008, 41(22), 8429–8435, DOI:10.1021/ma801725w.
- Ru, E. C. Le, E. Blackie, M. Meyer and P. G. Etchegoin, Surface Enhanced Raman Scattering Enhancement Factors:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A Comprehensive Study, J. Phys. Chem. C, 2007, 111(37), 13794–13803, DOI:10.1021/jp0687908.
A Comprehensive Study, J. Phys. Chem. C, 2007, 111(37), 13794–13803, DOI:10.1021/jp0687908. - J. Yuan, C. Sun, X. Guo, T. Yang, H. Wang, S. Fu, C. Li and H. Yang, A Rapid Raman Detection of Deoxynivalenol in Agricultural Products, Food Chem., 2017, 221, 797–802, DOI:10.1016/j.foodchem.2016.11.101.
- R. Gillibert, M. N. Triba and M. L. de la Chapelle, Surface Enhanced Raman Scattering Sensor for Highly Sensitive and Selective Detection of Ochratoxin A, Analyst, 2017, 143(1), 339–345, 10.1039/C7AN01730H.
- H. Wei, A. McCarthy, J. Song, W. Zhou and P. J. Vikesland, Quantitative SERS by Hot Spot Normalization – Surface Enhanced Rayleigh Band Intensity as an Alternative Evaluation Parameter for SERS Substrate Performance, Faraday Discuss., 2017, 205, 491–504, 10.1039/c7fd00125h.
- J. Cao, C. Lu, J. Zhuang, M. Liu, X. Zhang, Y. Yu and Q. Tao, Multiple Hydrogen Bonding Enables the Self-Healing of Sensors for Human–Machine Interactions, Angew. Chem., Int. Ed., 2017, 56(30), 8795–8800, DOI:10.1002/anie.201704217.
- J. Zhou, H. Lin, X.-F. Cheng, J. Shu, J.-H. He, H. Li, Q.-F. Xu, N.-J. Li, D.-Y. Chen and J.-M. Lu, Ultrasensitive and Robust Organic Gas Sensors through Dual Hydrogen Bonding, Mater. Horiz., 2019, 6(3), 554–562, 10.1039/C8MH01098F.
- E. Piletska, K. Karim, R. Coker and S. Piletsky, Development of the Custom Polymeric Materials Specific for Aflatoxin B1 and Ochratoxin A for Application with the ToxiQuant T1 Sensor Tool, J. Chromatogr. A, 2010, 1217(16), 2543–2547, DOI:10.1016/j.chroma.2009.11.091.
Footnote |
| † Electronic supplementary information (ESI) available: Vibrational band assignments for each mycotoxin, spectra of each mycotoxin without an affinity agent, and isothermal titration calorimetry studies. See DOI: 10.1039/d0ma00608d |
| This journal is © The Royal Society of Chemistry 2020 |