 Open Access Article
Open Access ArticleProduction, surface modification, physicochemical properties, biocompatibility, and bioimaging applications of nanodiamonds
Tirusew Tegafawa,
Shuwen Liua,
Mohammad Yaseen Ahmada,
Abdullah Khamis Ali Al Saidi a,
Dejun Zhaoa,
Ying Liua,
Huan Yuea,
Sung-Wook Nam
a,
Dejun Zhaoa,
Ying Liua,
Huan Yuea,
Sung-Wook Nam b,
Yongmin Chang*b and
Gang Ho Lee
b,
Yongmin Chang*b and
Gang Ho Lee *a
*a
aDepartment of Chemistry, College of Natural Sciences, Kyungpook National University, Taegu 41566, South Korea. E-mail: ghlee@mail.knu.ac.kr; Fax: +82-53-950-6330; Tel: +82-53-950-5340
bDepartment of Molecular Medicine, School of Medicine, Kyungpook National University, Taegu 41944, South Korea. E-mail: ychang@knu.ac.kr; Tel: +82-53-420-5471
First published on 3rd November 2023
Abstract
Nanodiamonds (ND) are chemically inert and stable owing to their sp3 covalent bonding structure, but their surface sp2 graphitic carbons can be easily homogenized with diverse functional groups via oxidation, reduction, hydrogenation, amination, and halogenation. Further surface conjugation of NDs with hydrophilic ligands can boost their colloidal stability and functionality. In addition, NDs are non-toxic as they are made of carbons. They exhibit stable fluorescence without photobleaching. They also possess paramagnetic and ferromagnetic properties, making them suitable for use as a new type of fluorescence imaging (FI) and magnetic resonance imaging (MRI) probe. In this review, we focused on recently developed ND production methods, surface homogenization and functionalization methods, biocompatibilities, and biomedical imaging applications as FI and MRI probes. Finally, we discussed future perspectives.
Introduction
Over the past decades, many studies have demonstrated the potential application of nanoparticles in biomedical imaging and clinical diagnosis.1–6 Before biomedical applications, they need to be refined to obtain good water solubility because most of them are water-insoluble and non-toxicity if they contain toxic elements via surface engineering.7,8 Among nanoparticles, carbon-based nanomaterials such as carbon dots, carbon nanotubes, and nanodiamonds (NDs) have attracted great attention due to their relatively low toxicity as they are made of carbons.9–11 Importantly, their attractive optical and magnetic properties may be useful for fluorescent imaging (FI) and magnetic resonance imaging (MRI) probes.9–11In particular, NDs as a metastable allotrope of carbons are especially attractive owing to their strong sp3 bonding structure, leading to superior chemical stability, inertness, and hardness.12,13 Although their core is chemically stable and inert, their surface is composed of graphitic sp2 carbons which can be easily conjugated with various functional molecules for biomedical applications.14 Several researchers have developed surface homogenization methods such as carboxylation,15 halogenation,16 hydrogenation,17 amination,18 hydroxylation,19 and graphitization.20 These functional groups can be further conjugated with hydrophilic ligands,21 peptides,22 proteins,23 nucleic acids,24 drugs,25 fluorescent dyes,26 cancer targeting ligands,27 dopamine derivatives,28 and additional imaging probes,29 which were conjugated directly or via linkers.30–33 NDs can be introduced with nitrogen vacancy (NV) defects as color centers via irradiation of high-energy particles (p+, He+, or e−) followed by high temperature annealing, making them promising candidates as FI probes.34,35 NDs can also act as MRI probes after conjugation with Gd(III)-complexes or Mn(II)-complexes on ND surfaces, extending their functionality to powerful hybrid MRI and FI probes.36–44 In addition, NDs themselves exhibit paramagnetic and ferromagnetic properties, which will be useful for use as MRI probes.45 Taking all these attractive features together, NDs have emerged as promising FI and MRI probes in single or multi-modal biomedical imaging applications.46
Currently, various production methods of NDs have been developed; these are detonation,47,48 fragmentation of high pressure and high temperature (HPHT) microdiamonds,49,50 laser ablation,51–53 chemical vapor deposition (CVD),54–56 chlorination of carbides,57 ultrasound cavitation,58 ion beam irradiation of graphites,59 and electron irradiation of carbon onions.60 Among them, NDs produced by first two methods are most common and commercially available.61,62 The first four production methods are briefly described in this review. Notably, the particle size and surface properties depend on the production method and thus, applications are somewhat tied to the production method.63
In this review, we concisely provided an insight into the recent advances in ND research such as the production and purification, surface homogenization and functionalization, physicochemical properties, biocompatibilities, and biomedical FI and MRI applications.
Production methods
Detonation
As shown in Fig. 1a,64 NDs are produced through explosion of a mixture of 2,4,6-trinitrotoluene and 1,3,5-trinitroperhydro-1,3,5-triazine in the presence of a combined atmosphere of N2, H2O, and CO2 in a metallic chamber. The explosion generates HPHT conditions (P > 10 GPa; T > 3000 K),64 resulting in ND crystals (called detonation NDs) via condensation of supersaturated carbon vapors.65,66 They consist of a ND core surrounded by a few layers of sp2 carbons. Their particle diameters typically range from 1 to 10 nm,65 with nearly spherical shape. However, they exhibit severe aggregations due to appreciable amount of sp2 surface carbons. They possess low nitrogen impurities which are needed to generate NVs as color centers. The NDs can be purified through oxidation with strong liquid oxidants such as HCl/HNO3, HClO4, H2SO4/HNO3, HF/HCl, K2Cr2O7/H2SO4, and HNO3/H2O2 (refs. 67 and 68) or with air or ozone at elevated temperatures (400–430 °C).69–72 These oxidations introduce oxygen-containing groups on ND surfaces such as hydroxyl, carbonyl, and carboxyl groups. A high energy balling73 or a salt-assisted ultrasonic de-aggregation74–76 can be used to separate aggregated NDs into individual NDs; the latter method is preferred because the former method leaves broken ball materials after milling. In addition, annealing at 500 °C followed by centrifugation of NDs in aqueous colloidal solution can be used to obtain de-aggregated NDs by removing surface graphite carbons on ND surfaces;77 even high-purity NDs of ∼1 nm can be obtained by controlling annealing temperature, whereas the former two methods allow only de-aggregations of NDs.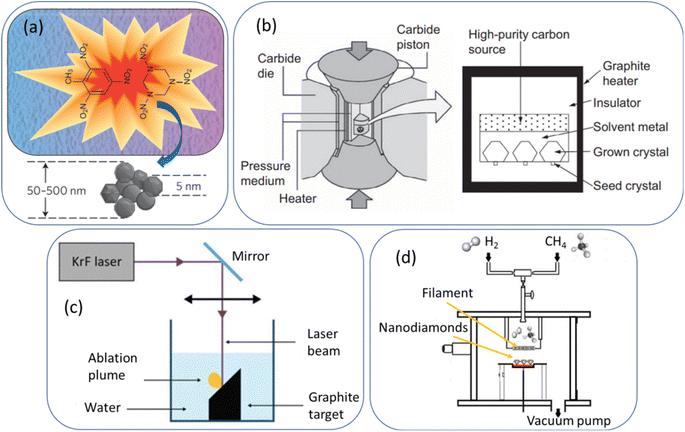 | ||
| Fig. 1 (a) Detonation production of nanodiamonds,64 (b) HPHT production of microdiamonds,78 (c) laser ablation production of nanodiamonds,53 and (d) CVD production of nanodiamonds.55 Reproduced with permissions from ref. 64 Copyright 2012 Springer-Nature, ref. 78 Copyright 2016 Elsevier, and ref. 53 Copyright 2018 Springer. | ||
Fragmentation of HPHT microdiamonds
As shown in Fig. 1b,78 graphites or nanotubes or fullerenes as carbon source in a reactor chamber are brought to a high pressure (5–6 GPa) and high temperature (1300–1700 °C).78 Iron and other metals such as cobalt and nickel as catalysts and small diamonds as crystallization seeds are provided. Under HPHT conditions achieved with a heater and carbide piston, carbon atoms are deposited on the diamond seed surfaces due to the temperature difference (20–50 °C) between the carbon source (high temperature side) and seed diamonds (lower temperature side) and exhibit transition from sp2 to sp3, making diamond seeds grow into bulk diamonds.78 The obtained diamonds are fragmented into NDs with size of 2–50 nm via bead-assisted sonic disintegration or mechanical ball milling (the final fragmented diamonds are simply called HPHT NDs). Finally, impurities such as graphites and metals are eliminated via purification process.79 Compared with detonation NDs, HPHT NDs have less amount of surface sp2 carbons, leading to less aggregation, non-spherical shape with sharp edges, and higher nitrogen impurities.Laser ablation
As shown in Fig. 1c,53 a pulsed laser beam is focused onto graphite target immersed in water.51–53,80,81 The laser pulse leads to the vaporization of carbon atoms to the surrounding liquid in the form of an ablation plume. Carbon atoms trapped inside the bubble under HPHT conditions (P = 1–10 GPa; T = 5000–6000 K)80 are subject to the formation of NDs with particle diameters of 3 to 15 nm51 or ∼100 nm,81 depending on laser power conditions. This method is more environmentally green and less hazardous compared with detonation and HPHT methods. In addition, high purity NDs with minimal metallic impurities can be obtained.CVD
As shown in Fig. 1d,55 a ND thin film can be obtained from the decomposition of methane in an excess of hydrogen gas in a plasma reactor.56,82 Jin-Woo et al. reported NDs of 4–6 nm using this method.82 Depending on the relative concentration of the gas mixture (CH4/H2), microdiamonds (>100 nm), NDs (10–100 nm), and ultrasmall (<10 nm) NDs in a film can be produced.56 For the synthesis mechanism of CVD NDs, a carbon-containing precursor (∼1% CH4 in H2) is introduced into a reaction chamber and then, plasma (a mixture of ˙H and ˙CHx radicals and electrons) is produced through microwave discharge or hot filament discharge.56,83 The released carbon atoms from the precursors form diamond nuclei in gas phase or on a substrate, serving as templates for further diamond growth. In addition, NDs can grow on prepared diamond seeds on a substrate.83 The diamonds grow slowly through the addition of carbon atoms onto the growing diamond lattice, ultimately yielding NDs.Comparison between different production methods
For detonation NDs, Osswald et al. found that the as-prepared (i.e., pristine) detonation NDs (d ∼5 nm) contained 77% sp2 carbons using soft X-ray absorption near-edge structure (XANES) spectroscopy, but after air oxidation, the amount of sp2 carbons was reduced to 5%.71 Stenclova et al. estimated 45% sp2 carbons for as-prepared detonation NDs (d = ∼5 nm) using high-resolution XPS.84 Stehlik et al. prepared sub-10 nm HPHT NDs and estimated 52% sp2 carbons for as-prepared HPHT NDs using XPS, but sp2 carbons decreased to 9% after air oxidation at 450 °C for 1–2 h.77 For CVD NDs, Kozakov et al. estimated 32–33% sp2 carbons using XPS.85 For laser ablation NDs, Xiao et al. produced NDs (3.1–3.6 nm) via laser ablation of coals and estimated 12% sp2 carbons using XPS.86Detonation NDs, HPHT NDs, and laser ablation NDs are produced under HPHT conditions and commercially available because they can be produced on a large scale at a reasonable cost. However, CVD NDs which are produced through nucleation reactions on a substrate, are not commercially available because the CVD synthesis takes a long time and thus, production cost is high.
Surface homogenization and hydrophilization
Surface homogenization
ND surfaces contain sp2 carbons and a mixture of hydroxyl, carbonyl, and carboxyl groups after production and purification (Fig. 2).87 Therefore, ND surfaces should be uniformly terminated with one type of functional groups to facilitate further conjugation with other ligands.31,88,89 As shown in Fig. 2, the surface homogenization includes carboxylation, hydroxylation, halogenation, amination, hydrogenation, and graphitization.87 Alternatively, silica can be coated on ND surfaces because the silica shell contains rich hydroxyl groups.90,91 Namely, core@shell structure of NDs such as ND@silica in aqueous media may be also used for surface homogenization of NDs with numerous –OH groups on silica surfaces.90,91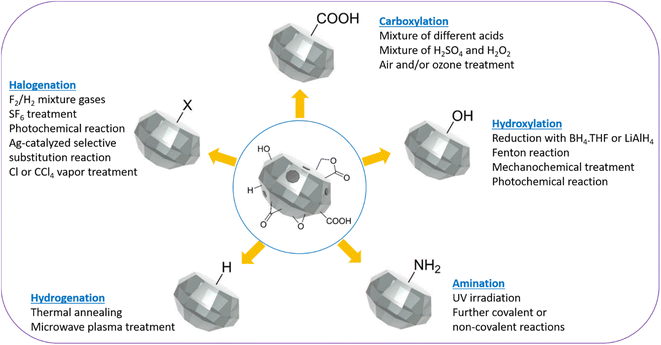 | ||
| Fig. 2 Schematic representation of homogenization of ND surfaces via carboxylation, hydroxylation, hydrogenation, amination, and halogenation.87 | ||
Surface hydrophilization
Surface homogenized NDs are not colloidally stable in aqueous phase and thus, should be further grafted with hydrophilic polymers such as polyethylene glycol (PEG),92,93 PEG derivatives,94,95 polyglycerol,39,96–98 polyethylenimine (PEI),99 and polydopamine.100 After this, hydrophilic polymer-grafted NDs can be further conjugated with diagnostic and therapeutic molecules for desired biomedical applications. ND–COOH can be conjugated with –NH2 groups of hydrophilic polymers via amide bond using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)/N-hydroxysuccinimide (NHS) coupling reaction.101–103 Or ND–COOH can be activated with thionyl chloride (SOCl2) to form acyl chloride, i.e., ND–COCl, which can directly react with amines of hydrophilic polymers to form amide bond.104–107 ND–NH2 can be conjugated with –COOH groups of hydrophilic polymers via amide bond using EDC/NHS coupling reaction.108–110 ND–OHs can be conjugated with acid chloride (ClO−) groups of hydrophilic polymers to form ester bond, i.e., ND–OOC–polymer111 or with silanes to form ND–O–Si–polymer.31 Silica coating can be considered as another method of surface hydrophilization of NDs.90 This is because silica and terminal –OH groups are hydrophilic. Bumb et al. used tetraethyl orthosilicate as silica precursor to obtain ND@silica in aqueous media,90 which was stable whereas uncoated NDs precipitated in aqueous media.Physicochemical properties
NDs can be characterized with various techniques.112–117 As shown in low- and high (inset)-resolution transmission electron microscope (TEM) images in Fig. 3a–d and Table 1, the diameters of NDs largely depend on production method. The detonation NDs contain a nearly spherical shape in severely aggregated forms with thick graphitic shell.64,65,116 NDs obtained via fragmentation of HPHT microdiamonds (simply, HPHT NDs) have irregular shapes with sharp edges, but are well dispersed.78,112 CVD NDs56,57,82 and laser ablation NDs51,52,81,117 are nearly spherical and well dispersed. The particle diameters of detonation NDs are very small (davg = 5 nm) and narrow (2–9 nm)65 (polydispersity index, PDI = 0.11–0.13).3 The particle diameters of HPHT NDs range from 2 to 50 nm, showing a broad size distribution because they are produced from fragmentation of HPHT microdiamonds.112 Laser ablation NDs exhibit the particle diameters of 3 to 15 nm51,52 or ∼100 nm,81 depending on laser power conditions. CVD diamonds exhibit microdiamonds (>100 nm), NDs (10–100 nm), and ultrasmall (<10 nm) NDs, depending on the relative concentration (CH4/H2).91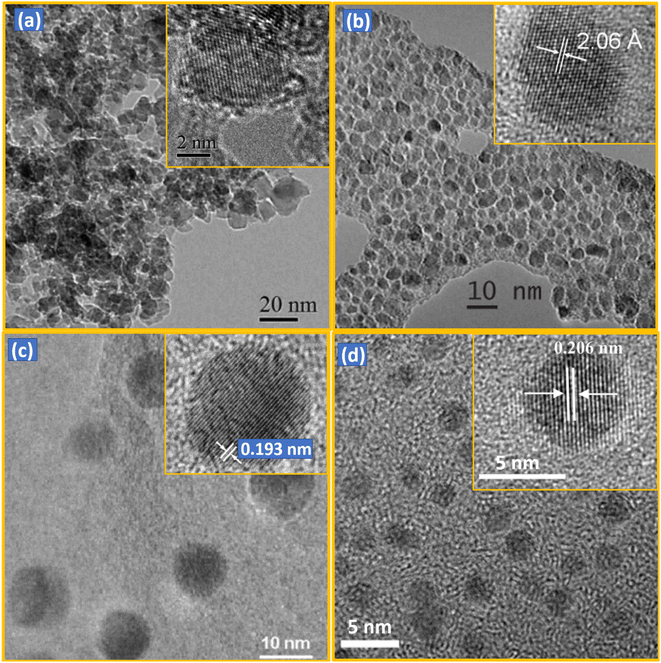 | ||
| Fig. 3 TEM images of NDs synthesized by different methods: (a) detonation (d = 4–5 nm),116 (b) HPHT after fragmentation (d = 2–7 nm; davg = 3.5 nm),112 (c) CVD (d = 1–10 nm),57 and (d) laser ablation (davg = 3.5 nm).117 Low- and high (inset)-resolution images are provided. Reproduced with permissions from ref. 116 Copyright 2012 Royal Society of Chemistry, ref. 112 Copyright 2009 IOP Science, ref. 57 Copyright 2003 American Institute of Physics, and ref. 117 Copyright 2015 American Chemical Society. | ||
| Production method | Particle diametera | Morphology | Application | Refs |
|---|---|---|---|---|
| a Particle diameter depends production method and conditions. | ||||
| Detonation | 1–10 nm | Nearly spherical, aggregated | Lubrication, composite filler | 64, 65, and 116 |
| Fragmentation of HPHT microdiamond | 2–50 nm | Irregular shape with sharp edges | Optical probe | 78 and 112 |
| CVD | 4–6 nm, 10–100 nm, >100 nm | Nearly spherical | Coating, abrasive | 56, 57, and 82 |
| Laser ablation | 3–15 nm, ∼100 nm | Nearly spherical | Magnetic probe | 51, 52, 81, and 117 |
NDs have a sp3 bonding structure. As observed in the X-ray diffraction (XRD) patterns (Fig. 4),114 NDs exhibit a face-centered cubic structure with the major peak at 2θ of 44° from the (111) diffraction plane along with two other peaks at 2θ of 75° and 91° from the (220) and (311) diffraction planes, respectively. The broad peaks are owing to small particle sizes of NDs.113,114
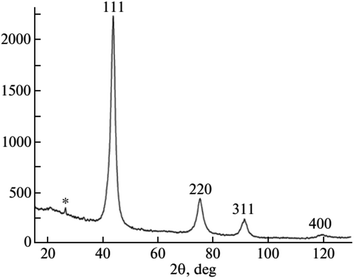 | ||
| Fig. 4 XRD pattern of NDs produced by laser ablation (d = 3–4 nm).114 Reproduced with permission from ref. 114 Copyright 2013 Springer. | ||
The Raman spectra of NDs produced by different methods exhibit a diamond peak from the C–C sp3 chemical bond at 1332.9 cm−1 (Fig. 5).115 The broad peak at ∼1580 cm−1 is due to graphite carbons on ND surfaces, which becomes stronger for smaller NDs owing to their higher surface to volume ratios.
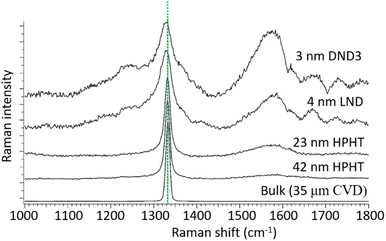 | ||
| Fig. 5 Raman spectra of NDs of different sizes under 355 nm excitation. Size in this figure is estimated by dynamic light scattering (DLS) for HPHT and DND3 NDs and by XRD for the LND NDs. The peak at 1332.9 cm−1 is due to the C–C sp3 chemical bond as indicated by the dotted line. The broad peak at ∼1580 cm−1 is due to graphite carbons on ND surfaces, which becomes stronger for smaller NDs owing to their higher surface to volume ratios.115 Reproduced with permission from ref. 115 Copyright 2017 Elsevier. | ||
The polydispersity index (PDI) of NDs defined as (σ/d)2 (σ = standard deviation of particle diameter distribution and d = average particle diameter) can be measured using a TEM or a dynamic light scattering (DLS) instrument.118 The DLS-PDI is more commonly used than TEM-PDI because DLS-PDI can be obtained from DLS. The DLS-PDI values of NDs synthesized by different methods are as follows. The detonation NDs exhibited PDI values ranging from 0.1 to 0.6,33,119 HPHT NDs32 and CVD NDs120 exhibited PDI values ranging from 0.1 to 0.3, and laser ablation NDs provided PDI values of 0.5 after glycidol surface coating.121 It should be noted that the PDI values are subject to variation based on experimental conditions and post-synthesis treatments.
Wilson et al. investigated stability of HPHT NDs in water and biological media such as 10% fetal bovine serum (FBS) in water, Dulbecco's modified eagle medium (DMEM; common cell culture medium), and DMEM with the addition of 10% v/v FBS (simply denoted as CM).122 1% antibiotic penicillin–streptomycin was added to both DMEM and CM media. DLS was used to investigate stability of NDs in water and biological media as a function of time. Hydrodynamic diameter did not change in water with time, only slightly increased in FBS and CM media with time, but severely increased in DMEM with time, resulting in precipitation of NDs owing to ND aggregation; therefore, stability of NDs in water, FBS, and CM was good, but poor in DMEM.
The surface zeta potential of NDs is important for colloidal stability in solution. The surface zeta potential of as-synthesized detonation NDs was slightly positive (∼1.5 mV) owing to –OH groups on the ND surfaces.123 However, surface zeta potential of detonation NDs became highly positive (>20 mV) or highly negative (<−20 mV) after surface modifications with different type of acids, respectively.124 On the other hand, the surface zeta potential of as-prepared HPHT NDs was highly negative (<−40 mV) owing to –COOH groups on the ND surfaces,125,126 but became positive after successive heat treatments in air (480 °C, 5 h), vacuum (0.001 mbar, 1000 °C, 2 h), and H2 (500 °C, 10 mbar, 5 h).126
Fluorescent properties
Color centers are crystal defects that absorb and emit photons while the crystal itself has no absorption; i.e., ND itself is colorless and transparent. The fluorescence of NDs is due to impurity atoms such as N and Si (mostly N) bound to structural vacancies. The most common color centers are the NVs which are created by irradiating NDs with high-energy particles (p+, He+, or e−), followed by vacuum annealing of NDs at high temperatures (600–800 °C);127–129 vacancies are first produced by irradiation and impurity atom vacancy centers are produced from migration of impurity atoms to the vacancies during heat treatment. Two types of NV centers are produced; neutral (NV0) emitting visible photons with slightly shorter wavelengths at 576 nm compared with those of negatively charged (NV−) emitting red photons;130–132 NV− centers are dominant at 638 nm and their quantum yield is ∼1.0. In addition, conjugation of octadecylamine to NDs allowed blue photon emissions.104Fig. 6a shows PL spectra of 100 nm HPHT NDs, showing two emissions, i.e., zero-phonon lines (ZPL) of NV0 and NV− centers at 576 and 638 nm, respectively; they were obtained with excitation wavelengths of 170 nm synchrotron far-UV source and 532 nm laser light, respectively.133 In addition, the positions of NV0 and NV− centers do not change with particle diameters whereas broad emission from graphitic carbons on ND surfaces are red-shifted with increasing ND size, as shown in Fig. 6b.122
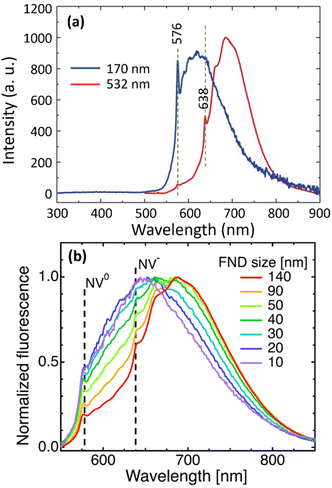 | ||
| Fig. 6 (a) Photoluminescence (PL) spectra of 100 nm HPHT NDs at 300 K with excitation wavelengths of 170 nm synchrotron source and 532 nm laser light, showing zero-phonon lines (ZPL) at 576 and 638 nm for NV0 and NV− centers, respectively.133 Vertical dotted lines indicate ZPL positions. (b) PL spectra as a function of HPHT ND diameters, showing no dependence of NV0 and NV− center positions on particle diameter (510 nm laser excitation).122 Reproduced with permissions from ref. 122 Copyright 2019 IOP Science and ref. 133 Copyright 2017 Wiley. | ||
Moreover, since the NV centers are incorporated deeply inside the ND matrix, their optical properties are not altered by surface coating.90,134,135 The exceptional photostability with no photobleaching of NV centers in NDs provides a greater advantage to emerge as an ideal candidate for long term imaging.131 Most brightly fluorescent NDs have been made from HPHT NDs because the HPHT NDs contain the highest amount of nitrogen impurities with a typical concentration of 100–200 ppm among NDs produced with different methods.112,127 A recent study showed that NV centers in detonation NDs can be enhanced up to 104 ppm through a HPHT sintering.136
NDs can be engineered to emit near-infrared fluorescence (NIRF). This property makes them useful in bio-imaging applications because NIR light can penetrate deeply into biological tissues. One of the commonly employed methods to induce NIRF in NDs is the introduction of silicon vacancy (SiV) centers (zero phonon line center = 738 nm) into NDs.137,138 The SiV centers are created by introducing silicon precursors as impurities into the diamond growth region for HPHT diamonds137 or detonation region for detonation NDs.138
Paramagnetic and ferromagnetic properties
Bulk diamonds are diamagnetic owing to sp3 bonding structure. However, NDs can take paramagnetic or ferromagnetic properties.139 Paramagnetism comes from graphitic surface layers in NDs.140 Shames et al. found a high concentration of radical like-paramagnetic surface spins (1020 spins per g).141,142 Fig. 7 shows electron paramagnetic resonance (EPR) spectra of 18 nm HPHT NDs showing that air oxidation of NDs reduces the amplitude of the broad component in the spectra owing to the removal of paramagnetic surface spins at the ND surfaces.45 A similar thing was observed in 50 nm NDs; i.e., the paramagnetic component from nitrogen impurity centers in ND crystalline core did not change during annealing while that from surface spins were significantly removed (Fig. 8).143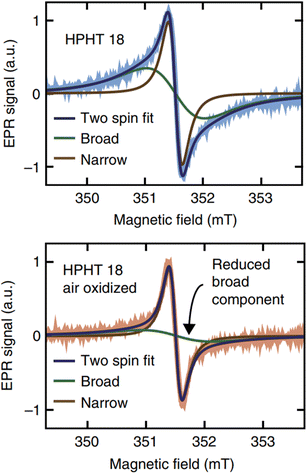 | ||
| Fig. 7 The top shows EPR spectra of HPHT 18 nm NDs in deionized water and the bottom shows the EPR spectra of the same NDs after air oxidation. Air oxidation removed the broad component arising from disordered dangling bonds at the ND surfaces while leaving the narrow component arising from lattice defects in the crystalline core unaffected.45 Reproduced with permission from ref. 45 Copyright 2017 Springer-Nature. | ||
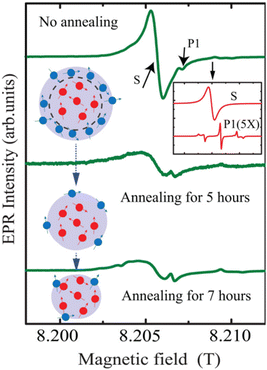 | ||
| Fig. 8 EPR analysis of 50 nm NDs before and after air annealing for 5 and 7 h. The inset on the top right shows contributions of nitrogen impurity centers (labeled as P1) and surface spins (labeled as S) to the EPR spectra. The EPR spectra under the annealing process are shown below. The red dots in the drawing represent the P1 centers and the blue dots represent surface spins.143 Reproduced with permission from ref. 143 Copyright 2020 American Institute of Physics. | ||
The ferromagnetism in NDs is not clearly understood up to now. Talapatra et al. observed ferromagnetism after they irradiated NDs with 15N and 12C ions; the results for 15N ion irradiation are provided in Fig. 9a and b.144 At low doses, the magnetization was independent of the irradiating species indicating that it mainly arises from structural deformation (i.e., generating sp2/sp3 defect structures) of the carbon bonds in NDs. However, 15N implants exhibited higher saturation magnetizations than 12C implants at higher doses (i.e., 5 × 1016 cm−2 in Fig. 9b whereas the magnetization was independent of the irradiating species up to 1016 cm−2 doses), which could be due to extensive defect generation or graphitization and, to some extent, incorporation of nitrogen in the graphitic network and formation of C–N bonds. Perevedentseva et al. prepared ferromagnetic NDs via thermal treatment of NDs produced by laser ablation in liquid (called RayND-M).139 They demonstrated that the surface structural modification could be the source of ferromagnetism in RayND-M (Fig. 10a) rather than elemental contents (Fe, Co, and Ni) as impurities because no such metals were detected from elemental analysis using energy dispersive X-ray spectroscopy (EDS) (Fig. 10b). For comparison, detonation NDs did not exhibit ferromagnetism (Fig. 10a).
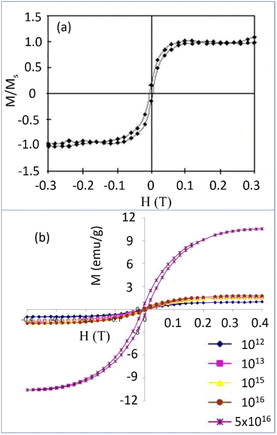 | ||
| Fig. 9 (a) Magnetization (M) versus applied field (H) curves for the detonation NDs (d = 4–5 nm) (Ms = saturation magnetization) with 15N dose at (a) 5 × 1014 cm−2 and (b) various doses: enhanced magnetizations were observed with doses above 5 × 1016 cm−2.144 Reproduced with permission from ref. 144 Copyright 2005 American Physical Society. | ||
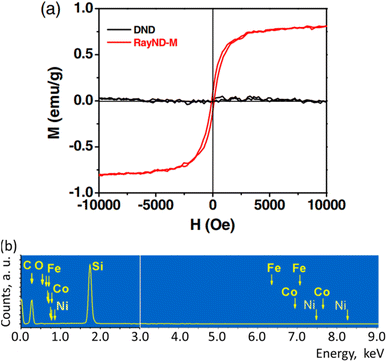 | ||
| Fig. 10 Magnetic properties of RayND-M (d = 4.3–4.5 nm). (a) Magnetization curve of RayND-M [in comparison with ordinary detonation ND (DND), d = 3–10 nm]. (b) EDS spectrum of RayND-M; Fe, Co, and Ni atoms as impurities were not detected.139 Reproduced with permission from ref. 139 Copyright 2018 SPIE. | ||
Setzer et al. investigated magnetic properties of natural and CVD diamonds after laser-cut.145 All of them exhibited ferromagnetism after laser-cut owing to surface graphite formation on diamond surfaces. The surface graphites formed on CVD diamond surfaces were <20 nm thick after laser-cut and reduced by 10% after laser polishing of the laser-cut surface. The zero-field-cooled (ZFC) and field-cooled (FC) magnetization curves of the laser-cut CVD diamonds before (sample #1a) and after laser polishing of the laser-cut surface (sample #1b) were measured at 0.01, 0.05, and 1 T applied fields between 2 and 380 K as shown in Fig. 11. The observed saturation magnetization values were 10 to 20 emu g−1 at 300 K, similar to those obtained with pure graphites, supporting that the ferromagnetism of NDs resulted from surface graphites formed on ND surfaces by laser-cut.
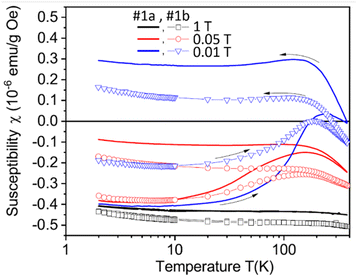 | ||
| Fig. 11 Zero-field-cooled and field-cooled curves of the CVD samples (#1a: samples after the laser-cut and #1b: after laser-polishing of the laser-cut surface) as a function of temperature between 2 and 380 K at applied fields of 0.01, 0.05, and 1 T.145 | ||
In vitro and in vivo biocompatibility
NDs are considered to be the most biocompatible form among carbons (fullerene, graphene, carbon nanotubes, and so on).146–151 The sp3 carbon core of NDs is chemically inert and non-toxic. Therefore, toxicity of the NDs mostly comes from surface materials including sp2 graphite carbons, functional groups, and metal impurities.152–156 NDs did not show any noticeable toxicity on human embryonic kidney tumor (HEK293T) cells up to concentration of 400 μg mL−1 [concentration (μg mL−1) = mass of NDs (μg)/volume of solution (mL)], confirming their non-toxicity.131 The viability of neuroblastoma cells after incubation with various concentrations of various types of NDs, carbon black (CB) nanoparticles, and cadmium oxide (CdO) submicroparticles were assessed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay (Fig. 12).150 The results exhibit that all NDs are non-toxic and less toxic than CB whereas CdO submicroparticles are highly toxic. The cellular toxicity of NDs on human embryonic kidney 293 (HEK293) cells was ranked in the order of ND-NH2 > ND-OH > ND-CO2H at a concentration of 200 μg mL−1 (Fig. 13),153 indicating that higher negative ND surface charges induce less cytotoxicity owing to less interaction between cells and NDs. Generally, large NDs (more than 100 nm) are more biocompatible than smaller NDs (5 nm).157 This is due to more active surface materials of smaller NDs compared with those of large NDs.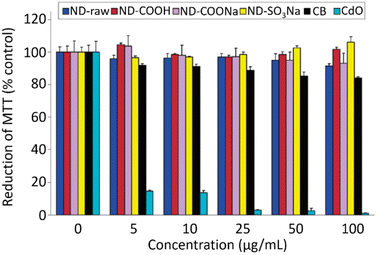 | ||
| Fig. 12 Cytotoxicity of neuroblastoma cells incubated with various types of NDs (d = 2–10 nm), carbon black (CB) nanoparticles (d = 20–30 nm), and CdO nanoparticles (submicrometer size) via MTT assay.150 Reproduced with permission from ref. 150 Copyright 2007 American Chemical Society. | ||
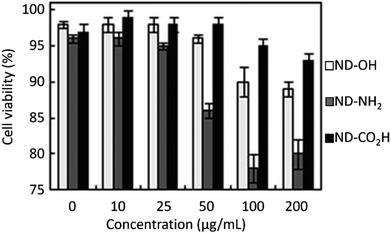 | ||
| Fig. 13 Cell cytotoxicity of NDs (davg = 4 nm) on HEK293 cells after normalization with respect to control cells that were not incubated with NDs. The control cells exhibited a cell viability of 97 ± 1% 24 h after incubation.153 Reproduced with permission from ref. 153 Copyright 2010 Royal Society of Chemistry. | ||
The clearance of NDs from the body depends on their particle diameter and surface properties. Small NDs (d < 3 nm) grafted with zwitterionic molecules can be excreted from the body through the renal system,158–160 whereas large NDs (d > 3 nm) grafted with polymers mostly accumulate in the body. NDs (d < 3 nm) may be excreted through the renal system within 1 h because inulin (3 nm) achieved 100% renal excretion with a blood half-life of only 9 min.158 However, large NDs (d > 3 nm) are mostly taken up by the reticuloendothelial system (RES), particularly in the liver and spleen, followed by the excretion through the hepatobiliary system.158 The excretion through the hepatobiliary system takes a long time. Yuan et al. investigated the biodistribution and fate of very large NDs (d = 50 nm) in vivo.161 They found that approximately 60% of injected NDs were entrapped in the liver and 8% in the lung at 30 min after intravenous injection in mouse. These accumulation values remained for 28 days after injection, implying that large NDs are not or negligibly excreted from the body.
Barone et al. conducted an in vivo histological evaluation of various rat organs including the liver, spleen, kidney, lung, and heart 12 weeks after intravenous injection of large NDs (hydrodynamic diameter = 800 nm) (Fig. 14).162 The analysis showed few changes in liver, spleen, kidney, lung, and heart after injection (60 mg of NDs per kg rat), showing no-toxicity of the NDs. All these in vitro and in vivo studies confirm that NDs are non-toxic.
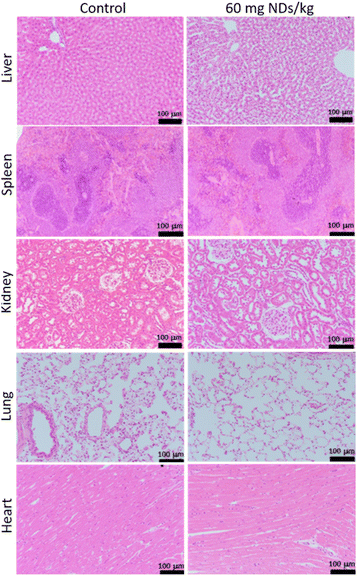 | ||
| Fig. 14 Histopathology of rats showing no toxicity of NDs 12 weeks after intravenous injection of NDs (hydrodynamic diameter = 800 nm) and 8 weeks after intravenous injection of PBS solution (control). Images of hematoxylin- and eosin-stained liver, spleen, kidney, lung, and heart. N = 3 for all groups. Injection dose (2 mL of PBS for control rats; 60 mg NDs per kg rat).162 | ||
Moore et al. conducted a comprehensive study on the biocompatibility of detonation NDs (hydrodynamic diameter = 52.7–59 nm) in non-human primates (i.e., monkeys) for 6 months and rats for 2 weeks by analyzing urine, body weight, histology, and blood.163 They did not find any adverse effects on various organs such as the liver, kidneys, and lung. This result supports the outstanding biocompatibility of NDs.
However, long-term toxicity (>1 year) of NDs should be carefully studied for clinical applications. For example, blood samples can be drawn and analyzed to detect any biomarkers that are indicative of potential inflammatory responses. In addition, routine welfare checks should be performed to monitor changes to the overall physical and mental health such as body weight, behavior, and signs of weakness of the animals. Furthermore, major organs should be collected and subjected to extensive histological analyses to detect any phenotypic changes.164 These long-term toxicity data are essential for U.S. Food and Drug Administration (FDA) approval for clinical applications.165
Specific functionalizations
There are many studies on specific functionalizations of NDs by conjugating specific ligands to NDs. Lake et al. synthesized Nup98 antibody-conjugated NDs to target Nup98, a component of the nucleoporin ring structures of the nuclear pore complex (NPC).166 The successful targeting of the Nup98 antibody-conjugated NDs onto the Nup98 of NPC was confirmed from TEM images and fluorescence imaging of NV centers of NDs. Leung et al. conjugated NDs with three ligands, i.e., an antisense oligonucleotide ANA4625, a human immounodeficiency virus TAT protein, and a nuclear localization signal (NLS) peptide in series in aqueous media.167 The ANA4625-NLS-TAT-NDs targeted the NPC and penetrated into cancer cells. They confirmed nuclear targeting through fluorescence imaging of NV centers of NDs. Nie et al. conjugated NDs with pH sensitive dextran to facilitate endocytosis of NDs inside HeLa cancer cells. They demonstrated cell internalization of NDs through fluorescence imaging of NV centers of NDs.168Biomedical imaging applications
FI
NDs are emerging as a new and promising imaging agent for long term in vitro and in vivo studies.34 NDs have defects as color centers that can emit fluorescence. ND fluorescence from NV centers is highly photostable with no photobleaching and quenching owing to protection by the robust diamond lattice.131,169 Chao et al. successfully observed fluorescence emission from NDs treated into human lung epithelial (A549) cells (Fig. 15), confirming potential of NDs as imaging agents.170 Igarashi et al. developed a background-free selective imaging protocol (SIP) to observe fluorescent NDs with nitrogen vacancy centers.171 To this end, they irradiated microwave (2.87 GHz) to remove autofluorescence intensity from biological molecules. Using this imaging technique, they observed background-free fluorescence from NDs fed into Caenorhabditis (C.) elegans (Fig. 16).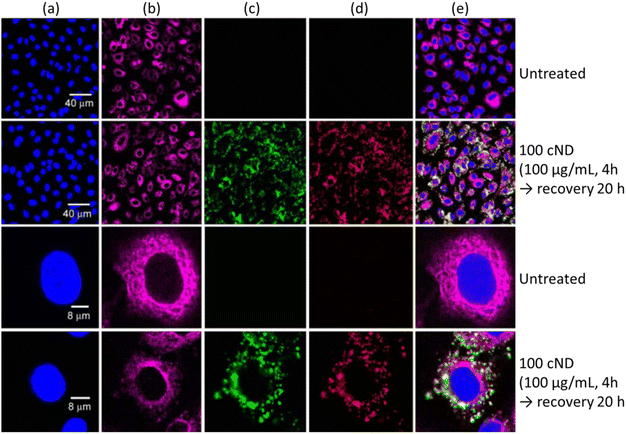 | ||
Fig. 15 Laser scanning confocal fluorescence images of A549 cells. (a) The cell nuclei were dyed with Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 to show the nucleus position (Ex: 351 nm, Em: 350–460 nm). (b) The cell tissue was dyed with anti-β-tubulin (Cy3) to show the cell cytoskeleton (Ex: 543 nm, Em: 550–615 nm). The cells treated with 100 nm NDs with excitation with (c) 488 nm (Em: 500–530 nm) and (d) 633 nm (Em: 640–720 nm). (e) Overlaid images of (a)–(d).170 Reproduced with permission from ref. 170 Copyright 2007 CellPress. 258 to show the nucleus position (Ex: 351 nm, Em: 350–460 nm). (b) The cell tissue was dyed with anti-β-tubulin (Cy3) to show the cell cytoskeleton (Ex: 543 nm, Em: 550–615 nm). The cells treated with 100 nm NDs with excitation with (c) 488 nm (Em: 500–530 nm) and (d) 633 nm (Em: 640–720 nm). (e) Overlaid images of (a)–(d).170 Reproduced with permission from ref. 170 Copyright 2007 CellPress. | ||
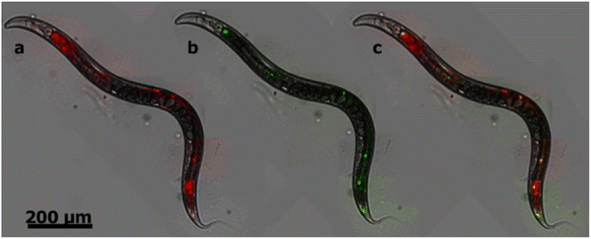 | ||
| Fig. 16 Bright-field fluorescence merged images of C. elegans fed with NDs (d = 50 nm) with nitrogen vacancy centers. (a) A conventional fluorescence image obtained without SIP as shown in red, resulting from background and NDs. (b) An image obtained by SIP as shown in green, resulting from only NDs. (c) The merged image of a and b; fluorescence from NDs is given in yellow whereas that from other sources remains red.171 Reproduced with permission from ref. 171 Copyright 2012 American Chemical Society. | ||
NDs can be used as in vivo tracking nanoprobes utilizing their fluorescence properties of NV centers. Mohan et al. found that the NDs (hydrodynamic diameter = ∼120 nm) enabled continuous imaging of the whole digestive system of the C. elegans via fluorescence imaging for several days after feeding them with NDs.172
NDs can be used as specific imaging agents after conjugation of specific targeting ligands with NDs. Han et al. synthesized hyaluronate (HA)-conjugated NDs (d = ∼100 nm) to use them for in vitro human liver cancer cell (HepG2) imaging and in vivo liver imaging utilizing the fact that the liver cells possess abundant HA receptors.173 Pham et al. synthesized rA27(aa 21–84)-conjugated NDs (d = ∼100 nm) to use them for human HeLa cancer cell imaging.174 As shown in Fig. 17a, rA27-conjugated NDs exhibited red fluorescence owing to their targeting to the glycosaminoglycans (GAGs) of HeLa cells, whereas rDA27-conjugated NDs as control showed negligible red fluorescence as shown in Fig. 17b because rDA27 is not specific to GAGs.
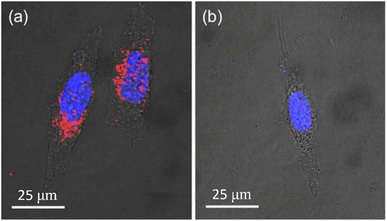 | ||
Fig. 17 Confocal microscopy fluorescence images of HeLa cells 2 h after incubation (d = ∼100 nm; 25 μg mL−1; 488 nm excitation) with (a) rA27-NDs and (b) rDA27-NDs as control. The red fluorescence in (a) is from targeted NDs, whereas negligible red fluorescence in (b) is owing to negligible targeting of NDs. The blue emission is due to fluorescence from Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342-stained cell nuclei.174 Reproduced with permission from ref. 174 Copyright 2017 American Chemical Society. 342-stained cell nuclei.174 Reproduced with permission from ref. 174 Copyright 2017 American Chemical Society. | ||
MRI
The paramagnetic and ferromagnetic properties of NDs are the potential sources for MRI. So far only the paramagnetic properties of NDs had been applied to MRI. Waddington et al. explored the Overhauser effect based on a proton–electron polarization transfer technique for MRI (called Overhauser-enhanced MRI: OMRI).45 This effect transfers spin polarization from paramagnetic impurities (i.e., NVs and unpaired electrons in sp2 graphite shell) in NDs to 1H proton spins in the surrounding water solution, creating MRI contrasts.45 As shown in phantom images (Fig. 18a–d), higher contrasts were observed with increasing ND concentration in solution at 6.5 mT MR field, demonstrating the capability of NDs as imaging probes in MRI.45 The same group applied the hyperpolarized 13C (1.1% natural abundance carbon content in ND) to MRI in vivo at 7 T MR field.175 They prepared hyperpolarized 13C NDs using a 2.88 T superconducting nuclear magnetic resonance (NMR) magnet. An MR contrast image was clearly observed in mouse after intrathoracic injection of hyperpolarized 2 μm-diamonds dispersed in water, as shown in Fig. 18e.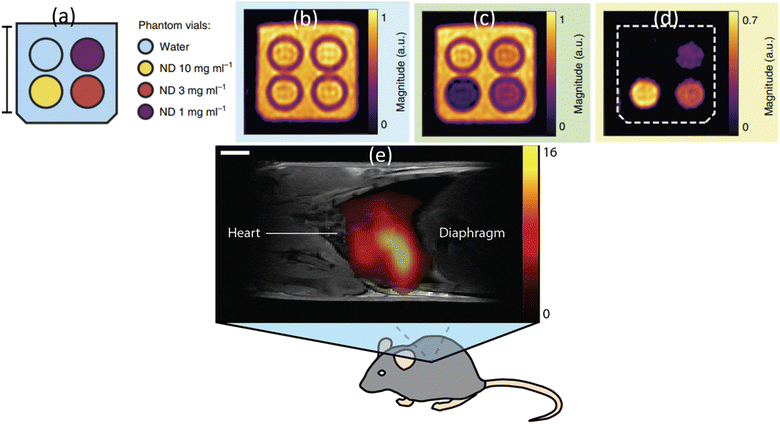 | ||
| Fig. 18 OMRI at 6.5 mT MR field. (a) A vial of deionized (DI) water (blue) and vials containing 18 nm HPHT NDs at concentrations of 10 mg mL−1 (yellow), 3 mg mL−1 (red) and 1 mg mL−1 (purple). The surrounding volume was filled with DI water (blue). Scale bar is 20 mm. (b) Phantom image obtained via standard balanced steady-state free precession (bSSFP) MRI. (c) OMRI bSSFP phantom image. (d) The difference between b and c.45 (e) A 1H:13C MR image at 7 T MR field for a mouse thorax after intrathoracic injection of hyperpolarized 2 μm diamonds. 1H MRI was used for mouse imaging and 13C MRI was used for hyperpolarized ND imaging: the scale bar is 3 mm.175 | ||
Recently, Lv et al. reported that microdiamonds containing NVs can be used as dual-modal optical and 13C MR imaging.176 They used 40 mg of 200 μm diamonds for phantom imaging and successfully performed dual optical and MR imaging, as shown in Fig. 19.
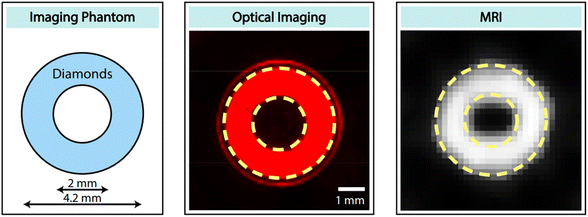 | ||
| Fig. 19 Dual-modal optical and 13C MR imaging. (left) Ring-shaped phantom filled with 40 mg of 200 μm diamonds. (middle) Fluorescence image with a 630 nm filter. (right) 13C MR image.176 Reproduced with permission from ref. 176 Copyright 2021 National Academy of Sciences USA. | ||
Conclusions and outlook
NDs have received a considerable interest owing to their non-toxicity, chemical inertness and hardness due to sp3 bonding structure, easy surface functionalization due to sp2 graphitic shell, optical properties suitable for use as FI probes, and paramagnetic and ferromagnetic properties suitable for use as MRI probes. NDs can be produced using various methods as reviewed here. In addition, they are commercially available at reasonable prices.Although pure NDs are colorless and optically transparent, NDs with NVs as color centers are fluorescent in the red region, which are extremely useful as FI probes. In addition, conjugation of octadecylamine to NDs allowed blue photon emissions. Their potential as FI probes has been demonstrated via in vitro and in vivo experiments. Compared with organic dyes and quantum dots (QDs), color centers of NDs are extremely photostable with no photobleaching and quenching owing to their location at the ND center and protection by strong sp3 structure. Very high quantum yields close to one supports the superiority of NDs as FI probes.
Pure NDs are diamagnetic owing to a closed-shell sp3 electronic structure. However, NDs are paramagnetic owing to defect structures such as NVs and sp2 surface graphitic carbons. NDs can be even ferromagnetic after thermal treatment or after ion irradiation with 15N and 12C. These paramagnetic and ferromagnetic properties of NDs make them useful as MRI probes. The paramagnetic properties had been successfully applied to MRI in vitro and in vivo whereas the ferromagnetic properties have not been applied to MRI so far. Considering that the ferromagnetic NDs can be applied as T2 MRI probes such as iron oxide nanoparticles, studies of ferromagnetic NDs as T2 MRI probes will be highly appreciated.
The fluorescent intensity of NDs is typically lower than those of organic dyes and quantum dots. Therefore, NDs with a high density of NV centers must be developed because their fluorescent intensity is proportional to the amount of NV centers per ND. On the other hand, for biomedical applications, small and size-controlled NDs (d < 3 nm) are preferred to ensure their renal excretion from the body, but small NDs have a small amount of NV centers, leading to reduced fluorescence intensity. Therefore, it will be extremely useful to develop size-controlled, small NDs (d < 3 nm) with a high density of NV centers. Although NDs do not cause biotoxicity in the body, they are not metabolized in the body. Therefore, synthesis of small and brightly fluorescent NDs on a large scale is a great future challenge.
Considering excellent optical and magnetic properties including outstanding biocompatibility,163 NDs are considered as potential dual-modal FI and MRI probes. ND surfaces can be easily conjugated with hydrophilic ligands for colloidal stability in aqueous media for biomedical applications. Further, ND surfaces can be conjugated with various functional molecules, antibodies, and drugs for further advanced applications. Therefore, NDs can serve as a new class of promising probes for theranostic biomedical applications in the future.
Abbreviations
| A549 | Human lung epithelial cell |
| bSSFP | Balanced steady-state free precession |
| C | Caenorhabditis |
| CB | Carbon black |
| CdO | Cadmium oxide |
| CVD | Chemical vapor deposition |
| DLS | Dynamic light scattering |
| DND3 | De-agglutinated detonation nanodiamond colloid |
| EDC | 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide |
| EDS | Energy dispersive X-ray spectroscopy |
| EPR | Electron paramagnetic resonance |
| HEK293 | Human embryonic kidney 293 cell |
| HPHT | High pressure and high temperature |
| TEM | Transmission electron microscope |
| FI | Fluorescence imaging |
| LND | Laser-synthesized ND |
| MRI | Magnetic resonance imaging |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| ND | Nanodiamond |
| NHS | N-Hydroxysuccinimide |
| NMR | Nuclear magnetic resonance |
| NV | Nitrogen vacancy |
| OMRI | Overhauser-enhanced MRI |
| PBS | Phosphate buffer solution |
| PEG | Plyethylene glycol |
| PEI | Polyethylenimine |
| QD | Quantum dot |
| SIP | Selective imaging protocol |
| HEK293T | Human embryonic kidney 293 tumor cell |
| XRD | X-ray diffraction |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Basic Science Research Program of the National Research Foundation (NRF) funded by the Korea government (Ministry of Science, and Information and Communications Technology: MSIT) (Basic Research Laboratory, No. 2021R1A4A1029433).Notes and references
- A. S. Thakor, J. V. Jokerst, P. Ghanouni, J. L. Campbell, E. Mittra and S. S. Gambhir, J. Nucl. Med., 2016, 57, 1833–1837 CrossRef CAS PubMed.
- J. Kim, N. Lee and T. Hyeon, Philos. Trans. R. Soc., A, 2017, 375, 20170022 CrossRef PubMed.
- E. B. Ehlerding, P. Grodzinski, W. Cai and C. H. Liu, ACS Nano, 2018, 12, 2106–2121 CrossRef CAS PubMed.
- R. M. Crist, S. S. K. Dasa, C. H. Liu, J. D. Clogston, M. A. Dobrovolskaia and S. T. Stern, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2021, 13, e1665 CAS.
- H. Dong, S.-R. Du, X.-Y. Zheng, G.-M. Lyu, L.-D. Sun, L.-D. Li, P.-Z. Zhang, C. Zhang and C.-H. Yan, Chem. Rev., 2015, 115, 10725–10815 CrossRef CAS PubMed.
- D. Kim, J. Kim, Y. I. Park, N. Lee and T. Hyeon, ACS Cent. Sci., 2018, 4, 324–336 CrossRef CAS PubMed.
- S. Sabella, R. P. Carney, V. Brunetti, M. A. Malvindi, N. A. Juffali, G. Vecchio, S. M. Janes, O. M. Bakr, R. Cingolani, F. Stellacci and P. P. Pompa, Nanoscale, 2014, 6, 7052–7061 RSC.
- L. S. Arias, J. P. Pessan, A. P. M. Vieira, T. M. T. de Lima, A. C. B. Delbem and D. R. Monteiro, Antibiotics, 2018, 7, 46 CrossRef PubMed.
- Y. H. Hu, Small, 2014, 10, 1451–1452 CrossRef CAS PubMed.
- T. Tegafaw, I. T. Oh, H. Cha, H. Yue, X. Miao, S. L. Ho, M. Y. Ahmad, S. Marasini, A. Ghazanfari, H. K. Kim, K. S. Chae, Y. Chang and G. H. Lee, J. Phys. Chem. Solids, 2018, 120, 96–103 CrossRef CAS.
- G. Hong, S. Diao, A. L. Antaris and H. Dai, Chem. Rev., 2015, 115, 10816–10906 CrossRef CAS PubMed.
- O. A. Shenderova and G. E. McGuire, Biointerphases, 2015, 10, 030802 CrossRef CAS PubMed.
- A. Shakun, J. Vuorinen, M. Hoikkanen, M. Poikelispää and A. Das, Composites, Part A, 2014, 64, 49–69 CrossRef CAS.
- M. B. A. Olia, P. S. Donnelly, L. C. L. Hollenberg, P. Mulvaney and D. A. Simpson, ACS Appl. Nano Mater., 2021, 4, 9985–10005 CrossRef.
- A. Wolcott, T. Schiros, M. E. Trusheim, E. H. Chen, D. Nordlund, R. E. Diaz, O. Gaathon, D. Englund and J. S. Owen, J. Phys. Chem. C, 2014, 118, 26695–26702 CrossRef CAS PubMed.
- A. Freedman, J. Appl. Phys., 1994, 75, 3112–3120 CrossRef CAS.
- J.-C. Arnault, T. Petit, H. Girard, A. Chavanne, C. Gesset, M. Sennour and M. Chaigneau, Phys. Chem. Chem. Phys., 2011, 13, 11481–11487 RSC.
- K.-I. Sotowa, T. Amamoto, A. Sobana, K. Kusakabe and T. Imato, Diamond Relat. Mater., 2004, 13, 145–150 CrossRef CAS.
- W.-W. Zheng, Y.-H. Hsieh, Y.-C. Chiu, S.-J. Cai, C.-L. Cheng and C. Chen, J. Mater. Chem., 2009, 19, 8432–8441 RSC.
- T. Petit, J.-C. Arnault, H. A. Girard, M. Sennour and P. Bergonzo, Phys. Rev. B: Condens. Matter Mater. Phys., 2011, 84, 233407 CrossRef.
- M. Lu, Y. K. Wang, J. Zhao, H. Lu, M. H. Stenzel and P. Xiao, Macromol. Rapid Commun., 2016, 37, 2023–2029 CrossRef CAS PubMed.
- S. Vial, C. Mansuy, S. Sagan, T. Irinopoulou, F. Burlina, J.-P. Boudou, G. Chassaing and S. Lavielle, ChemBioChem, 2008, 9, 2113–2119 CrossRef CAS PubMed.
- B.-M. Chang, H.-H. Lin, L.-J. Su, W.-D. Lin, R.-J. Lin, Y.-K. Tzeng, R. T. Lee, Y. C. Lee, A. L. Yu and H.-C. Chang, Adv. Funct. Mater., 2013, 23, 5737–5745 CrossRef CAS.
- C. Gaillard, H. A. Girard, C. Falck, V. Paget, V. Simic, N. Ugolin, P. Bergonzo, S. Chevillard and J. C. Arnault, RSC Adv., 2014, 4, 3566–3572 RSC.
- K.-K. Liu, W.-W. Zheng, C.-C. Wang, Y.-C. Chiu, C.-L. Cheng, Y.-S. Lo, C. Chen and J.-I. Chao, Nanotechnology, 2010, 21, 315106 CrossRef PubMed.
- H.-D. Wang, Q. Yang, C. Hui Niu and I. Badea, Diamond Relat. Mater., 2012, 26, 1–6 CrossRef CAS.
- J. Neburkova, F. Sedlak, J. Z. Suchanova, L. Kostka, P. Sacha, V. Subr, T. Etrych, P. Simon, J. Barinkova, R. Krystufek, H. Spanielova, J. Forstova, J. Konvalinka and P. Cigler, Mol. Pharm., 2018, 15, 2932–2945 CrossRef CAS.
- A. Barras, J. Lyskawa, S. Szunerits, P. Woisel and R. Boukherroub, Langmuir, 2011, 27, 12451–12457 CrossRef CAS PubMed.
- J. Slegerova, M. Hajek, I. Rehor, F. Sedlak, J. Stursa, M. Hruby and P. Cigler, Nanoscale, 2015, 7, 415–420 RSC.
- C. Presti, J. G. Alauzun, D. Laurencin and P. H. Mutin, Langmuir, 2014, 30, 9239–9245 CrossRef CAS PubMed.
- A. Krüger, Y. Liang, G. Jarre and J. Stegk, J. Mater. Chem., 2006, 16, 2322–2328 RSC.
- Y. Wu, A. Ermakova, W. Liu, G. Pramanik, T. M. Vu, A. Kurz, L. McGuinness, B. Naydenov, S. Hafner, R. Reuter, J. Wrachtrup, J. Isoya, C. Förtsch, H. Barth, T. Simmet, F. Jelezko and T. Weil, Adv. Funct. Mater., 2015, 25, 6576–6585 CrossRef CAS.
- R. Kaur, J. M. Chitanda, D. Michel, J. Maley, F. Borondics, P. Yang, R. E. Verrall and I. Badea, Int. J. Nanomed., 2012, 7, 3851–3866 CAS.
- C. C. Fu, H. Y. Lee, K. Chen, T. S. Lim, H. Y. Wu, P. K. Lin, P. K. Wei, P. H. Tsao, H. C. Chang and W. Fann, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 727–732 CrossRef CAS PubMed.
- S. Claveau, J.-R. Bertrand and F. Treussart, Micromachines, 2018, 9, 247 CrossRef.
- A. M. Panich, M. Salti, S. D. Goren, E. B. Yudina, A. E. Aleksenskii, A. Y. Vul’ and A. I. Shames, J. Phys. Chem. C, 2019, 123, 2627–2631 CrossRef CAS.
- L. M. Manus, D. J. Mastarone, E. A. Waters, X.-Q. Zhang, E. A. S. Sikma, K. W. MacRenaris, D. Ho and T. J. Meade, Nano Lett., 2010, 10, 484–489 CrossRef CAS PubMed.
- N. Rammohan, K. W. MacRenaris, L. K. Moore, G. Parigi, D. J. Mastarone, L. M. Manus, L. M. Lilley, A. T. Preslar, E. A. Waters, A. Filicko, C. Luchinat, D. Ho and T. J. Meade, Nano Lett., 2016, 16, 7551–7564 CrossRef CAS.
- L. Zhao, A. Shiino, H. Qin, T. Kimura and N. Komatsu, J. Nanosci. Nanotechnol., 2015, 15, 1076–1082 CrossRef CAS PubMed.
- A. M. Panich, A. I. Shames, N. A. Sergeev, V. Y. Osipov, A. E. Alexenskiy and A. Y. Vul’, J. Phys. Chem. C, 2016, 120, 19804–19811 CrossRef CAS.
- A. M. Panich, A. I. Shames, O. Medvedev, V. Y. Osipov, A. E. Aleksenskiy and A. Y. Vul’, Appl. Magn. Reson., 2009, 36, 317–329 CrossRef CAS.
- W. Hou, T. B. Toh, L. N. Abdullah, T. W. Z. Yvonne, K. J. Lee, I. Guenther and E. K.-H. Chow, Nanomed.: Nanotechnol. Biol. Med., 2017, 13, 783–793 CrossRef CAS PubMed.
- A. I. Shames, A. M. Panich, V. Y. Osipov, A. E. Aleksenskiy, A. Y. Vul’, T. Enoki and K. Takai, J. Appl. Phys., 2010, 107, 014318 CrossRef.
- A. M. Panich, A. Altman, A. I. Shames, V. Y. Osipov, A. E. Aleksenskiy and A. Y. Vul’, J. Phys. D: Appl. Phys., 2011, 44, 125303 CrossRef.
- D. E. J. Waddington, M. Sarracanie, H. Zhang, N. Salameh, D. R. Glenn, E. Rej, T. Gaebel, T. Boele, R. L. Walsworth, D. J. Reilly and M. S. Rosen, Nat. Commun., 2017, 8, 15118 CrossRef PubMed.
- N. Prabhakar and J. M. Rosenholm, Curr. Opin. Colloid Interface Sci., 2019, 39, 220–231 CrossRef CAS.
- K. V. Volkov, V. V. Danilenko and V. I. Elin, Combust. Explos. Shock Waves, 1990, 26, 366–368 CrossRef.
- V. Y. Dolmatov, A. N. Ozerin, I. I. Kulakova, O. O. Bochechka, N. M. Lapchuk, V. Myllymäki and A. Vehanen, Russ. Chem. Rev., 2020, 89, 1428–1462 CrossRef CAS.
- I. M. Abdullahi, M. Langenderfer, O. Shenderova, N. Nunn, M. D. Torelli, C. E. Johnson and V. N. Mochalin, Carbon, 2020, 164, 442–450 CrossRef CAS PubMed.
- J.-P. Boudou, J. Tisler, R. Reuter, A. Thorel, P. A. Curmi, F. Jelezko and J. Wrachtrup, Diamond Relat. Mater., 2013, 37, 80–86 CrossRef CAS.
- D. Amans, A.-C. Chenus, G. Ledoux, C. Dujardin, C. Reynaud, O. Sublemontier, K. M. Varlot and O. Guillois, Diamond Relat. Mater., 2009, 18, 177–180 CrossRef CAS.
- S. Hu, J. Sun, X. Du, F. Tian and L. Jiang, Diamond Relat. Mater., 2008, 17, 142–146 CrossRef CAS.
- L. Basso, F. Gorrini, N. Bazzanella, M. Cazzanelli, C. Dorigoni, A. Bifone and A. Miotello, Appl. Phys. A, 2018, 124, 72 CrossRef.
- H.-Y. Kim, D.-S. Kim and N.-M. Hwang, RSC Adv., 2021, 11, 5651–5657 RSC.
- E. Tamburri, R. Carcione, F. Vitale, A. Valguarnera, S. Macis, M. Lucci and M. L. Terranova, Adv. Mater. Interfaces, 2017, 4, 1700222 CrossRef.
- P. W. May, M. N. R. Ashfold and Y. A. Mankelevich, J. Appl. Phys., 2007, 101, 053115 CrossRef.
- S. Welz, Y. Gogotsi and M. J. McNallan, J. Appl. Phys., 2003, 93, 4207–4214 CrossRef CAS.
- A. K. Khachatryan, S. G. Aloyan, P. W. May, R. Sargsyan, V. A. Khachatryan and V. S. Baghdasaryan, Diamond Relat. Mater., 2008, 17, 931–936 CrossRef CAS.
- T. L. Daulton, M. A. Kirk, R. S. Lewis and L. E. Rehn, Nucl. Instrum. Methods Phys. Res. Sect. B, 2001, 175–177, 12–20 CrossRef CAS.
- F. Banhart and P. M. Ajayan, Nature, 1996, 382, 433–435 CrossRef CAS.
- I. Rehor and P. Cigler, Diamond Relat. Mater., 2014, 46, 21–24 CrossRef CAS.
- A. P. Puzyr, A. E. Burova, V. S. Bondar, C. K. Rhee, W. H. Rhee and K. C. Hwang, J. Korean Powder Metall. Inst., 2011, 18, 297–302 CrossRef.
- A. Brož, L. Bačáková, P. Štenclová, A. Kromka and Š. Potocký, Beilstein J. Nanotechnol., 2017, 8, 1649–1657 CrossRef.
- V. N. Mochalin, O. Shenderova, D. Ho and Y. Gogotsi, Nat. Nanotechnol., 2012, 7, 11–23 CrossRef CAS PubMed.
- V. Pichot, B. Risse, F. Schnell, J. Mory and D. Spitzer, Sci. Rep., 2013, 3, 2159 CrossRef PubMed.
- V. Y. Dolmatov, Russ. Chem. Rev., 2001, 70, 607–626 CrossRef CAS.
- S. P. Hong, S. W. Ha and S. W. Lee, Diamond Relat. Mater., 2018, 81, 27–32 CrossRef CAS.
- S. Ha, S. P. Hong, M. Lee, S. Lee and S. W. Lee, Mater. Today Commun., 2019, 21, 100571 CrossRef CAS.
- O. Shenderova, A. Koscheev, N. Zaripov, I. Petrov, Y. Skryabin, P. Detkov, S. Turner and G. Van Tendeloo, J. Phys. Chem. C, 2011, 115, 9827–9837 CrossRef CAS.
- J. Ackermann and A. Krueger, Nanoscale, 2019, 11, 8012–8019 RSC.
- S. Osswald, G. Yushin, V. Mochalin, S. O. Kucheyev and Y. Gogotsi, J. Am. Chem. Soc., 2006, 128, 11635–11642 CrossRef CAS PubMed.
- C. Bradac and S. Osswald, Carbon, 2018, 132, 616–622 CrossRef CAS.
- A. Krüger, F. Kataoka, M. Ozawa, T. Fujino, Y. Suzuki, A. E. Aleksenskii, A. Y. Vul’ and E. Ōsawa, Carbon, 2005, 43, 1722–1730 CrossRef.
- S. Stehlik, J. Henych, P. Stenclova, R. Kral, P. Zemenova, J. Pangrac, O. Vanek, A. Kromka and B. Rezek, Carbon, 2021, 171, 230–239 CrossRef CAS.
- S. Gupta, B. Evans, A. Henson and S. B. Carrizosa, Materials, 2017, 10, 1292 CrossRef.
- A. Pentecost, S. Gour, V. Mochalin, I. Knoke and Y. Gogotsi, ACS Appl. Mater. Interfaces, 2010, 2, 3289–3294 CrossRef CAS PubMed.
- S. Stehlik, M. Varga, M. Ledinsky, V. Jirasek, A. Artemenko, H. Kozak, L. Ondic, V. Skakalova, G. Argentero, T. Pennycook, J. C. Meyer, A. Fejfar, A. Kromka and B. Rezek, J. Phys. Chem. C, 2015, 119, 27708–27720 CrossRef CAS PubMed.
- M. Kasu, Prog. Cryst. Growth Charact. Mater., 2016, 62, 317–328 CrossRef CAS.
- R. Kumar, S. J. Yoon, K. G. Lee, P. Pal, R. P. Pant, C. K. Suman, S. R. Dhakate, R. Kumar, D. K. Avasthi and D. K. Singh, RSC Adv., 2016, 6, 47164–47173 RSC.
- F. Gorrini, M. Cazzanelli, N. Bazzanella, R. Edla, M. Gemmi, V. Cappello, J. David, C. Dorigoni, A. Bifone and A. Miotello, Sci. Rep., 2016, 6, 35244 CrossRef CAS PubMed.
- L. Yang, P. W. May, L. Yin, J. A. Smith and K. N. Rosser, Diamond Relat. Mater., 2007, 16, 725–729 CrossRef CAS.
- J.-W. Park, K.-S. Kim and N.-M. Hwang, Carbon, 2016, 106, 289–294 CrossRef CAS.
- J. E. Butler and A. V. Sumant, Chem. Vap. Deposition, 2008, 14, 145–160 CrossRef CAS.
- P. Stenclova, V. Celedova, A. Artemenko, V. Jirasek, J. Jira, B. Rezek and A. Kromka, RSC Adv., 2017, 7, 38973–38980 RSC.
- A. T. Kozakov, A. G. Kochur, N. Kumar, K. Panda, A. V. Nikolskii and A. V. Sidashov, Appl. Surf. Sci., 2021, 536, 147807 CrossRef CAS.
- J. Xiao, P. Liu and G. W. Yang, Nanoscale, 2015, 7, 6114–6125 RSC.
- H.-S. Jung and K. C. Neuman, Nanomaterials, 2021, 11, 153 CrossRef CAS PubMed.
- T. Ando, J. Tanaka, M. Ishii, M. Kamo, Y. Sato, N. Ohashi and S. Shimosaki, J. Chem. Soc. Faraday Trans., 1993, 89, 3105–3109 RSC.
- M. Börsch, R. Reuter, G. Balasubramanian, R. Erdmann, F. Jelezko and J. Wrachtrup, Proc. SPIE, 2009, 7183, 71832N CrossRef.
- A. Bumb, S. K. Sarkar, N. Billington, M. W. Brechbiel and K. C. Neuman, J. Am. Chem. Soc., 2013, 135, 7815–7818 CrossRef CAS PubMed.
- N. Prabhakar, T. Näreoja, E. von Haartman, D. Ş. Karaman, H. Jiang, S. Koho, T. A. Dolenko, P. E. Hänninen, D. I. Vlasov, V. G. Ralchenko, S. Hosomi, I. I. Vlasov, C. Sahlgren and J. M. Rosenholm, Nanoscale, 2013, 5, 3713–3722 RSC.
- D. Wang, Y. Tong, Y. Li, Z. Tian, R. Cao and B. Yang, Diamond Relat. Mater., 2013, 36, 26–34 CrossRef CAS.
- A. S. Barnard, Nanoscale, 2017, 9, 70–74 RSC.
- X. Zhang, C. Fu, L. Feng, Y. Ji, L. Tao, Q. Huang, S. Li and Y. Wei, Polymer, 2012, 53, 3178–3184 CrossRef CAS.
- G. Yang, W. Long, W. Yan, H. Huang, M. Liu, H. Ouyang, Y. Feng, L. Liu, X. Zhang and Y. Wei, J. Drug Delivery Sci. Technol., 2020, 57, 101644 CrossRef CAS.
- L. Zhao, Y.-H. Xu, T. Akasaka, S. Abe, N. Komatsu, F. Watari and X. Chen, Biomaterials, 2014, 35, 5393–5406 CrossRef CAS PubMed.
- J.-P. Boudou, M.-O. David, V. Joshi, H. Eidi and P. A. Curmi, Diamond Relat. Mater., 2013, 38, 131–138 CrossRef CAS.
- L. Zhao, T. Takimoto, M. Ito, N. Kitagawa, T. Kimura and N. Komatsu, Angew. Chem., Int. Ed., 2011, 50, 1388–1392 CrossRef CAS PubMed.
- M. Chen, X.-Q. Zhang, H. B. Man, R. Lam, E. K. Chow and D. Ho, J. Phys. Chem. Lett., 2010, 1, 3167–3171 CrossRef CAS.
- S. Qin, M. Cui, S. Qiu, H. Zhao, L. Wang and A. Zhang, RSC Adv., 2018, 8, 3694–3704 RSC.
- S. P. Santana, N. F. Salazar, A. S. Sainz, E. S. Campa, A. B. Estrella, A. A. Molina, R. Melendrez, M. P. Montero and R. Riera, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2018, 9, 015013 Search PubMed.
- Y. Dong, R. Cao, Y. Li, Z. Wang, L. Li and L. Tian, RSC Adv., 2015, 5, 82711–82716 RSC.
- L. Li, L. Tian, W. Zhao, F. Cheng, Y. Li and B. Yang, RSC Adv., 2016, 6, 36407–36417 RSC.
- V. N. Mochalin and Y. Gogotsi, J. Am. Chem. Soc., 2009, 131, 4594–4595 CrossRef CAS PubMed.
- P. Karami and A. Shojaei, Polym. Int., 2017, 66, 557–565 CrossRef CAS.
- D. G. Lim, K. H. Kim, E. Kang, S. H. Lim, J. Ricci, S. K. Sung, M. T. Kwon and S. H. Jeong, Int. J. Nanomed., 2016, 11, 2381–2395 Search PubMed.
- D. Wang, Y. Li, Z. Tian, R. Cao and B. Yang, Ther. Delivery, 2014, 5, 511–524 CrossRef CAS PubMed.
- D. Meziane, A. Barras, A. Kromka, J. Houdkova, R. Boukherroub and S. Szunerits, Anal. Chem., 2012, 84, 194–200 CrossRef CAS PubMed.
- L. Marcon, Z. Kherrouche, J. Lyskawa, D. Fournier, D. Tulasne, P. Woisel and R. Boukherroub, Chem. Commun., 2011, 47, 5178–5180 RSC.
- A. Barras, S. Szunerits, L. Marcon, N. M. Dupont and R. Boukherroub, Langmuir, 2010, 26, 13168–13172 CrossRef CAS PubMed.
- A. Krueger and T. Boedeker, Diamond Relat. Mater., 2008, 17, 1367–1370 CrossRef CAS.
- J.-P. Boudou, P. A. Curmi, F. Jelezko, J. Wrachtrup, P. Aubert, M. Sennour, G. Balasubramanian, R. Reuter, A. Thorel and E. Gaffet, Nanotechnology, 2009, 20, 235602 CrossRef PubMed.
- V. Kuzmin, K. Safiullin, G. Dolgorukov, A. Stanislavovas, E. Alakshin, T. Safin, B. Yavkin, S. Orlinskii, A. Kiiamov, M. Presnyakov, A. Klochkov and M. Tagirov, Phys. Chem. Chem. Phys., 2018, 20, 1476–1484 RSC.
- M. V. Baidakova, Y. A. Kukushkina, A. A. Sitnikova, M. A. Yagovkina, D. A. Kirilenko, V. V. Sokolov, M. S. Shestakov, A. Y. Vul’, B. Zousman and O. Levinson, Phys. Solid State, 2013, 55, 1747–1753 CrossRef CAS.
- V. I. Korepanov, H.-o. Hamaguchi, E. Osawa, V. Ermolenkov, I. K. Lednev, B. J. M. Etzold, O. Levinson, B. Zousman, C. P. Epperla and H.-C. Chang, Carbon, 2017, 121, 322–329 CrossRef CAS.
- S. S. Batsanov, S. M. Gavrilkin, A. S. Batsanov, K. B. Poyarkov, I. I. Kulakova, D. W. Johnson and B. G. Mendis, J. Mater. Chem., 2012, 22, 11166–11172 RSC.
- J. Xiao, P. Liu, L. Li and G. Yang, J. Phys. Chem. C, 2015, 119, 2239–2248 CrossRef CAS.
- K. N. Clayton, J. W. Salameh, S. T. Wereley and T. L. Kinzer-Ursem, Biomicrofluidics, 2016, 10, 054107 CrossRef PubMed.
- C. Fryer, P. Murray and H. Zhang, Nanomaterials, 2022, 12, 4196 CrossRef CAS PubMed.
- C. D. C. Wachesk, C. R. Hurtado, R. F. B. D. O. Correia, D. B. Tada, G. Vasconcelos, E. J. Corat and V. J. T. Airoldi, Biomed. J. Sci. Tech. Res., 2022, 41, 32695–32709 Search PubMed.
- D. Maziukiewicz, R. Mrówczyński, S. Jurga and B. F. Grześkowiak, Diamond Relat. Mater., 2022, 128, 109308 CrossRef CAS.
- E. R. Wilson, L. M. Parker, A. Orth, N. Nunn, M. Torelli, O. Shenderova, B. C. Gibson and P. Reineck, Nanotechnology, 2019, 30, 385704 CrossRef CAS PubMed.
- W. Cao, X. Peng, X. Chen, X. Wang, F. Jin, Q. Li, H. Chen, C. Jiang, Z. Ye and X. Xing, J. Mater. Sci., 2017, 52, 1856–1867 CrossRef CAS.
- X. Xu, Z. Yu, Y. Zhu and B. Wang, Diamond Relat. Mater., 2005, 14, 206–212 CrossRef CAS.
- L. Zhao, Y. Nakae, H. Qin, T. Ito, T. Kimura, H. Kojima, L. Chan and N. Komatsu, Beilstein J. Org. Chem., 2014, 10, 707–713 CrossRef PubMed.
- L. Gines, S. Mandal, A. I. Ahmad, C.-L. Cheng, M. Sow and O. A. Williams, Nanoscale, 2017, 9, 12549–12555 RSC.
- Y.-R. Chang, H.-Y. Lee, K. Chen, C.-C. Chang, D.-S. Tsai, C.-C. Fu, T.-S. Lim, Y.-K. Tzeng, C.-Y. Fang, C.-C. Han, H.-C. Chang and W. Fann, Nat. Nanotechnol., 2008, 3, 284–288 CrossRef CAS PubMed.
- J. Havlik, V. Petrakova, I. Rehor, V. Petrak, M. Gulka, J. Stursa, J. Kucka, J. Ralis, T. Rendler, S.-Y. Lee, R. Reuter, J. Wrachtrup, M. Ledvina, M. Nesladek and P. Cigler, Nanoscale, 2013, 5, 3208–3211 RSC.
- G. Dantelle, A. Slablab, L. Rondin, F. Lainé, F. Carrel, P. Bergonzo, S. Perruchas, T. Gacoin, F. Treussart and J. F. Roch, J. Lumin., 2010, 130, 1655–1658 CrossRef CAS.
- Y. Y. Hui, Y.-R. Chang, N. Mohan, T.-S. Lim, Y.-Y. Chen and H.-C. Chang, J. Phys. Chem. A, 2011, 115, 1878–1884 CrossRef CAS PubMed.
- S.-J. Yu, M.-W. Kang, H.-C. Chang, K.-M. Chen and Y.-C. Yu, J. Am. Chem. Soc., 2005, 127, 17604–17605 CrossRef CAS PubMed.
- L. Rondin, G. Dantelle, A. Slablab, F. Grosshans, F. Treussart, P. Bergonzo, S. Perruchas, T. Gacoin, M. Chaigneau, H.-C. Chang, V. Jacques and J.-F. Roch, Phys. Rev. B: Condens. Matter Mater. Phys., 2010, 82, 115449 CrossRef.
- H.-C. Lu, Y.-C. Peng, S.-L. Chou, J.-I. Lo, B.-M. Cheng and H.-C. Chang, Angew. Chem., Int. Ed., 2017, 56, 14469–14473 CrossRef CAS PubMed.
- E. von Haartman, H. Jiang, A. A. Khomich, J. Zhang, S. A. Burikov, T. A. Dolenko, J. Ruokolainen, H. Gu, O. A. Shenderova, I. I. Vlasov and J. M. Rosenholm, J. Mater. Chem. B, 2013, 1, 2358–2366 RSC.
- S. Sotoma, F.-J. Hsieh, Y.-W. Chen, P.-C. Tsai and H.-C. Chang, Chem. Commun., 2018, 54, 1000–1003 RSC.
- P. G. Baranov, A. A. Soltamova, D. O. Tolmachev, N. G. Romanov, R. A. Babunts, F. M. Shakhov, S. V. Kidalov, A. Y. Vul’, G. V. Mamin, S. B. Orlinskii and N. I. Silkin, Small, 2011, 7, 1533–1537 CrossRef CAS PubMed.
- W. Liu, M. N. A. Alam, Y. Liu, V. N. Agafonov, H. Qi, K. Koynov, V. A. Davydov, R. Uzbekov, U. Kaiser, T. Lasser, F. Jelezko, A. Ermakova and T. Weil, Nano Lett., 2022, 22, 2881–2888 CrossRef CAS PubMed.
- Y. Makino, T. Mahiko, M. Liu, A. Tsurui, T. Yoshikawa, S. Nagamachi, S. Tanaka, K. Hokamoto, M. Ashida, M. Fujiwara, N. Mizuochi and M. Nishikawa, Diamond Relat. Mater., 2021, 112, 108248 CrossRef CAS.
- E. Perevedentseva, A. Karmenyan, Y. C. Lin, C. Y. Song, Z. R. Lin, A. I. Ahmed, C. C. Chang, S. Norina, V. Bessalova, N. Perov, O. Levinson, B. Zousman and C.-L. Cheng, J. Biomed. Opt., 2018, 23, 091404 Search PubMed.
- V. Y. Osipov, A. I. Shames, T. Enoki, K. Takai, M. V. Baidakova and A. Y. Vul’, Diamond Relat. Mater., 2007, 16, 2035–2038 CrossRef CAS.
- A. I. Shames, A. M. Panich, W. Kempiński, A. E. Alexenskii, M. V. Baidakova, A. T. Dideikin, V. Y. Osipov, V. I. Siklitski, E. Osawa, M. Ozawa and A. Y. Vul, J. Phys. Chem. Solids, 2002, 63, 1993–2001 CrossRef CAS.
- I. Kratochvílova, J. Šebera, P. Ashcheulov, M. Golan, M. Ledvina, J. Míčova, F. Mravec, A. Kovalenko, D. Zverev, B. Yavkin, S. Orlinskii, S. Záliš, A. Fišerová, J. Richter, L. Šefc and J. Turánek, J. Phys. Chem. C, 2014, 118, 25245–25252 CrossRef.
- Z. Peng, J. Dallas and S. Takahashi, J. Appl. Phys., 2020, 128, 054301 CrossRef CAS.
- S. Talapatra, P. G. Ganesan, T. Kim, R. Vajtai, M. Huang, M. Shima, G. Ramanath, D. Srivastava, S. C. Deevi and P. M. Ajayan, Phys. Rev. Lett., 2005, 95, 097201 CrossRef CAS PubMed.
- A. Setzer, P. D. Esquinaz, S. Buga, M. T. Georgieva, T. Reinert, T. Venus, I. Eestrela-Lopis, A. Ivashenko, M. Bondarenko, W. Böhlmann and J. Meijer, Materials, 2022, 15, 1014 CrossRef CAS PubMed.
- A. M. Schrand, L. Dai, J. J. Schlager, S. M. Hussain and E. Osawa, Diamond Relat. Mater., 2007, 16, 2118–2123 CrossRef CAS.
- C. Cheng, A. E. Porter, K. Muller, K. Koziol, J. N. Skepper, P. Midgley and M. Welland, J. Phys. Conf. Ser., 2009, 151, 012030 CrossRef.
- Y. Zhu, J. Li, W. Li, Y. Zhang, X. Yang, N. Chen, Y. Sun, Y. Zhao, C. Fan and Q. Huang, Theranostics, 2012, 2, 302–312 CrossRef CAS PubMed.
- V. Paget, J. A. Sergent, R. Grall, S. A. Morel, H. A. Girard, T. Petit, C. Gesset, M. Mermoux, P. Bergonzo, J. C. Arnault and S. Chevillard, Nanotoxicology, 2014, 8, 46–56 CrossRef CAS PubMed.
- A. M. Schrand, H. Huang, C. Carlson, J. J. Schlager, E. Ōsawa, S. M. Hussain and L. Dai, J. Phys. Chem. B, 2007, 111, 2–7 CrossRef CAS PubMed.
- X. Zhang, W. Hu, J. Li, L. Tao and Y. Wei, Toxicol. Res., 2012, 1, 62–68 CrossRef CAS.
- X. Zhang, J. Yin, C. Kang, J. Li, Y. Zhu, W. Li, Q. Huang and Z. Zhu, Toxicol. Lett., 2010, 198, 237–243 CrossRef CAS PubMed.
- L. Marcon, F. Riquet, D. Vicogne, S. Szunerits, J.-F. Bodart and R. Boukherroub, J. Mater. Chem., 2010, 20, 8064–8069 RSC.
- N. Dworak, M. Wnuk, J. Zebrowski, G. Bartosz and A. Lewinska, Carbon, 2014, 68, 763–776 CrossRef CAS.
- K. Solarska, A. Gajewska, W. Kaczorowski, G. Bartosz and K. Mitura, Diamond Relat. Mater., 2012, 21, 107–113 CrossRef CAS.
- A. P. Puzyr, D. A. Neshumayev, S. V. Tarskikh, G. V. Makarskaya, V. Y. Dolmatov and V. S. Bondar, Diamond Relat. Mater., 2004, 13, 2020–2023 CrossRef CAS.
- K. Turcheniuk and V. N. Mochalin, Nanotechnology, 2017, 28, 252001 CrossRef CAS PubMed.
- M. Longmire, P. L. Choyke and H. Kobayashi, Nanomedicine, 2008, 3, 703–717 CrossRef CAS PubMed.
- H. S. Choi, W. Liu, P. Misra, E. Tanaka, J. P. Zimmer, B. I. Ipe, M. G. Bawendi and J. V. Frangioni, Nat. Biotechnol., 2007, 25, 1165–1170 CrossRef CAS PubMed.
- S. Rojas, J. D. Gispert, R. Martin, S. Abad, C. Menchon, D. Pareto, V. M. Victor, M. Alvaro, H. Garcia and J. R. Herance, ACS Nano, 2011, 5, 5552–5559 CrossRef CAS PubMed.
- Y. Yuan, Y. Chen, J.-H. Liu, H. Wang and Y. Liu, Diamond Relat. Mater., 2009, 18, 95–100 CrossRef CAS.
- F. C. Barone, C. Marcinkiewicz, J. Li, Y. Feng, M. Sternberg, P. I. Lelkes, D. Rosenbaum-Halevi, J. A. Gerstenhaber and G. Z. Feuerstein, Int. J. Nanomed., 2019, 14, 1163–1175 CrossRef CAS PubMed.
- L. Moore, J. Yang, T. T. H. Lan, E. Osawa, D.-K. Lee, W. D. Johnson, J. Xi, E. K.-H. Chow and D. Ho, ACS Nano, 2016, 10, 7385–7400 CrossRef CAS PubMed.
- J. C. Hsu, Z. Tang, O. E. Erenina, A. M. Soflas, T. Lammers, J. F. Lovell, C. Zavaleta, W. Cai and D. P. Cormode, Nat. Rev. Method Primers, 2023, 3, 30 CrossRef CAS.
- L. M. Liz-Marzán, A. E. Nel, C. J. Brinker, W. C. W. Chan, C. Chen, X. Chen, D. Ho, T. Hu, K. Kataoka, N. A. Kotov, W. J. Parak and M. M. Stevens, ACS Nano, 2022, 16, 13257–13259 CrossRef.
- M. P. Lake and L.-S. Bouchard, PLoS One, 2017, 12, e0179295 CrossRef PubMed.
- H. M. Leung, M. S. Chan, L. S. Liu, S. W. Wong, T. W. Lo, C. H. Lau, C. Tin and P. K. Lo, ACS Sustainable Chem. Eng., 2018, 6, 9671–9681 CrossRef CAS.
- L. Nie, Y. Zhang, L. Li, P. van Rijn and R. Schirhagl, Nanomaterials, 2021, 11, 1837 CrossRef CAS PubMed.
- P. Reineck, M. Capelli, D. W. M. Lau, J. Jeske, M. R. Field, T. Ohshima, A. D. Greentree and B. C. Gibson, Nanoscale, 2017, 9, 497–502 RSC.
- J.-I. Chao, E. Perevedentseva, P.-H. Chung, K.-K. Liu, C.-Y. Cheng, C.-C. Chang and C.-L. Cheng, Biophys. J., 2007, 93, 2199–2208 CrossRef CAS PubMed.
- R. Igarashi, Y. Yoshinari, H. Yokota, T. Sugi, F. Sugihara, K. Ikeda, H. Sumiya, S. Tsuji, I. Mori, H. Tochio, Y. Harada and M. Shirakawa, Nano Lett., 2012, 12, 5726–5732 CrossRef CAS PubMed.
- N. Mohan, C.-S. Chen, H.-H. Hsieh, Y.-C. Wu and H.-C. Chang, Nano Lett., 2010, 10, 3692–3699 CrossRef CAS PubMed.
- H. H. Han, H. Kang, S. Kim, R. Pal, A. T. N. Kumar, H. S. Choi and S. K. Hahn, RSC Adv., 2021, 11, 23073–23081 RSC.
- M. D. Pham, C. P. Epperla, C.-L. Hsieh, W. Chang and H.-C. Chang, Anal. Chem., 2017, 89, 6527–6534 CrossRef CAS PubMed.
- D. E. J. Waddington, T. Boele, E. Rej, D. R. McCamey, N. J. C. King, T. Gaebel and D. J. Reilly, Sci. Rep., 2019, 9, 5950 CrossRef PubMed.
- X. Lv, J. H. Walton, E. Druga, F. Wang, A. Aguilar, T. McKnelly, R. Nazaryan, F. L. Liu, L. Wu, O. Shenderova, D. B. Vigneron, C. A. Meriles, J. A. Reimer, A. Pines and A. Ajoy, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2023579118 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |
