Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The identification of peptides by nanoLC-MS/MS from human surface tooth enamel following a simple acid etch extraction†
Nicolas Andre
Stewart
 a,
Gabriela Ferian
Molina
b,
João Paulo
Mardegan Issa
b,
Nathan Andrew
Yates
cd,
Mark
Sosovicka
e,
Alexandre Rezende
Vieira
f,
Sergio Roberto Peres
Line
g,
Janet
Montgomery
h and
Raquel Fernanda
Gerlach
*b
a,
Gabriela Ferian
Molina
b,
João Paulo
Mardegan Issa
b,
Nathan Andrew
Yates
cd,
Mark
Sosovicka
e,
Alexandre Rezende
Vieira
f,
Sergio Roberto Peres
Line
g,
Janet
Montgomery
h and
Raquel Fernanda
Gerlach
*b
aSchool of Pharmacy and Biomolecular Sciences, University of Brighton, Brighton, UK
bDepartment of Morphology, Physiology, and Basic Pathology, School of Dentistry of Ribeirão Preto, University of São Paulo, FORP/USP. Address: Avenida do Café, S/N, 14040-904 Ribeirão Preto, SP, Brazil. E-mail: rfgerlach@forp.usp.br
cDepartment of Cell Biology, University of Pittsburgh School of Medicine, Pittsburgh, PA 15261, USA
dBiomedical Mass Spectrometry Center, University of Pittsburgh Schools of the Health Sciences, Pittsburgh, PA 15213, USA
eDepartment of Oral and Maxillofacial Surgery, School of Dental Medicine, University of Pittsburgh, 3501 Terrace Street, Pittsburgh, PA 15261, USA
fDepartment of Oral Biology, School of Dental Medicine, University of Pittsburgh. 3501 Terrace Street, Pittsburgh, PA 15261, USA
gDepartment of Morphology, Dental School of Piracicaba, University of Campinas, FOP/UNICAMP, Brazil
hDepartment of Archaeology, Durham University, Dawson Building, South Road, Durham, DH1 3LE, UK
First published on 15th June 2016
Abstract
Tooth enamel is the hardest, densest and most mineralized tissue in vertebrates. This is due to the high crystallinity of enamel. During enamel formation, proteins responsible for mineralization are degraded by proteases, which results in mature enamel having less than 1% proteinaceous material, mostly as peptides. Many toxicological studies have taken advantage of the stability of tooth enamel to study heavy metal exposure, however few studies have been successful in identifying peptides from the enamel, especially from a single tooth. Furthermore, amelogenin, the most abundant protein involved in tooth development, is expressed from both the X and Y chromosomes and is dimorphic. Sequencing of the gender dimorphic peptide regions may be useful in determining gender, especially when no other biomaterial is available nor intact DNA remains. In light of this, a method employing nanoflow liquid chromatography (nanoLC) electrospray ionization tandem mass spectrometry (MS/MS) was used to analyse peptides released through an acid etch of the enamel from individual teeth. Two approaches were investigated, one with trypsin digest following acid etch and one without. Peptide identification was accomplished using typical proteomics methodology by searching against the human proteome. Peptides from the major enamel structural proteins were identified including amelogenin isoforms, ameloblastin, and enamelin. Furthermore, Y-chromosome-specific amelogenin peptides were also detected in mature enamel. Peptides were identified from the enamel of single teeth on present-day and archaeological samples in a non-destructive and minimally invasive method by nanoLC-MS/MS. The identification of tooth enamel specific peptides with this approach allows for its potential applications in forensic analysis and archaeological studies.
Introduction
Dental enamel is the outermost tissue that covers the tooth crown. It is the hardest, densest, and most calcified tissue in toothed vertebrates (96% by weight).1 Because of its unique physical characteristics, enamel is the best preserved tissue from remains which contain teeth. Its mostly unaltered structure is often used for phylogenetic classification of species based, for instance, on morphological features such as the Hunter-Schreger bands.2,3 The epi-illumination of enamel and the differential spread of light through groups of enamel rods has recently been shown to produce light-and-dark patterns resembling a digital impression.4 Toxicological studies have taken advantage of the stability of tooth enamel to study heavy metal exposure.5,6 While enamel exhibits these rich morphological features and important toxicological information in its “rocky matter”, it does not contain DNA due to a lack of cells. Mature enamel, however, does contain a small amount of proteins (<1%). These proteins are mainly the heterogeneous amelogenins (AMELX and AMELY, comprising 90% of the immature enamel matrix),7 ameloblastin (AMBN), enamelin (ENAM), matrix metalloproteinase 20 (MMP20), and kallikrein 4 (KLK4),1 as characterized from immature enamel.8 Most of these proteins are enamel-specific and they are thought to play a significant role in the formation of mature enamel, having evolved from independent genes ∼500 million years ago.9 Biochemical characterization of enamel has been done, mostly from abundant porcine material in order to purify sufficient amounts of protein10 or by expressing and purifying the cloned proteins from bacteria.11 Peptides can be recovered from thousand-year-old to million-year-old remains, for example, enamel-specific peptides were recovered from 1100 year-old mummy teeth from which one peptide was identified by matrix assisted laser desorption ionization tandem time-of-flight (MALDI-TOF/TOF) MS,12 bone proteins were recovered from a femur of a 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 year-old mammoth,13 and collagen has been obtained from mastodon and dinosaur bone.14,15 The ability to recover peptides from old to ancient samples has significant value in many fields where the study of proteins preserved from the past could shed light into diet, lifestyle, and on evolution of the proteins themselves, since evidence of such changes may not be encoded in present DNA.15–17 Schweitzer et al.,18 previously discussed the challenges of obtaining biomolecules from ancient material, and mentions many advantages of using ancient peptides/proteins over DNA for evolutionary phylogenetic analyses.
000 year-old mammoth,13 and collagen has been obtained from mastodon and dinosaur bone.14,15 The ability to recover peptides from old to ancient samples has significant value in many fields where the study of proteins preserved from the past could shed light into diet, lifestyle, and on evolution of the proteins themselves, since evidence of such changes may not be encoded in present DNA.15–17 Schweitzer et al.,18 previously discussed the challenges of obtaining biomolecules from ancient material, and mentions many advantages of using ancient peptides/proteins over DNA for evolutionary phylogenetic analyses.
Previously we have shown the feasibility of identifying peptides from mature enamel from fully calcified human and porcine teeth using MALDI-TOF/TOF MS.19 However, single tooth acid-etchings did not provide sufficient material for MS analysis. We were also successful in identifying two N-terminal peptides from AMELX from >1000 years old mummy teeth, but this required etching of the whole crown.12 Workflows using nano-flow liquid chromatography tandem mass spectrometry (nanoLC-MS/MS) followed by protein database searching has become a routine approach in the field of proteomics as it is a sensitive and selective method for detecting and identifying low levels of peptides from complex mixtures.
In this report, we describe the recovery of enamel-specific peptides from an acid etch sampling of single teeth and identification of unique peptides by nanoLC-MS/MS. Two sampling methods are presented; one with reductive alkylation of cysteines followed by a trypsin digest and one without these steps. The first method was performed on 6 teeth (3 male, 3 female) and the second method was performed on 4 teeth (2 male, 2 female) and 2 teeth (1 male, 1 female) originating from an archeological site c. 600–900 AD. This approach has great potential in its application to the many different fields including forensics, paleontology and archaeology, where enamel, due to its unique properties, may be the only remaining source of unaltered preserved tissue.
Materials & methods
All chemicals and materials were of reagent grade unless otherwise mentioned. The first set of human teeth (3 male, 3 female) were obtained from patients whose 3rd molars were being extracted for various reasons at the Dental Surgery Clinic at the University of Pittsburgh following ethical guidelines under the University of Pittsburgh Institutional Review Board, protocol number: IRB 0511110. The second set of teeth (2 male, 2 female) were attained from patients attending the Dental Surgery Clinic at the Dental School of Ribeirão Preto of the University of São Paulo (Faculdade de Odontologia de Ribeirão Preto, FORP/USP), as approved by the Institutional Review Board protocol number CAAE 0229412.0.0000.5419. Two teeth (one female and one male) were obtained from skeletons from a mid to late Anglo-Saxon cemetery (c. 600–900 AD) in Seaham, UK.Sample preparation: acid etch method
In both experimental sets each tooth provided one sample for analysis. Teeth were freed from any macroscopic soft tissue. The enamel crown was first washed with 3% H2O2 for 5 min, etched in 1 mL 10% (v/v) HCl for 1 min followed by a second etch of 10% HCl containing protease inhibitors: phenanthroline, N-ethylmaleimide, and phenylmethylsulfonyl fluoride; added just prior to use all at 1.0 mM (Sigma-Aldrich, St. Louis, MI, USA) in the cap of a separate microcentrifuge tube for 5 min. The crown was then washed twice with 500 μL ddH2O (milliQ water, Millipore). The initial 1 min etch was discarded and samples from the second etch (5 min acid etch) were dried by vacuum centrifugation (Speed-Vac, Thermo Scientific) and used for analysis. Samples were desalted using a pipet tip packed with reversed-phase resin (POROS R2 50 μm, Life Technologies, NY, USA); 50 μL of resin/tip, to retain proteins/peptides. Elution from resin was accomplished using 50 μL of acetonitrile at 50% (v/v) containing formic acid at 0.2% (v/v) (both from Sigma, HPLC grade). Tryptic digestion of the enamel peptides was carried out on three male and three female samples (first set of teeth). Each dried sample was resuspended in 20 μL water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile (50% v/v, 50 mM ammonium bicarbonate). Five μL of DTT (45 mM, Sigma, HPLC grade) were added to the solution, which was then incubated for 1 hour in the dark at 56 °C. Afterwards, five μL of iodoacetamide (Sigma) at 100 mM were added to the sample, which was incubated for another hour in the dark at room temperature. Samples were then diluted 5 times with 100 mM ammonium bicarbonate solution, and trypsin (Trypsin Gold, Mass Spectrometry grade, Promega, Madison, WI, USA) was added to the solution, which was incubated at 37 °C in the dark for 22 hours. Five μL of formic acid (PA, 98%, Sigma) were added to stop the reaction, and samples were passed again through a tip column with POROS R2 resin. Elution of peptides from the resin was carried out using 50% (v/v) methanol containing 5% (v/v) acetic acid. Samples were dried under vacuum centrifugation (Speed Vac, Savant Thermo Scientific) and redissolved in 50 μL of 0.1% trifluoroacetic acid ddH2O prior to MS analysis.
acetonitrile (50% v/v, 50 mM ammonium bicarbonate). Five μL of DTT (45 mM, Sigma, HPLC grade) were added to the solution, which was then incubated for 1 hour in the dark at 56 °C. Afterwards, five μL of iodoacetamide (Sigma) at 100 mM were added to the sample, which was incubated for another hour in the dark at room temperature. Samples were then diluted 5 times with 100 mM ammonium bicarbonate solution, and trypsin (Trypsin Gold, Mass Spectrometry grade, Promega, Madison, WI, USA) was added to the solution, which was incubated at 37 °C in the dark for 22 hours. Five μL of formic acid (PA, 98%, Sigma) were added to stop the reaction, and samples were passed again through a tip column with POROS R2 resin. Elution of peptides from the resin was carried out using 50% (v/v) methanol containing 5% (v/v) acetic acid. Samples were dried under vacuum centrifugation (Speed Vac, Savant Thermo Scientific) and redissolved in 50 μL of 0.1% trifluoroacetic acid ddH2O prior to MS analysis.
NanoLC-MS/MS
Two microliters of each sample from the first sample set were analyzed by reversed phase nanoLC-MS/MS using a high performance liquid chromatography (HPLC) system (Ultimate 3000, Dionex) equipped with a static flow-splitter, and binary solvent system (solvent A: 0.1% formic acid in HPLC grade water; solvent B: 0.1% formic acid in acetonitrile) coupled to a linear ion trap (LTQ XL, Thermo Scientific). Samples were loaded on the column (75 μm ID, 15 μm tip packed with 10.5 cm of Reprosil-PUR C18, 3 μm particle size, 120 Å pore size, PicoChip, New Objective) for 8 min in 2% solvent A at a flow rate of 0.5 μL min−1, flow was split post sample loop at 8.5 min and chromatography was performed using a linear gradient program (8.5–50 min, 2–40% B, 50–51 min, 40–95% B; 51–52 min, 95% B; 52–52.5 min, 95–2% B, 52.5–55, 2% B). The data-dependent acquisition mode was used to collect MS/MS spectra for the most intense ions (up to 5) from the preceding full-scan mass spectrum (350–1800 m/z) for a total acquisition time of 60 min.One microliter injections of each sample from the second sample set (no trypsin digest) and five microliter injections of the archeological sample set were subjected to reversed phase nanoLC-MS/MS (nanoRS U3000, Thermo Scientific), and binary solvent system (solvent A, 0.1% formic acid, 3% DMSO in HPLC grade water; solvent B, 0.1% formic acid, 3% DMSO in acetonitrile) coupled to a hybrid linear ion trap orbitrap (Orbitrap XL, Thermo Scientific). Peptides were loaded onto a C18 trapping cartridge (Pepmap100C18; 0.3 × 5 mm ID; 5 μm particle size) for 5 min at a flow-rate of 5 μL min−1 in 0.1% TFA loading buffer. Peptides were separated on an analytical column (25 cm × 75 μm; 5 μm particle size, C18 PepMap100) with a flow rate of 300 nL min−1 and a gradient of 0 to 30% solvent B over 40 min, 30% to 70% solvent B over 5 min, 70% to 90% solvent B over 5 min, held constant at 90% for 10 min, 90% to 0% in one min and equilibrated at 0% for 10 min. Nanospray was performed with a 10 μm uncoated silica tip emitter (New Objective, FS360-20-10-N-20).
The MS was operated in data-dependent MS/MS mode in which each full MS scan was collected in the orbitrap, precursor ion range of 300–1600 m/z (R = 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 @ 400 m/z), followed by up to eight MS/MS scans performed in the linear ion trap where the most abundant peptide molecular ions were selected for collision-induced dissociation (CID), using a normalized collision energy of 35%. Total MS acquisition time was 72 min.
000 @ 400 m/z), followed by up to eight MS/MS scans performed in the linear ion trap where the most abundant peptide molecular ions were selected for collision-induced dissociation (CID), using a normalized collision energy of 35%. Total MS acquisition time was 72 min.
Database searches
Data from the first set of samples was searched against the human proteome (Uniprot 02/2013, 87![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 656 entries) with no enzyme constraint, methionine oxidation as variable modification, using average mass with a peptide tolerance of 1.4 Da and a MS/MS tolerance of 0.5 Da using Mascot (v2.4.0, Matrix Science Ltd.). Filtering the data was performed using Scaffold (version 4.2.0, Proteome Software Inc., Portland, OR). Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides.
656 entries) with no enzyme constraint, methionine oxidation as variable modification, using average mass with a peptide tolerance of 1.4 Da and a MS/MS tolerance of 0.5 Da using Mascot (v2.4.0, Matrix Science Ltd.). Filtering the data was performed using Scaffold (version 4.2.0, Proteome Software Inc., Portland, OR). Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides.
For the second set and archaeological sets of samples, data was searched against the human proteome (UniprotKB, 10/2015, canonical and isoform, 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 035 entries) using MaxQuant (Version 1.5.1.2) employing default search settings, methionine oxidation as variable modification, unspecific digestion mode, with a first search peptide tolerance of 20 ppm and a main search peptide tolerance of 4.5 ppm.
035 entries) using MaxQuant (Version 1.5.1.2) employing default search settings, methionine oxidation as variable modification, unspecific digestion mode, with a first search peptide tolerance of 20 ppm and a main search peptide tolerance of 4.5 ppm.
Results and discussion
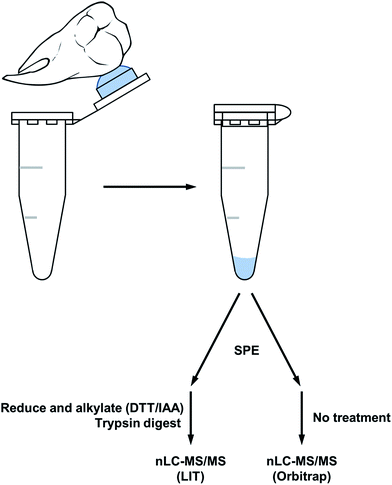
A simple acid etch methodology (see Fig. 1) was applied to two sample sets of teeth to produce peptides for nanoLC-MS/MS analysis. The first sample which was reduced and alkylated followed by an overnight trypsin digest, produced peptides predominantly specific to tooth enamel proteins (see Table 1). Peptides derived from the X/Y isoforms of amelogenin made up the bulk of peptides identified. Other tooth specific peptides identified were from osteopontin and the structural proteins ameloblastin and enamelin. Non-tooth specific peptides were also identified from serum albumin, hemoglobin and collagen and this may be due to the natural lifetime contact of the teeth with the oral cavity. The presence of collagen and osteopontin may also have originated from the root's cementum during the etching procedure as the acid solution tends to spread over the tooth during the etching and may have come into contact with the root.| Peptide count | Accession numbers | Protein name |
|---|---|---|
| 95 | Q99217 | Amelogenin, X isoforms |
| 17 | Q9NP70, Q9NP70-2 | Ameloblastin |
| 10 | P02768 | Serum albumin |
| 9 | P68871 | Hemoglobin subunit beta |
| 8 | Q9NRM1 | Enamelin |
| 7 | P08123 | Collagen alpha-2(I) chain |
| 7 | P69905 | Hemoglobin subunit alpha |
| 6 | Q99218-1 | Amelogenin, Y isoform |
| 6 | P02452 | Collagen alpha-1(I) chain |
| 6 | P10451-5 | Isoform 5 of osteopontin |
| 5 | P01009 | Alpha-1-antitrypsin |
| 2 | Q99217-2 | Isoform 2 of amelogenin, X isoform |
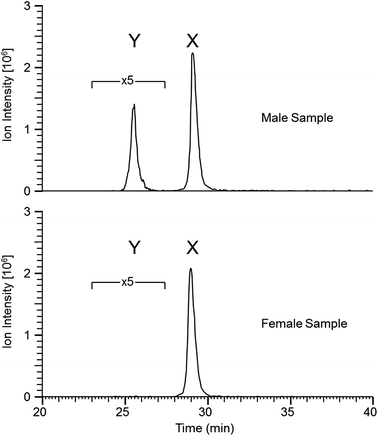
Similar results were obtained from the second set of teeth with the reductive alkylation and trypsin steps omitted (see Table 2); with the exception that osteopontin was not identified. Results from the first set (with trypsin digest) produced additional peptide identifications for amelogenin which are likely the result of the cleavage C-terminal to lysine 24 (AMELX) by trypsin (e.g. ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) .WYQSIRPP.Y). Although the use of trypsin increased the variety of peptides identified it did not greatly improve sequence coverage. Overall, from both methods, the list of proteins is small (a dozen or less) indicative of the nature of the sample. Peptides specific to the Y isoform of amelogenin were identified; a significant finding in this study, allowing the possibility to determine sex from enamel sampling alone. The sequence coverage of amelogenin is shown in Fig. 2 with the dimorphic peptides identified highlighted (all peptide sequences identified for amelogenin isoforms are shown in ESI Table 1†). Of note, the identified peptides originated from two regions of the protein sequence; the tyrosine-rich amelogenin polypeptide (TRAP) N-terminal region (AA1-45) and the hydrophilic charge containing C-terminus (AA165-180) region. Peptides from the central region of the protein, which includes a histidine-rich coil-domain region (AA46-125) and the PXX repeat domain region (AA126-164)20 were not identified. The loss of the central domains is thought to be the result of proteolytic processing during maturation of enamel by matrix metalloproteinase 20 (MMP20) and kallikrein 4 (KLK4).21 The dimorphic sequences of AMELY found in the enamel samples analyzed in this study are: YEVLTPLKWYQSMIRPPYS, WYQSMIRPPY, WYQSMIRPPYS, SMIRPPY and LRPLPPILPDLHLEAWPATDK (dimorphic amino acids are shown in bold, annotated CID spectra are shown in ESI Fig. 1–3 and highlighted in ESI Tables 1 and 2†). Peptides belonging to isoform Y of amelogenin were only identified in male samples. This does not appear to be a result from non-identification from the database searches but a real absence of these as shown by the reconstructed ion chromatogram (RIC) for peptide SIRPPYPPSY ([M + 2H]2+ = 540.2796 m/z) from isoform X compared to the identified dimorphic peptide SM(ox)IRPPY ([M + 2H]2+ = 440.2233 m/z) from isoform Y (see Fig. 3). The amelogenin peptides identified (ESI Tables 1 and 2†) have sequences which agree with predicted cleavage products of KLK4 determined from recombinant porcine amelogenin substrates and fluorogenic peptide substrates.22 Using this simple method to produce enamel derived peptides, without the need of a trypsin digest, different depths of the enamel could be probed by varying the time of exposure to acid. This method may be useful in the identification of inherited diseases of the enamel when DNA material is unavailable, such as in Amelogenesis Imperfecta (AI), where one cause of AI is a mutation in the gene encoding for AMELX.
.WYQSIRPP.Y). Although the use of trypsin increased the variety of peptides identified it did not greatly improve sequence coverage. Overall, from both methods, the list of proteins is small (a dozen or less) indicative of the nature of the sample. Peptides specific to the Y isoform of amelogenin were identified; a significant finding in this study, allowing the possibility to determine sex from enamel sampling alone. The sequence coverage of amelogenin is shown in Fig. 2 with the dimorphic peptides identified highlighted (all peptide sequences identified for amelogenin isoforms are shown in ESI Table 1†). Of note, the identified peptides originated from two regions of the protein sequence; the tyrosine-rich amelogenin polypeptide (TRAP) N-terminal region (AA1-45) and the hydrophilic charge containing C-terminus (AA165-180) region. Peptides from the central region of the protein, which includes a histidine-rich coil-domain region (AA46-125) and the PXX repeat domain region (AA126-164)20 were not identified. The loss of the central domains is thought to be the result of proteolytic processing during maturation of enamel by matrix metalloproteinase 20 (MMP20) and kallikrein 4 (KLK4).21 The dimorphic sequences of AMELY found in the enamel samples analyzed in this study are: YEVLTPLKWYQSMIRPPYS, WYQSMIRPPY, WYQSMIRPPYS, SMIRPPY and LRPLPPILPDLHLEAWPATDK (dimorphic amino acids are shown in bold, annotated CID spectra are shown in ESI Fig. 1–3 and highlighted in ESI Tables 1 and 2†). Peptides belonging to isoform Y of amelogenin were only identified in male samples. This does not appear to be a result from non-identification from the database searches but a real absence of these as shown by the reconstructed ion chromatogram (RIC) for peptide SIRPPYPPSY ([M + 2H]2+ = 540.2796 m/z) from isoform X compared to the identified dimorphic peptide SM(ox)IRPPY ([M + 2H]2+ = 440.2233 m/z) from isoform Y (see Fig. 3). The amelogenin peptides identified (ESI Tables 1 and 2†) have sequences which agree with predicted cleavage products of KLK4 determined from recombinant porcine amelogenin substrates and fluorogenic peptide substrates.22 Using this simple method to produce enamel derived peptides, without the need of a trypsin digest, different depths of the enamel could be probed by varying the time of exposure to acid. This method may be useful in the identification of inherited diseases of the enamel when DNA material is unavailable, such as in Amelogenesis Imperfecta (AI), where one cause of AI is a mutation in the gene encoding for AMELX.
| Peptide count | Accession numbers | Protein name |
|---|---|---|
| 125 | Q99217 | Amelogenin, X isoforms |
| 28 | Q9NRM1 | Enamelin |
| 9 | P68871 | Hemoglobin subunit beta |
| 8 | P02769 | Serum albumin |
| 4 | Q9NP70, Q9NP70-2 | Ameloblastin |
| 3 | Q15149 | Plectin |
| 2 | H0YA46; A1E959 | Odontogenic ameloblast-associated protein |
| 1 | P69905 | Hemoglobin subunit alpha |
| 1 | Q99218-1 | Amelogenin, Y isoform |
 | ||
| Fig. 2 Aligned amino acid sequence of isoform X and Y of amelogenin with coverage and dimorphic peptides highlighted. | ||
It is widely accepted that amelogenins are transcribed from both the X and the Y chromosomes, but that only 10% of the transcripts originate from chromosome Y.23 Jobling, et al., studied samples from 45 males with deletions in the short arms of the Y chromosome, in which AMELY is deleted, and although these teeth had a normal appearance, they suggest that this deletion may be of functional significance.24
Information on the presence of protein transcribed and translated from the AMELY gene is still lacking. Our study suggests that AMELY specific transcripts are translated into protein and some of these peptides remain in the mature enamel and the significance of the Y-specific transcripts with respect to function can be further studied.
The central region of mammalian amelogenin was described by Sire, et al.,25 to be highly variable in sequence when compared to that of reptiles, however the N- and C-terminal regions were highly conserved (over 250 million years of evolution). This implies that these sequence regions of amelogenin are evolutionarily critical. The feasibility to obtain peptides from these highly conserved N- and C-terminal sequences directly from mature enamel using the acid etch method described herein, may contribute in the study of evolution.
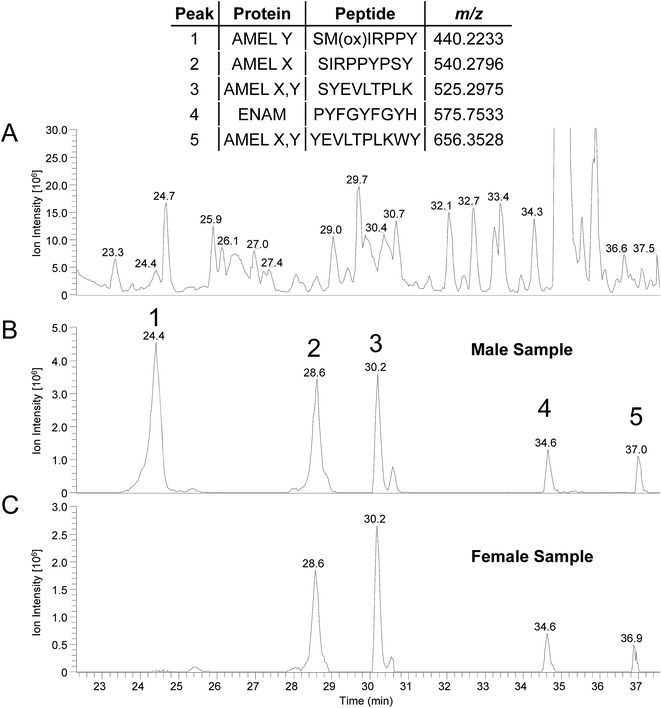
Amelogenin peptides have been found in mummy teeth,12 suggesting that peptides may be preserved inside dental enamel, protected by the hardest of all mammal tissues. Since many species have dimorphism in the amelogenin gene, and since DNA rarely survives more than 10–15 thousand years, the use of enamel peptides may open a window into the past to determine the gender of ancient humans and possibly fossils. To test whether the method can be used on “old” archeological samples we applied the direct acid etch method to two teeth; one male one female, recovered from an Anglo-Saxon cemetery (c. 600–900 AD, Seaham, UK). Similar results to those of “present-day” samples were obtained, as shown in Table 3, where the majority of peptides identified originate from the predominant enamel proteins; amelogenin, ameloblastin and enamelin. The peptide SM(ox)IRPPY ([M + 2H]2+ = 440.2233 m/z) from amelogenin isoform Y was not identified in the male sample from the database search (peptides identified are shown in ESI Table 3†). This is thought to be due to the “unspecific” enzyme search parameter, for when a search is performed using kallikrein like specificity26 it is identified (see MS/MS spectrum; ESI Fig. 4,† which is identical to ESI Fig. 2†). An example of a few peptides from the two archaeological samples is shown in Fig. 4, as the RICs of their corresponding m/z. Again peptide SM(ox)IRPPY is not present in the female sample, however it seems to be in higher abundance compared to the present day samples (Fig. 3); this difference in intensity may be indicative of increased oxidation of the methionine, due to its age or other factors which must be investigated. From these results, it is evident that this method can be applied to archaeological samples and we are currently pursuing this, but results may differ from “present-day” samples and further study is required to characterize these variations. Also, further work would be required to implement this method to assess in the field of forensics/archaeology.
| Peptide count | Accession numbers | Protein name | Sex |
|---|---|---|---|
| 11 | Q99217, Q99218-1 | Amelogenin, X isoforms | Male |
| 4 | Q9NRM1 | Enamelin | Male |
| 3 | Q99217-2 | Isoform 2 of amelogenin, X isoform | Male |
| 3 | Q9NP70, Q9NP70-2 | Ameloblastin | Male |
| 9 | Q99217, Q99218-1 | Amelogenin, X isoforms | Female |
| 5 | Q9NRM1 | Enamelin | Female |
| 2 | Q9NP70, Q9NP70-2 | Ameloblastin | Female |
During the preparation of this manuscript, Castiblanco et al., identified peptides from human tooth enamel without the use of trypsin digest using LC-MS/MS and a Mascot database search.27 However, their methodology involved the cutting of the tooth crown and removing the enamel under a stereomicroscope followed by grinding in liquid nitrogen. Results from our method clearly demonstrate that this intricate process is avoidable if not unnecessary and in comparison our method is minimally invasive; a key feature in the preserving of precious archeological samples.
In conclusion, by means of a simple acid etch technique followed by nanoLC-MS/MS, peptides specific to enamel proteins were identified; isoforms X and Y of amelogenin, ameloblastin and enamelin. This offers the possibilities for studying both present-day and older specimens, since due to its hard and dense properties dental enamel is ideal for preserving such peptides. The etching from one tooth provided ample material to easily identify peptides specific to tooth enamel including sex specific peptides from amelogenin isoform Y.
Acknowledgements
We would like to thank Dr Steve M. Sweet at the Genome Damage and Stability Center, Sussex University for access to the nanoLC-Orbitrap MS instrument. This study was supported by the State of São Paulo Research Foundation (FAPESP, São Paulo, SP, Brazil, Grant 2011/23963-3) and the (Brazilian) National Research Council (CNPq, Brasilia, DF, Brazil). Some of this work used the University of Pittsburgh Cancer Institute shared resources that are supported in part by award P30CA047904. We thank the Laboratorio Nacional de Biociências (LNBio) at LNLS, in Campinas, Brazil, for the use of Mass Spectrometers. We are grateful to Niall Hammond (Archaeo-Environment Ltd) and Northern Archaeological Associates for access to the Seaham skeletal collection excavated in 1999 and to Tessi Loeffelmann for sexing the individuals in the course of her MSc dissertation in 2014 ‘Of doomed and transient men: the Anglo-Saxon skeletons of Seaham in their context’ at Durham University.References
- A. Nanci and A. R. Ten Cate, Ten Cate's oral histology: development, structure, and function, Mosby, St. Louis, Mo, London, 7th edn, Antonio Nanci, 2008 Search PubMed.
- S. R. P. Line and L. P. Bergqvist, J. Vertebr. Paleontol., 2005, 25, 924–928 CrossRef.
- S. R. Line and P. D. Novaes, Brazilian Journal of Morphological Sciences, 2005, 22, 67–72 Search PubMed.
- L. L. Ramenzoni and S. R. Line, Proc. Biol. Sci., 2006, 273, 1155–1158 CrossRef PubMed.
- G. R. de Almeida, C. S. Guerra, J. E. Tanus-Santos, F. Barbosa Jr and R. F. Gerlach, Environ. Res., 2008, 107, 264–270 CrossRef PubMed.
- G. R. Costa de Almeida, C. de Sousa Guerra, G. de Angelo Souza Leite, R. C. Antonio, F. Barbosa Jr, J. E. Tanus-Santos and R. F. Gerlach, Sci. Total Environ., 2011, 409, 1799–1805 CrossRef CAS PubMed.
- S. J. Brookes, C. Robinson, J. Kirkham and W. A. Bonass, Arch. Oral Biol., 1995, 40, 1–14 CrossRef CAS PubMed.
- J. Moradian-Oldak, Front. Biosci., 2012, 17, 1996–2023 CrossRef.
- K. Kawasaki and K. M. Weiss, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4060–4065 CrossRef CAS PubMed.
- H. Limeback and A. Simic, Arch. Oral Biol., 1990, 35, 459–468 CrossRef CAS PubMed.
- J. Svensson Bonde and L. Bulow, PLoS One, 2012, 7, e33269 Search PubMed.
- I. M. Porto, H. J. Laure, R. H. Tykot, F. B. de Sousa, J. C. Rosa and R. F. Gerlach, Eur. J. Oral Sci., 2011, 119(1), 83–87 CrossRef PubMed.
- E. Cappellini, L. J. Jensen, D. Szklarczyk, A. Ginolhac, R. A. da Fonseca, T. W. Stafford, S. R. Holen, M. J. Collins, L. Orlando, E. Willerslev, M. T. Gilbert and J. V. Olsen, J. Proteome Res., 2012, 11, 917–926 CrossRef CAS PubMed.
- J. D. San Antonio, M. H. Schweitzer, S. T. Jensen, R. Kalluri, M. Buckley and J. P. Orgel, PLoS One, 2011, 6, e20381 CAS.
- J. M. Asara, M. H. Schweitzer, L. M. Freimark, M. Phillips and L. C. Cantley, Science, 2007, 316, 280–285 CrossRef CAS PubMed.
- M. Buckley, M. Collins, J. Thomas-Oates and J. C. Wilson, Rapid Commun. Mass Spectrom., 2009, 23, 3843–3854 CrossRef CAS PubMed.
- M. Buckley, N. D. Melton and J. Montgomery, Rapid Commun. Mass Spectrom., 2013, 27, 531–538 CrossRef CAS PubMed.
- M. H. Schweitzer, E. R. Schroeter and M. B. Goshe, Anal. Chem., 2014, 86, 6731–6740 CrossRef CAS PubMed.
- I. M. Porto, H. J. Laure, F. B. de Sousa, J. C. Rosa and R. F. Gerlach, J. Archaeol. Sci., 2011, 38, 3596–3604 CrossRef.
- X. Zhang, B. E. Ramirez, X. Liao and T. G. Diekwisch, PLoS One, 2011, 6, e24952 CAS.
- J. D. Bartlett, ISRN Dentistry, 2013, 2013, 684607 CrossRef PubMed.
- T. Nagano, A. Kakegawa, Y. Yamakoshi, S. Tsuchiya, J. C. Hu, K. Gomi, T. Arai, J. D. Bartlett and J. P. Simmer, J. Dent. Res., 2009, 88, 823–828 CrossRef CAS PubMed.
- E. C. Salido, P. H. Yen, K. Koprivnikar, L. C. Yu and L. J. Shapiro, Am. J. Hum. Genet., 1992, 50, 303–316 CAS.
- M. A. Jobling, I. C. Lo, D. J. Turner, G. R. Bowden, A. C. Lee, Y. Xue, D. Carvalho-Silva, M. E. Hurles, S. M. Adams, Y. M. Chang, T. Kraaijenbrink, J. Henke, G. Guanti, B. McKeown, R. A. van Oorschot, R. J. Mitchell, P. de Knijff, C. Tyler-Smith and E. J. Parkin, Hum. Mol. Genet., 2007, 16, 307–316 CrossRef CAS PubMed.
- J. Y. Sire, S. Delgado, D. Fromentin and M. Girondot, Arch. Oral Biol., 2005, 50, 205–212 CrossRef CAS PubMed.
- https://merops.sanger.ac.uk/cgi-bin/pepsum?id=S01.251 .
- G. A. Castiblanco, D. Rutishauser, L. L. Ilag, S. Martignon, J. E. Castellanos and W. Mejia, Eur. J. Oral Sci., 2015, 123, 390–395 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra05120k |
| This journal is © The Royal Society of Chemistry 2016 |