 Open Access Article
Open Access ArticleCharacterization of organic species and functional groups in pollen, fungi, algae, and bacteria bioaerosols†
Palina
Bahdanovich
 ab,
Kevin
Axelrod
ab,
Kevin
Axelrod
 ab,
Andrey Y.
Khlystov
ab,
Andrey Y.
Khlystov
 ab and
Vera
Samburova
ab and
Vera
Samburova
 *ab
*ab
aDesert Research Institute, 2215 Raggio Parkway, Reno, NV 89512, USA. E-mail: vera.samburova@dri.edu
bUniversity of Nevada, Reno, 1664 N Virginia St, Reno, NV 89557, USA
First published on 22nd August 2024
Abstract
Though the importance of bioaerosols is increasing with the changing climate, very little is known about the chemistry of bioaerosols, their atmospheric fate, and chemical composition. This paper is focused on the characterization of chemical functional groups of four atmospherically relevant bioaerosols: pollen (lodgepole pine and rabbitbrush), fungi (western gall rust), bacteria (Pedobacter and hay bacillus), and algae (spirulina). For this purpose, the proton nuclear magnetic resonance (1H-NMR) technique was used on water-soluble extracts of the selected bioaerosols, while quantitative analysis of individual organic species (saccharides, amino acids, and fatty acids) was performed using gas chromatography mass spectrometry (GC-MS), ultra-high performance liquid chromatography (UPLC-MS), and UV-Vis-NIR (ultraviolet-visible-infrared) spectrophotometry. The obtained 1H-NMR results revealed major contributions from aliphatic protons in Bacillus (50.2%) and Pedobacter (57.0%) bacteria, western gall rust fungus (39.7%), spirulina algae (73.8%), and rabbitbrush pollen (31.3%). Protons from saccharides were dominant in lodgepole pine pollen (27.6%). The quantitative analysis shows that the saccharide glucose is common among the analyzed bioaerosols, as well as proline, leucine, isoleucine, alanine, and phenylalanine amino acids, and palmitic, oleic, linoleic, linolenic, and stearic fatty acids (except in Bacillus bacteria). Concentrations of analyzed saccharides ranged between 2.01 μg mg−1 of dry mass (in Bacillus bacteria) and 183.54 μg mg−1 (in lodgepole pine pollen), followed by amino acids (from 2.57 μg mg−1 in western gall rust fungus to 21.38 μg mg−1 in Bacillus bacteria), and fatty acids (from 0.05 μg mg−1 in Bacillus bacteria to 25.82 μg mg−1 in lodgepole pine pollen). Comparison of 1H-NMR and quantitative analyses showed a good correlation (R2 = 0.608) between the saccharide segment of 1H-NMR bioaerosol spectra and individual saccharide analysis.
Environmental significanceLimited information is available regarding the chemical composition of bioaerosols (airborne pollen, fungi, algae, bacteria), despite their growing importance with the changing climate. In this study, selected bioaerosols were extracted with water and then characterized for their chemical components using various analytical techniques (1H-NMR, GC-MS, UPLC-MS, UV-Vis-NIR). The presented results give valuable insight into the contributions of saccharides, amino acids, fatty acids, and various functional groups in bioaerosols, thereby advancing the field of bioaerosol chemistry and their applications. |
1 Introduction
Bioaerosols are particles, such as pollen, fungi, bacteria, microalgae, and their dispersal units and fragments, emitted into the atmosphere from biological sources.1–3 These particles range from tens of nanometers to a few hundred micrometers in diameter.4–6 Bioaerosols can represent a significant portion of the atmospheric aerosol load,7–9 being emitted into the atmosphere in thousands of tera-grams (Tg) per year, with concentrations fluctuating based on region and season.9,10 Recent estimates of bioaerosol contribution to atmospheric particulate matter (PM) indicate that bioaerosols can account for 16.5% of PM2.5 and 16.3% of PM10.11 Due to their aerodynamic size and structure, bioaerosols can be transferred by wind and other mechanisms, and thus may play a role in atmospheric chemical and physical processes. Although the atmospheric abundance of bioaerosols is roughly 30% in urban and rural environments,1 several studies have found that some bioaerosols, such as pollen, bacteria, and fungi, can be effective cloud condensation nuclei (CCN) and ice nucleating particles (INP).5,12–17 For example, Bauer et al. (2003)18 analyzed cloud water and atmospheric aerosol samples in the Austrian mountains and determined that the main identified species (A. agilis and S. echinoides) were CCN active. Huffman et al. (2013)8 performed DNA analysis of bioaerosol samples collected in the central Rocky Mountains (Colorado, USA) and showed that some bacteria and fungi can play a significant role in the precipitation formation, especially during rain events, which can trigger a large emission of bioaerosols from the forest. Another study highlighted the importance of fungal spores in cloud formation processes on local and regional scales.16 Further, a recent field study by Cornwell and colleagues determined that the major source of INPs between −12 and −20 °C were bioaerosols.19 Microorganisms (i.e., bacteria, fungi, and algae) have been found as high as the stratosphere, where they endure extreme atmospheric conditions, such as short wavelength radiation, desiccation, and very low temperatures.20 Airborne biological particles can also be transported long distances, even intercontinentally.21–23Like other atmospheric aerosols, bioaerosols significantly affect human health,24,25 including respiratory irritation due to allergens,26,27 exposure to bacteria, pathogens, and possible inhalation of neurotoxins.28 Recent studies have reported that a changing climate fuels algal blooms29 and increases concentrations of pollen.30 The amplified harmful algal blooms can emit toxins via lake spray aerosol,29 which can negatively impact humans and the environment.31 In addition to airborne toxin emission, some algae species can produce allergic reactions like that of pollen, such as respiratory issues and reactions on the skin.32 Dramatic changes in pollen concentrations and longer pollen seasons negatively impact public health.30 According to the Center for Disease Control and Prevention, 24.4 million people suffer from seasonal pollen allergies in the U.S.33 These concerns and lack of studies on contribution of bioaerosols to atmospheric processes create an urgency to study bioaerosols, especially their chemical composition and atmospheric fate.
In the present study, several bioaerosols were selected considering their relevance, abundance in the atmosphere, and availability: lodgepole pine and rabbitbrush pollens (Pinus contorta and Ericameria nauseosa), western gall rust fungus (Endocronartium harknessii), Pedobacter and hay bacillus bacteria (Pedobacter sp. and Bacillus subtilis), and spirulina algae (Arthrospira platensis). Lodgepole pine and rabbitbrush are two dominant producers of pollen in western North America.34,35 Pine pollen species have been detected in air samples in the Arctic (transported from other locations to the pine-free area of the Arctic), which confirms their presence in the atmosphere.36 Western gall rust is a fungus that affects hard pine trees in western and northern North America.37,38 This fungus produces galls on pine trees, which contain orange spores39 that can easily be dispersed through the atmosphere.40 Although there is limited literature available regarding Western gall rust, airborne fungal spore counts can range from 1000 to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 spores per cubic meter of air,9,41 which highlights their contribution to the atmospheric aerosol load. Pedobacter and hay bacillus are commonly found in soil42–44 and several Bacillus species have been identified in thousands of PM2.5 aerosol samples across the U.S.45 Spirulina is an incredibly adaptable algae that can thrive in extreme conditions and is commonly found in soil, freshwater, and saltwater, among other aquatic habitats.46 To our knowledge, there have been no studies conducted on airborne spirulina algae, however, algae and other microorganisms (i.e., bacteria and fungi) can become airborne by aerosolization,47,48 and even survive stratospheric conditions.20
000 spores per cubic meter of air,9,41 which highlights their contribution to the atmospheric aerosol load. Pedobacter and hay bacillus are commonly found in soil42–44 and several Bacillus species have been identified in thousands of PM2.5 aerosol samples across the U.S.45 Spirulina is an incredibly adaptable algae that can thrive in extreme conditions and is commonly found in soil, freshwater, and saltwater, among other aquatic habitats.46 To our knowledge, there have been no studies conducted on airborne spirulina algae, however, algae and other microorganisms (i.e., bacteria and fungi) can become airborne by aerosolization,47,48 and even survive stratospheric conditions.20
To characterize chemical functional groups of water-soluble bioaerosol extracts, the proton nuclear magnetic resonance (1H-NMR) spectroscopy technique was selected for the present study. 1H-NMR has been widely used for analysis of atmospheric organic aerosols, especially those of anthropogenic origin.49 While research surrounding analysis of airborne pollen particles and water-soluble aerosols by means of 1H-NMR has been published,49–52 limited research was conducted on the chemistry of other types of biological aerosols. Several studies on the chemical composition of bioaerosols reported that bioaerosols generally contain common organic compounds like saccharides, amino acids,50,53,54 carbohydrates,55,56 proteins,57 fatty acids, and lipids.58 Saccharides such as glucose and sucrose can comprise anywhere from 4.0 to 29% (total dry weight) in pollen,53,59 while amino acid content ranges from 0.29 to 15% (total dry weight).53 Recent studies found that sub pollen particles (∼0.60–∼2.5 μm)60 are largely composed of starch.13,61 In our recent study we found that the starch content in bioaerosols can range from 0.045 (in hay bacillus bacteria) to 0.43% (in western gall rust fungus) of total dry weight.56 However, there is still a large knowledge gap on chemical composition and the atmospheric transformation of these bioaerosols and their fragments.1,50,62 Therefore, the goal of the present study is to characterize the chemical composition and functional groups of organic compounds in the aqueous extracts of the selected bioaerosols. For this purpose, the bioaerosols were lysed to represent the fragmentation that can occur in the atmosphere.1,631H-NMR spectroscopy results were compared with the data obtained using quantitative analyses such as gas chromatography-mass spectrometry (GC-MS) and ultra-high performance liquid chromatography-mass spectrometry (UPLC-MS). The starch content of the selected bioaerosols was assessed with ultraviolet-visible-infrared (UV-Vis-NIR) spectroscopy.56,64
2 Experimental section
2.1. Standards and materials
Deuterated dimethyl sulfoxide (DMSO-d6) (99.9%) and deuterium oxide (D2O) (99.96%) were used as solvents in this study and were purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA, USA). Standards were purchased from Sigma-Aldrich, Co. (St. Louis, MO, USA) and can be found in Table S1 of the ESI.† Sodium hydroxide (NaOH) pellets were purchased from Ward's Science (Rochester, NY, USA). Ultra-high purity water (≥18 MΩ cm−1) used in this study was dispensed by Elga Veolia PURELAB Chorus 1 water purification system (Woodridge, IL, USA). For GC-MS sample derivatization, N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) with 1% trimethylchlorosilane (TMCS) (Thermo Fisher Scientific, Waltham, MA, USA) and pyridine (Sigma-Aldrich) were used. Eluents such as toluene, acetonitrile (ACN), and HPLC-grade water were purchased from Thermo Fisher Scientific. Saccharide standards were purchased from Sigma-Aldrich, Cambridge Isotope Laboratories, Inc., and Accustandard (New Haven, CT, USA).UPLC-MS eluents include HPLC-grade ACN (Fisher Scientific) and ultra-high purity water. Formic acid (Fisher Scientific, Fair Lawn, NJ, USA) and ammonium formate (Sigma-Aldrich) were used as additives in the eluent for amino acid analysis. Amino acid and fatty acid standards were purchased from Sigma-Aldrich and Toronto Research Chemicals (North York, ON, Canada). A collection of calibration standards for saccharide, amino acid, and fatty acid analysis can be found in ESI (Table S2).† Soluble potato starch (≥95% purity) was purchased from Sigma-Aldrich. Class 1b ethyl alcohol (Fisher Scientific), 1 N sodium hydroxide solution (Fisher Scientific), and 1 M hydrochloric acid (HCl) (Sigma-Aldrich) were used for starch and fatty acid preparation. Potassium iodide and iodine were obtained from Ward's Science.
2.2. Bioaerosol species
Bioaerosols were either purchased or collected in the field (see Table 1). Samples consist of 15 mg (dry weight) of each bioaerosol for 1H-NMR analysis (samples weighed using the Cahn C-33 microbalance, Cerritos, CA, USA). For GC- and UPLC-MS analysis, bioaerosol samples were weighed at 3 mg each. Arthrospira platensis (commonly known as Spirulina) was commercially purchased from Amazon.com, Inc. (Seattle, WA, USA) in freeze-dried form. Endocronartium harknessii (commonly known as western gall rust) was collected directly from infected pine trees in Mount Shasta, CA, USA. Ericameria nauseosa (Rubber-Rabbit Brush) and Pinus contorta (Lodgepole Pine) pollen were collected directly from plants in various areas near Reno, NV, USA (see Table 1 for specific locations). Lodgepole pine pollen was collected throughout a period of three years (July 2020; June 2021; May 2022). Pedobacter sp. and Bacillus subtilis were cultured at the Molecular Microbial Ecology Genomics Laboratory, Desert Research Institute, Reno, NV USA. Freshly cultured bacteria samples were lyophilized (freeze-fried) at −40 °C for 24 hours using the Thermo Micro Modulyo 115 freeze dryer system (Asheville, NC, USA). Bioaerosols are referred to using their common name throughout this study.| Bioaerosol | Common name | Botanical name | Origin |
|---|---|---|---|
| Pollen | Lodgepole pine | Pinus contorta | Collected July-5-2020 in North Lake Tahoe, NV, USA (2020 season) |
| Collected June-22-2021 at Mt. Rose Highway Summit, NV, USA (2021 season) | |||
| Collected May-23-2022 in Reno, NV, USA (2022 season) | |||
| Rabbitbrush | Ericameria nauseosa | Collected October-8-2019 in Reno, NV, USA | |
| Fungus | Western gall rust | Endocronartium harknessii | Collected May-31-2021 in Mt. Shasta, CA, USA |
| Bacteria | Hay bacillus; Pedobacter | Bacillus subtilis; Pedobacter sp | Cultured in the Molecular Microbial Ecology and Genomics Lab at the Desert research Institute, NV, USA and freeze-dried on February-13-2022 |
| Microalgae | Spirulina | Arthrospira platensis | Aquaculture farms; purchased from Amazon, Inc. |
2.3 1H-NMR analysis
1H-NMR analysis was performed using the Agilent Technologies, Inc. 500 MHz PremiumCompact + NMR (Santa Clara, CA, USA) at 25 °C, using VnmrJ software.65 The spectra were acquired in the range of 0.0–14.0 parts per million (ppm). For 1H-NMR characterization of functional groups of the selected bioaerosols (Table 1) and for assigning chemical shifts, standards were prepared, and 1H-NMR spectra acquired for the compounds of the following chemical classes: fatty acids, amino acids, triacylglycerols (TAG), saccharides, sugar alcohols, starch, polycyclic aromatic hydrocarbons (PAH's), oxygenated PAH's, aldehydes, aliphatic alcohols, and aromatic alcohols (see Table S1† for specific compounds). Dry standards were desiccated in the presence of NaOH pellets (to reduce water signal in 1H-NMR spectra) prior to addition of 0.75 mL DMSO-d6 or D2O. Standards were dissolved in DMSO-d6 or D2O (see Table S1†) directly for each trial and sonicated for five min at 30 °C. Standard solutions were then transferred to 5 mm high-throughput 8′′ standard series NMR tubes (Norell, Morganton, NC, USA and Wilmad LabGlass, Vineland, NJ, USA). Three replicates of saccharides and fatty acids were prepared separately to determine consistency of sample preparation. Standards were run for 64 scans to assign chemical shifts and major functional groups for analysis of the bioaerosol sample spectra (Table 2).| Segment | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| a Weak signal. | ||||||||
| Spectral shift (ppm) | 0.5–2.0 | 2.0–2.7 | 2.7–3.7 | 3.7–4.1 | 4.1–5.7 | 6.0–6.6 | 6.6–8.3 | 9.0–14 |
| Aliphatic CH | α-carbon O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH1,2 CH1,2 |
H in saccharide ring | Alcohol CH | All OH in saccharide ring except –O–CH(OH)- | OH in saccharide ring –O–CH(OH)- | Aromatic-Ha | Ar–OHa | |
| Aliphatic CH2 | α-Carbon CH-COO | CH–OH in saccharide alcohol | Alcohol OH | Glycerol OH | CONH, CONH2a | COOHa | ||
| Aliphatic CH3 | α to amine H | C–H in saccharide CH2-OH | H in saccharide ring | CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2 CH2 |
C(O)Ha | |||
| NH, NH2 | α-carbon CH2C-NH2a | Alcohol OH | ||||||
| OH in TAG | ||||||||
Bioaerosol samples (see Table 1) were hydrated with ultra-high purity water (Elga Veolia PURELAB Chorus) for a concentration of 10 mg mL−1. Samples were thoroughly lysed (Bertin Instruments Precellys 2 mL Lysing Kit, Rockville, MD, USA) for one min, in intervals of 20 seconds at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 revolutions per min (RPM) using the Bertin Instruments Minilys Personal Homogenizer (Rockville, MD, USA) to encourage compound extraction from bioaerosol cells and fragmentation of the bioaerosols. The vials containing the homogenized mixtures were put on ice for one min between each 20 second interval. Then, the samples were centrifuged for two min at 10
000 revolutions per min (RPM) using the Bertin Instruments Minilys Personal Homogenizer (Rockville, MD, USA) to encourage compound extraction from bioaerosol cells and fragmentation of the bioaerosols. The vials containing the homogenized mixtures were put on ice for one min between each 20 second interval. Then, the samples were centrifuged for two min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm to separate the supernatant from the pellet that did not dissolve in the solution. The supernatant was filtered through a 0.45 μm pore size syringe filter (hydrophilic polytetrafluoroethylene (PTFE) membrane, 25 mm diameter, Foxx Life Sciences (EZFlow, Salem, NH, USA) and Thermo Fisher Scientific Inc. (Titan 3) (Rockwood, TN, USA)). To prepare the samples for 1H-NMR analysis, the supernatant was evaporated using ultra-high purity nitrogen gas (Pierce Reacti-Vap Evaporating Unit Model 18
000 rpm to separate the supernatant from the pellet that did not dissolve in the solution. The supernatant was filtered through a 0.45 μm pore size syringe filter (hydrophilic polytetrafluoroethylene (PTFE) membrane, 25 mm diameter, Foxx Life Sciences (EZFlow, Salem, NH, USA) and Thermo Fisher Scientific Inc. (Titan 3) (Rockwood, TN, USA)). To prepare the samples for 1H-NMR analysis, the supernatant was evaporated using ultra-high purity nitrogen gas (Pierce Reacti-Vap Evaporating Unit Model 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780, Rockford, IL, USA) and fully dried in a vacuum oven (Precision Scientific Inc, Chicago, IL, USA) overnight at room temperature (20–22 °C). Once the sample was dry, 0.75 mL of DMSO-d6 was added to all vials. DMSO-d6 was chosen as the solvent for our samples due to its common use as a solvent in 1H-NMR spectroscopy and ability to display OH functional groups in the spectra.66 To promote dissolution of the samples in DMSO-d6, each mixture was sonicated for five min at 30 °C. Three analytical replicates of lodgepole pine pollen (2022 season), western gall rust fungus, and spirulina algae samples were prepared separately to ensure sample preparation consistency. Each bioaerosol sample (in DMSO-d6) was run for 256 scans.
780, Rockford, IL, USA) and fully dried in a vacuum oven (Precision Scientific Inc, Chicago, IL, USA) overnight at room temperature (20–22 °C). Once the sample was dry, 0.75 mL of DMSO-d6 was added to all vials. DMSO-d6 was chosen as the solvent for our samples due to its common use as a solvent in 1H-NMR spectroscopy and ability to display OH functional groups in the spectra.66 To promote dissolution of the samples in DMSO-d6, each mixture was sonicated for five min at 30 °C. Three analytical replicates of lodgepole pine pollen (2022 season), western gall rust fungus, and spirulina algae samples were prepared separately to ensure sample preparation consistency. Each bioaerosol sample (in DMSO-d6) was run for 256 scans.
0.75 mL of pure DMSO-d6 and D2O were analyzed to determine the reference peaks of the solvents. Standard and analyte 1H-NMR spectra were analyzed using MestReNova software.67 The chemical shift range for this study was set to 0 to 14 ppm. Each spectrum was phased and referenced at DMSO-d6 (2.50 ppm; for bioaerosol samples) and D2O (4.79 ppm; for several standards) solvent peaks. To determine the quality of the shims, the tetramethylsilane (TMS) peak (internal standard used in 1H-NMR)68 width (at 0.0 ppm) of each spectrum was under 1 Hz at half height of the peak. The baseline was corrected at the 3rd polynomial order. The H2O peak, which was present in all DMSO-d6 samples and standards, was suppressed at 3.32 ppm, using the convolution method and a selectivity parameter of 64. This suppressed area was not integrated. DMSO-d6 is hygroscopic,69 thus water peaks are present when DMSO-d6 is used as a solvent in 1H-NMR, despite desiccation of the samples. For the D2O standard spectra, the solvent (D2O) peak was cut from 4.75 to 4.85 ppm.
Spectra were integrated based on chemical shift ranges found in previous 1H-NMR studies49,50,66,70,71 and by analysis of standard compounds in the present study (Tables 2 and S1†). However, some of the studies' segments are incomplete or broad, thus for the present research, selected standards (Table S1†) were analyzed, and 1H-NMR segments were determined (see Fig. S1†). Chemical shift ranges were set for the following chemical groups: C–H (aliphatic groups, such as –CH, –CH2, –CH3) at 0.5–2.0 ppm,66 α-carbon (such as O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH1,2, CH–COO:, and α to amine H) and amines (NH and NH2) at 2.0–2.7 ppm,66,71 protons in saccharides at 2.7–3.7 ppm, H–C–O (oxygenated aliphatics, alcohol CH and OH, and saccharide H) at 3.7–4.1 ppm,49 –OH (carbonyls and –OH found in saccharide, glycerol, alcohol, and TAG molecules) at 4.1–5.7 ppm,50 OH in saccharides at 6.0–6.6 ppm, Ar–H (aromatics) and amides (CONH and CONH2) at 6.6–8.3 ppm,66,70,71 and Ar-OH, –CHO, –COOH (oxygenated aromatics, aldehydes, and carboxylic acids) at 9.0–14.0 ppm (ref. 70) (see Table 2). The oxygenated aromatic and carboxylic acid segment (segment 8) was not used for analysis in this study, as bioaerosol spectra had weak signals in this region. A total of seven segments were assigned and used, where the majority of bioaerosol signals reside. After subtraction of the solvent peaks, absolute integration values were summed from 0.5–8.3 ppm, with the first segment (aliphatic at 0.5–2.0 ppm) normalized to one (1). Each chemical shift range was divided by the total and recalculated as a percentage for percent distribution of functional groups (see the Results section). An NMR Solvent Data Chart72 was used to determine locations of solvent peaks (see Table S3†).
CH1,2, CH–COO:, and α to amine H) and amines (NH and NH2) at 2.0–2.7 ppm,66,71 protons in saccharides at 2.7–3.7 ppm, H–C–O (oxygenated aliphatics, alcohol CH and OH, and saccharide H) at 3.7–4.1 ppm,49 –OH (carbonyls and –OH found in saccharide, glycerol, alcohol, and TAG molecules) at 4.1–5.7 ppm,50 OH in saccharides at 6.0–6.6 ppm, Ar–H (aromatics) and amides (CONH and CONH2) at 6.6–8.3 ppm,66,70,71 and Ar-OH, –CHO, –COOH (oxygenated aromatics, aldehydes, and carboxylic acids) at 9.0–14.0 ppm (ref. 70) (see Table 2). The oxygenated aromatic and carboxylic acid segment (segment 8) was not used for analysis in this study, as bioaerosol spectra had weak signals in this region. A total of seven segments were assigned and used, where the majority of bioaerosol signals reside. After subtraction of the solvent peaks, absolute integration values were summed from 0.5–8.3 ppm, with the first segment (aliphatic at 0.5–2.0 ppm) normalized to one (1). Each chemical shift range was divided by the total and recalculated as a percentage for percent distribution of functional groups (see the Results section). An NMR Solvent Data Chart72 was used to determine locations of solvent peaks (see Table S3†).
2.4. GC-MS analysis
Selected bioaerosols, excluding Pedobacter bacteria (due to lack of specimen), rabbit brush, and 2020 season lodgepole pine pollen (saccharide content determined by Axelrod et al. (2021)53) (see Table 1), were prepared and extracted following the same blueprint as those for 1H-NMR (lysing, centrifuge, and syringe filtering). For GC-MS saccharide analysis, the additional step of chemical derivatization via silylation73 after filtration was conducted. Samples were prepared at a concentration of 3 mg mL−1, lysed, centrifuged, filtered with 0.45 μm pore size syringe filters, and placed into deactivated vials (Waters Co., Milford, MA, USA). Lodgepole pine pollen (2022) was prepared seperately in three replicates (3 mg mL−1) for preparation error and statistical purposes. For derivatization, 20 μL of known concentration internal standard (glucose-d7) was added to 50 μL of filtered bioaerosol sample solution (total 70 μL volume prior to evaporation), then evaporated to dryness with nitrogen gas. Then, 50 μL of ACN was added to the evaporated samples following sonication (no heat) for 10 min. 50 μL of 99.8% pyridine and 150 μL BSTFA (with 1% TMCS) were added, and the samples were heated for 2 hours at 65 °C. After heating, 50 μL of toluene was added to the samples. The Varian CP-3800 gas chromatograph coupled with the Varian 4000 Ion Trap Mass Spectrometer were used for GC-MS analysis. A 30 meter DB-5MS 5% phenylmethylsilicone fused silica capillary column was used for the chromatographic separation of analytes (Agilent Technologies, Santa Clara, CA, USA), with a temperature ramp from 50 to 320 °C. This saccharide analysis used five calibration points, with a range of 5–100 ng μL−1, and an R2 range of 0.969–0.998. Detailed GC-MS methodology for saccharide analysis can be found in Axelrod et al. (2021).532.5. UV-Vis-NIR starch analysis
The starch UV-Vis-NIR quantitative method was described previously in Bahdanovich et al. (2022).56 Briefly, bioaerosols were heated for 24 hours at 105 °C. 0.1 mL of 95% ethanol and 1 mL NaOH (1 N) solution were added to 10 mg of bioaerosol (dry weight). The mixture was kept at 4 °C for 24 hours. Then, the volume of the mixture was adjusted to 10 mL with ultra-high purity water for a concentration of 1000 μg mL−1 and kept at 4 °C for 16–18 hours. The acidity of the solution was adjusted to pH 6 with 1 M HCl. An iodine reagent (0.2% concentration) was prepared to detect starch in bioaerosol samples by combining 20 mg iodine and 200 mg potassium iodide, then adjusting to 10 mL with ultra-high purity water. 0.1 mL of this reagent solution was added to each bioaerosol sample. The PerkinElmer Lambda 1050 UV/Vis/NIR Spectrophotometer (Waltham, MA, USA) was used for starch analysis, with 3.5 mL UV quartz cuvettes (FireflySci, Inc., Northport, NY, USA). This method used six calibration levels (1–100 ng μL−1), and the R2 of the calibration was 0.999. The starch method detection limit (MDL) was 0.22 ng μL−1.2.6. UPLC-MS analysis
Bioaerosol samples were prepared at 3 mg mL−1 (for both amino acid and fatty acid analysis), using ultra-high purity water. Sample preparation followed the same blueprint as for 1H-NMR (Section 2.3). After lysing, centrifuging, and filtration with 0.45 μm pore size syringe filters, the samples were analyzed for amino acids on a Waters Acquity UPLC tandem the Waters MicroMass Quattro Micro API MS system (Waters Co., Milford, MA, USA). The column installed in the UPLC was the Waters Acquity UPLC BEH Amide 1.7 μm 2.1 × 150 mm (Waters, Co.). Column temperature was set at 35 °C, with an injection volume of 5 μL, and a flow of 0.4 mL min−1. The samples were run at room temperature. This method used positive ionization, a desolvation temperature of 300 °C, and a desolvation flow of 350 L h−1. Eluents used for this purpose were water and ACN, with additives ammonium formate and formic acid. The capillary voltage was set to 3.00 kV, and the cone voltage varied from 20 to 60 V. The total run time was 18 min, and the quantification mode was single ion recording (SIR). Six calibration levels in the range of 0.5–50 ng μL−1 were used, resulting in an R2 value range of 0.972–0.999. MDL values of amino acids range from 0.009 to 0.26 ng μL−1. Detailed UPLC-MS methodology for amino acid analysis can be found in Axelrod et al. (2021).53Existing literature shows that pollen74,75 and algae76,77 species share common fatty acids: myristic (C14:0), palmitic (C16:0), stearic (C18:0), oleic (C18:1), linoleic (C18:2), and linolenic (C18:3) acids. There is a lack of information on fatty acids present in the fungus selected for this study (western gall rust). These seven fatty acids were chosen as standards for this analysis, with nonadecanoic acid (C19:0) as the internal standard. Saponification, or alkaline hydrolysis, is a well described and efficient method for the extraction of fatty acids from lipids in biological samples.78–80 Bioaerosols were prepared as described above (Section 2.3), and after drying with nitrogen gas, were saponified with ethanolic NaOH (0.1 M NaOH in ethanol). 1 mL of the ethanolic NaOH was added to vials containing the dried bioaerosols and sonicated at 60 °C for 1 hour. Afterward, the samples were dried with nitrogen gas and resuspended in equal parts ACN and water (850 μL total), then neutralized from pH 12 to pH 7 with 12 μL of 1 M HCl.
A previously optimized method for UPLC-MS quantification of fatty acids was adapted, but modified, for our analysis.76 Fatty acids of bioaerosols were analyzed using the Waters Aquity Class I UPLC (Waters, Co.) tandem the Waters Xevo TQ-S MS (Waters, Co.). A reverse phase Waters BEH-C18 2.1 mm × 50 mm column (Waters, Co.) was used for this purpose, set to 40 °C. The MS method used negative ionization with a desolvation temperature of 500 °C and a desolvation flow of 700 L h−1. The cone voltage was set at 35 V, and the capillary voltage at 2.00 kV. The LC used an injection volume of 10 μL, a flow of 0.3 mL min−1 and the sample temperature was set to 20 °C. The inlet method used ACN (A) and water (B) as eluents, with the elution method as fellows: 30% A 70% B initially, 75% A 25% B over 10 min, then 100% A 0% B over 12 min, and 30% A 70% B over eight min. The total run time was 30 min, and data was quantified using SIR quantification mode. Six calibration points (0.05–5 ng μL−1) were used and resulted in an R2 range of 0.965–0.997. The MDL range for fatty acids is 0.003–0.10 ng μL−1. The uncertainty of sample preparation was determined by using three replicates of bioaerosol samples prepared separately, from which standard deviations were calculated. The water-soluble fraction of bioaerosol extracts ranged from 9.8% to 22.3%.
3 Results and discussion
3.1. 1H-NMR analysis
Fig. 1 shows the percent distribution of bioaerosol functional groups' protons detected with 1H-NMR for Bacillus and Pedobacter bacteria species, western gall rust fungus, spirulina algae, 2020 (July), 2021 (June), and 2022 (May) lodgepole pine pollen, and rabbitbrush pollen. Overall, results from bioaerosol 1H-NMR analysis show aliphatic protons (segment 1), protons in saccharides (segment 3), and various OH (segment 5) as the major segments (aliphatic protons – up to 73.8% in spirulina, saccharide protons – up to 36.1% in 2022 lodgepole pine pollen, various –OH – up to 27.2% in rabbitbrush pollen). The percent distribution varies for each type of bioaerosol, as well as between the different bacteria and pollen species.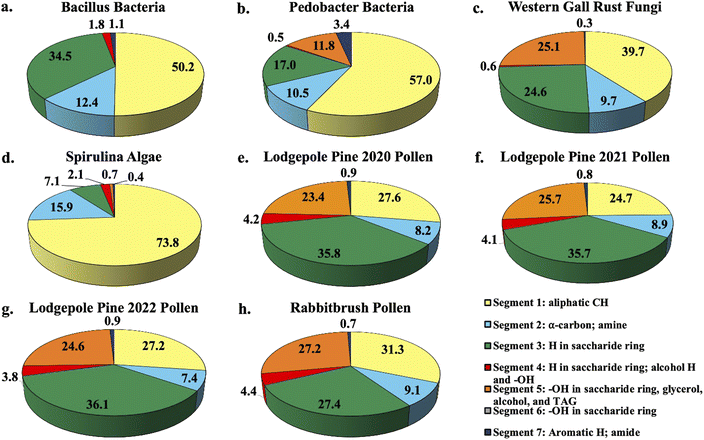 | ||
| Fig. 1 Pie chart percent distribution of functional groups in select bioaerosol extracts (water extracts resuspended in DMSO-d6), analyzed with 1H-NMR spectroscopy. Each pie slice corresponds to chemical shift segments 1 through 7 (see legend). Details on each segment can be found in Table 2. | ||
1H-NMR analysis of bacteria functional groups (Fig. 1a and b) show the largest contribution (50.2% in Bacillus, 57.0% in Pedobacter) from the aliphatic proton (segment 1; Fig. 1a and b). This may be due to the presence of fatty acids, which have long aliphatic chains in their molecules.66 The bacteria species also show a proton signal (12.4% in Bacillus, 10.5% in Pedobacter) that represents α-carbon (segment 2) of fatty acids or amino acids. To assess the presence of saccharides in our bioaerosol aqueous extracts, we selected segment 3 since it is mainly responsible for protons in saccharide molecules (see Table 2). Bacillus (34.5%) has nearly twice as much of the saccharide proton signal (segment 3) as Pedobacter (17.0%). The largest difference in total proton signal among the two bacteria samples was observed for segment 5, which is responsible for OH (in saccharides, glycerol, alcohol, and triglycerides) and CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2 functional groups (see Table 2). This indicates that the chemical contribution of organic compounds with these functional groups varies between the analyzed bacteria species. The signals of aromatic and amide protons (segment 7) were found to be low in bacteria (1.1% in Bacillus, 3.4% in Pedobacter) relative to other assigned segments. To our knowledge, the functional groups of Bacillus and Pedobacter species in aqueous extracts have not been studied in detail, except for some of their products. However, several studies81,82 used solid-state 13C and 15N CPMAS NMR and reported specific signals for amine, aliphatic, α-carbon, and saccharide functional groups in S. aureus and E. coli bacteria, which were also observed in our study. Further, one study which evaluated biosurfactant production from Bacillus subtilis found aliphatic, α-carbon, and amide signals in their 1H-NMR analysis of the biosurfactant,83 while another study by Mohapatra et al. (2017)84 determined the polyhydroxybutyrate produced by the same bacterium contained aliphatic, carbonyl, and HC
CH2 functional groups (see Table 2). This indicates that the chemical contribution of organic compounds with these functional groups varies between the analyzed bacteria species. The signals of aromatic and amide protons (segment 7) were found to be low in bacteria (1.1% in Bacillus, 3.4% in Pedobacter) relative to other assigned segments. To our knowledge, the functional groups of Bacillus and Pedobacter species in aqueous extracts have not been studied in detail, except for some of their products. However, several studies81,82 used solid-state 13C and 15N CPMAS NMR and reported specific signals for amine, aliphatic, α-carbon, and saccharide functional groups in S. aureus and E. coli bacteria, which were also observed in our study. Further, one study which evaluated biosurfactant production from Bacillus subtilis found aliphatic, α-carbon, and amide signals in their 1H-NMR analysis of the biosurfactant,83 while another study by Mohapatra et al. (2017)84 determined the polyhydroxybutyrate produced by the same bacterium contained aliphatic, carbonyl, and HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH signals when analyzed by 13C NMR. 1H-NMR analysis of a biopolymer of Pedobacter sp. in DMSO-d6 by Beltrani et al. (2015)85 showed signals belonging to aliphatic protons and sugars, which were also identified in our analysis.
CH signals when analyzed by 13C NMR. 1H-NMR analysis of a biopolymer of Pedobacter sp. in DMSO-d6 by Beltrani et al. (2015)85 showed signals belonging to aliphatic protons and sugars, which were also identified in our analysis.
The western gall rust fungus has a high contribution from aliphatic protons (39.7%) and an α-carbon proton contribution of 9.7% (Fig. 1c). Approximately a quarter of protons signals were from saccharides (24.6%), while 25.1% of the total signal of the fungus extract were from OH and CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2 functional groups. Western gall rust is unique in its proton percent distribution and does not follow the pattern of any of the other bioaerosols (see Fig. 1). To our knowledge, western gall rust has not been studied for its chemical composition.
CH2 functional groups. Western gall rust is unique in its proton percent distribution and does not follow the pattern of any of the other bioaerosols (see Fig. 1). To our knowledge, western gall rust has not been studied for its chemical composition.
Distribution of proton chemical shifts in spirulina shows the largest contribution from aliphatic protons (73.8%), followed by α-carbon protons (15.9%), and saccharide proton signals (7.1%) (Fig. 1d). OH and CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2 protons contributed only 0.7%, while aromatic and amide proton signals were only 0.4%. Functional groups and chemical composition of spirulina have been previously studied from biofuel and nutrition perspectives, though different methods were used.58,86–88 For example, Sarpal et al. (2016)88 conducted a study on biodiesel potential of various types of algae, including spirulina. Although their study was focused on fatty acid and lipid extraction, the spirulina 1H-NMR spectrum shows strong aliphatic signals, as well as α-carbon, saccharide protons, and OH and CH
CH2 protons contributed only 0.7%, while aromatic and amide proton signals were only 0.4%. Functional groups and chemical composition of spirulina have been previously studied from biofuel and nutrition perspectives, though different methods were used.58,86–88 For example, Sarpal et al. (2016)88 conducted a study on biodiesel potential of various types of algae, including spirulina. Although their study was focused on fatty acid and lipid extraction, the spirulina 1H-NMR spectrum shows strong aliphatic signals, as well as α-carbon, saccharide protons, and OH and CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2. Additionally, a study by Rajasekar et al. (2019),86 which examined the composition of sulfated polysaccharide isolated from spirulina, showed similar peaks to the Sarpal et al. (2016) study. Kumar et al. (2014)58 developed a method that produces a stable extract of spirulina algae for oil extraction using solid-state 13C CPMAS NMR, which reveals signals for carbohydrates, carboxyl, and amide carbons. Our results are comparable to the existing studies, and although completely different extraction methods and techniques were used, similar functional groups are present overall (aliphatic, α-carbon, saccharides/carbohydrates, OH and CH
CH2. Additionally, a study by Rajasekar et al. (2019),86 which examined the composition of sulfated polysaccharide isolated from spirulina, showed similar peaks to the Sarpal et al. (2016) study. Kumar et al. (2014)58 developed a method that produces a stable extract of spirulina algae for oil extraction using solid-state 13C CPMAS NMR, which reveals signals for carbohydrates, carboxyl, and amide carbons. Our results are comparable to the existing studies, and although completely different extraction methods and techniques were used, similar functional groups are present overall (aliphatic, α-carbon, saccharides/carbohydrates, OH and CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2, and amides).
CH2, and amides).
1H-NMR analysis of three lodgepole pine pollen extracts (2020, 2021, 2022 – Fig. 1e, f and g) reveals the presence of strong aliphatic proton signals (24.7–27.6%) as well as α-carbon protons (7.4–8.9%). Unlike the bacteria, fungus, and algae analysis, for lodgepole pine pollen, saccharide protons have the largest contribution (35.7–36.1%). The third largest segment is attributed to the –OH and CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2 functional groups (23.4–25.7%). Proton distribution varies slightly (between 0.02 and 2.9%) between the 2020, 2021, and 2022 lodgepole pine pollen samples, collected in the same area. These small differences are also reflected in the quantitative analysis of individual saccharides (discussed below in Section 3.2). Comparable to the other bioaerosols (bacteria, fungus, and algae), the amide proton signal in lodgepole pine pollen is weak, at 0.8–0.9%.
CH2 functional groups (23.4–25.7%). Proton distribution varies slightly (between 0.02 and 2.9%) between the 2020, 2021, and 2022 lodgepole pine pollen samples, collected in the same area. These small differences are also reflected in the quantitative analysis of individual saccharides (discussed below in Section 3.2). Comparable to the other bioaerosols (bacteria, fungus, and algae), the amide proton signal in lodgepole pine pollen is weak, at 0.8–0.9%.
In the case of rabbitbrush pollen (Fig. 1h), we see a similar pattern of the distribution of the proton chemical shift segments as in lodgepole pine pollen, however, there is some variation. For example, the aliphatic proton signal in rabbitbrush is 31.3%, while the α-carbon signal is 0.9% larger than in lodgepole pine pollen. Protons from saccharides contribute to 27.4%, which is 8.5% less than lodgepole pine. Like lodgepole pine pollen, rabbitbrush contains a weak amide proton signal (0.7%), but a high signal from –OH and CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2 functional groups (27.2%). Several studies on 1H-NMR analysis of aqueous pollen extracts (resuspended in deuterated solvents) found fatty and amino acid proton signals in the aliphatic region, and sucrose (and other saccharides) in the saccharide proton regions of the spectra.89,90 The results from these studies are in good agreement with our findings, as we have high aliphatic and saccharide proton signals in our pollen samples. For characterization of the specific compounds that may be responsible for the protons analyzed with 1H-NMR (i.e., aliphatic, α-carbon, H and OH in saccharides), we conducted analysis of individual compounds: starch, saccharides, amino acids, and fatty acids.
CH2 functional groups (27.2%). Several studies on 1H-NMR analysis of aqueous pollen extracts (resuspended in deuterated solvents) found fatty and amino acid proton signals in the aliphatic region, and sucrose (and other saccharides) in the saccharide proton regions of the spectra.89,90 The results from these studies are in good agreement with our findings, as we have high aliphatic and saccharide proton signals in our pollen samples. For characterization of the specific compounds that may be responsible for the protons analyzed with 1H-NMR (i.e., aliphatic, α-carbon, H and OH in saccharides), we conducted analysis of individual compounds: starch, saccharides, amino acids, and fatty acids.
Based on our segment assignment (see Table 1), we used segment 5 as a representative segment for polar functional groups (OH groups found in glycerol, alcohol, and triglyceride molecules). Analysis of the polar segment among all analyzed species, shows that the most polar bioaerosols are both pollen species and western gall rust (lodgepole pine 2020–23.4%; lodgepole pine 2021–25.7%; lodgepole pine 2022–24.6%; rabbitbrush – 27.2%; western gall rust – 25.1%) in comparison with spirulina and Pedobacter, which are only about 0% and 11.8% polar, respectively. These polar functional groups are not detected in the Bacillus spectra, most likely due to a limited amount extracted from Bacillus. These differences in polarity may significantly affect the solubility of these bioaerosols in water in the atmosphere. For instance, we know from existing literature that pollen grains rupture in the presence of water.61
3.2. Saccharide analysis
The distribution of saccharides in the tested bioaerosols (analyzed with GC-MS) is presented in Fig. 2, in μg mg−1 of dry bioaerosol mass. The two analyzed pollen species (lodgepole pine and rabbitbrush) were found to have the highest total saccharide concentrations (95.3–183.5 μg mg−1 in lodgepole pine samples; 87.2 μg mg−1 in rabbitbrush). The secondary y-axis represents the percentage of total analyzed saccharides per milligram of dry bioaerosol mass. The most common saccharide in all analyzed bioaerosols was glucose (from 5.8% of the total analyzed saccharides in Bacillus bacteria to 49.8% in rabbitbrush pollen). Sucrose was also present in all samples (from 3.6% in spirulina algae to 87.2% in 2022 lodgepole pine pollen), except for rabbitbrush pollen.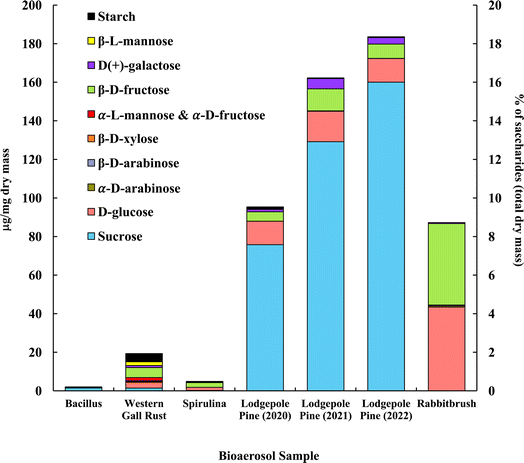 | ||
| Fig. 2 Saccharide and starch concentration (in μg per mg dry mass) of selected bioaerosols. The secondary axis shows the percentage of saccharides per milligram of dry mass. Standard deviations range from 0.004 μg mg−1 in galactose (rabbitbrush) to 12.92 μg mg−1 in sucrose (lodgepole pine 2021) (see Table S4† for details). α-L-Mannose and α-D-fructose are presented together due to their co-elution during gas chromatography separation. | ||
Analysis of saccharides in Bacillus (Fig. 2) reveals only sucrose (71.9%) and glucose (5.8%), which is most likely due to low concentrations of analyzed species in the water extract. Western gall rust shows a mix of saccharides, with the most abundant ones being β-D-fructose (26.9%) and glucose (15.9%). Other saccharides in western gall rust were found in low concentrations: α- and β-D-arabinose (1.2% and 0.9%, respectively), β-D-xylose (1.5%), α-L-mannose/α-D-fructose (8.7%), D-galactose (5.4%), β-L-mannose (10.0%), and sucrose (7.3%). Our results show that β-D-fructose (50.0%) is the most dominant saccharide in spirulina, followed by glucose (33.3%) and sucrose (3.6%). Our results are in good agreement with Brown et al. (1991) study,91 where glucose was also one of the major saccharides found in 16 various microalgae species, though spirulina was not analyzed. Another study that did analyze spirulina for saccharide content, also determined that glucose contributed to ∼54% of total saccharides, followed by rhamnose at ∼22%.92 Meanwhile, Chaiklahan et al. (2013)93 study showed rhamnose as the major contributor (∼54%) to overall saccharide concentrations in spirulina, followed by glucose (∼14%). Fructose was not analyzed in either study.
Lodgepole pine pollen shows particularly high concentrations of sucrose (79.4 to 87.2%), followed by glucose (6.7 to 12.8%) and β-D-fructose (4.0 to 7.0%). Low levels were observed for galactose (1.4 to 3.3%), β-L-mannose (0.2 to 0.3%), and β-D-arabinose (0.04 to 0.1%). Lodgepole pine pollen saccharide concentrations vary from May (2022) to June (2021) and July (2020); with May (2022) having the highest overall saccharide concentration (183.54 μg mg−1 total analyzed saccharides), as well as the highest sucrose value (87.2% of the total analyzed saccharides). Overall, July 2020 lodgepole pine has 6.7% less saccharides than in June 2021, and June 2021 lodgepole pine pollen has 2.1% less than that of May 2022. The variation of saccharide content in lodgepole pine pollen may be explained by the collection of the pollen at different times of the season, in different years. Previous studies showed that saccharide concentrations vary seasonally in pollen, and the highest saccharide concentration is typically found earlier in the pollen season.94 Furthermore, Fu et al. (2012)94 also found that sucrose had the highest contribution earlier in the pollen season. This justifies the differences of lodgepole pine pollen samples (July 2020, June 2021, May 2022) in our results, as the May 2022 sample was collected earlier in the season and contains more saccharides overall and has a higher sucrose concentration than the other samples.
The dominant saccharides in rabbitbrush pollen were glucose (49.8%) and β-D-fructose (48.4%), followed by galactose, α-D-arabinose, and β-D-arabinose (<1.0%). α-D-xylose, α-lactose, and trehalose were also analyzed in the selected bioaerosol samples; however, their concentrations were below the detection limit. Common saccharides found in various pollen species in Axelrod et al. (2021)53 study were β-D-fructose and glucose, which were also found in the pollen samples selected for this study. Fu et al. (2012)94 reported sucrose as the dominant saccharide among all tested pollen samples, however, in our study, only lodgepole pine pollen had sucrose as the major saccharide. As such, sucrose may not be a suitable tracer for airborne pollen analysis (e.g., source apportionment), as Fu et al. concluded.
Starch content analyzed with UV-Vis-NIR spectrophotometry56 is presented together with saccharides, in Fig. 2, to show a total distribution of mono- (i.e., glucose, etc.), di- (i.e., sucrose), and poly- (i.e., starch) saccharides in the chosen bioaerosols. The results showed that starch contributes to 22.3% of total analyzed saccharides in Bacillus, 22.2% in western gall rust, 13.1% in spirulina, and 1.0% 2020 lodgepole pine.
3.3. Amino acid analysis
Fig. 3 shows the distribution of analyzed amino acids (analyzed with UPLC-MS) in selected bioaerosols (Table 1). The concentration and composition of amino acids among bioaerosols vary significantly. We found that proline, leucine, isoleucine, alanine, and phenylalanine are the only common amino acids between all the tested bioaerosols, among 20 amino acids. Bacillus bacteria (21.4 μg mg−1) and rabbitbrush pollen (18.1 μg mg−1) show the highest concentrations of total analyzed amino acids overall. Bacillus, Pedobacter, and spirulina show the highest values of glutamic acid (64.5%, 14.8%, and 49.1% of total analyzed amino acids, respectively), while both pollen species are rich in proline (lodgepole pine – 38.6%, rabbitbrush – 83.2%).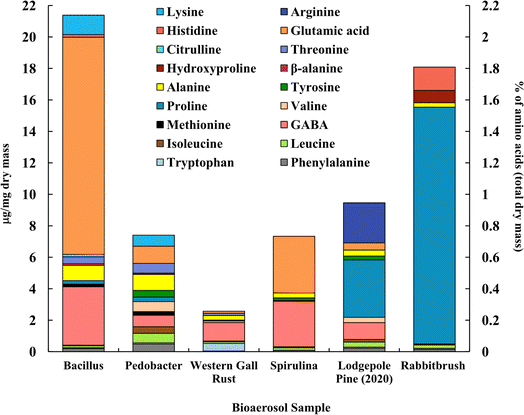 | ||
| Fig. 3 Amino acid concentration (in μg per mg dry mass) of selected bioaerosols (primary axis). The secondary axis shows the percentage of total amino acids per milligram of dry mass. Standard deviations range from ± 0.001 μg mg−1 in β-alanine (Bacillus) to ± 1.770 μg mg−1 in proline (lodgepole pine 2020) (see Table S4† for details). | ||
High concentrations of glutamic acid (64.5%) and γ-aminobutyric acid (GABA, 17.4%) were found in Bacillus bacteria, followed by lysine (5.8%) and alanine (4.5%). Pedobacter bacteria also shows high concentrations of glutamic acid (14.8%) and GABA (9.8%) but has higher values of alanine (13.7%). Overall, Bacillus and Pedobacter bacteria have very distinct distributions of amino acids. GABA (45.8%), tryptophan (19.0%), and alanine (11.1%) are the dominant amino acids in western gall rust, and other amino acids (phenylalanine, tryptophan, leucine, isoleucine, valine, proline, alanine, threonine, and glutamic acid) were found in relatively small amounts (<6%) (see Table S4† for exact concentrations and standard deviations). To our knowledge, these are the first available results regarding western gall rust amino acid composition and distribution. Glutamic acid (49.1%) and GABA (39.4%) alone were found to compose 88.5% of the total analyzed amino acids in spirulina. A study by Vendruscolo et al. (2018)95 found that glutamic acid was the most abundant amino acid in four microalgae species, while Bashir et al. (2016)96 determined that the major amino acids found in spirulina were glutamic acid followed by leucine, which our data shows only a small amount of. Similarly, Andreeva et al. (2021)97 showed that spirulina's main amino acids are glutamic acid, followed by alanine, which is also minimal in our results.
In addition to proline (38.6%), lodgepole pine pollen amino acid analysis shows significant amounts of GABA (11.3%) and arginine (26.9%). Ozler et al. (2009)98 analyzed pollen of a different pine species (P. nigra) and found that the major contributor is also proline and has a less significant contribution of arginine, which agrees with our results. While the amino acid profile of rabbitbrush pollen also shows a high concentration of proline (83.2%), it also contains histidine (8.2%) and hydroxyproline (4.3%). Based on our analysis of amino acids in bioaerosols, glutamic acid may be a suitable atmospheric tracer for Bacillus bacteria and spirulina algae, while proline may be a better tracer for rabbitbrush and lodgepole pine pollens, as these amino acids are found in the highest concentrations in these bioaerosols. Glycine and serine amino acids were also analyzed but remained below the detection limit.
3.4. Fatty acid analysis
Fig. 4 shows the distribution of ten analyzed fatty acids in selected bioaerosols (analyzed with UPLC-MS). All analyzed bioaerosols, except Bacillus (which contained only myristic acid), show a variety of fatty acids. Lodgepole pine pollen (25.82 μg mg−1), western gall rust fungus (10.63 μg mg−1) and rabbitbrush pollen (7.72 μg mg−1) have the highest total concentration of the analyzed fatty acids. Linoleic and palmitic acids were found in the highest concentrations in both pollen species (lodgepole pine and rabbitbrush).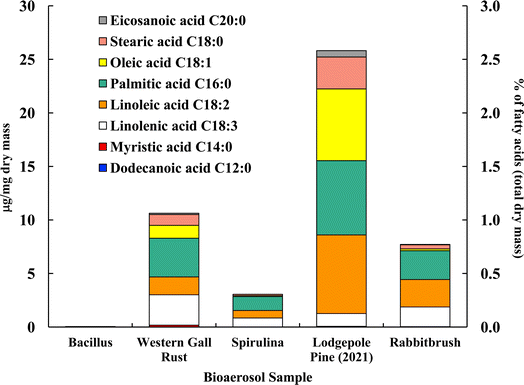 | ||
| Fig. 4 Fatty acid concentration (in μg per mg dry mass) of selected bioaerosols. The secondary axis shows the percentage of fatty acids per milligram of dry mass. Fatty acids in Bacillus were below detection limit. Standard deviations range from 0.006 μg mg−1 in eicosanoic acid (spirulina) to 1.03 μg mg−1 in palmitic acid (lodgepole pine 2021) (see Table S4† for details). | ||
Analysis of Bacillus bacteria revealed that most fatty acids were below detection limit, except for myristic acid (0.05 μg mg−1). The dominant fatty acids in western gall rust are linolenic (26.6% of total analyzed fatty acids) and palmitic (34.0%). Linoleic (15.7%), oleic (11.3%), and stearic (9.5%) acids are also present in lower concentrations. The dominant fatty acid found in spirulina was palmitic acid (42.6%), followed by linolenic (27.7%), linoleic (22.6%), stearic (4.2%), and oleic (2.9%) acids. Our results are comparable to Samburova et al. (2013)76 study, where the most abundant fatty acids found between four strains of another algae, Dunaliella, were palmitic, linolenic, linoleic, and oleic acids.
Both pollen species (lodgepole pine and rabbitbrush) contain the same variation of fatty acids (except dodecanoic acid) in different concentrations. Major fatty acids found in lodgepole pine pollen are linoleic (28.5%), palmitic (26.9%), and oleic (26.0%) acids, while the main fatty acids in rabbitbrush are palmitic (34.9%), linoleic (33.2%), and linolenic (23.8%) acids. Erdyneeva et al. (2021)99 found that dominant fatty acids in three Pinus species were palmitic and linoleic acids, which were also the major fatty acids present in both of our pollen samples.
According to a review by Kaneda et al. (1977),100 the most common fatty acids among living organisms are palmitic, oleic, linoleic, linolenic, and stearic acids.101 This combination of fatty acids is seen all analyzed bioaerosols, except for Bacillus (where most of the fatty acids were below detection limit). Although spirulina and Bacillus show 73.8 and 50.2% of the aliphatic functional group in their 1H-NMR proton distribution, these bioaerosol species have the lowest concentrations, according to our fatty acid analysis via UPLC-MS. This may be due to the low mass of Bacillus and spirulina that was available for quantitative analysis, thus, most fatty acids were below detection limit for Bacillus samples.
3.5. Correlation analysis
To assess if 1H-NMR analysis of chemically complex bioaerosol extracts can reflect quantitative analysis of organic species, we ran correlations between the aliphatic, saccharide, and amide segments (1H-NMR analysis) and quantitative analysis of the organic compounds (GC-MS, UPLC-MS, and UV-Vis-NIR analysis) (Table S5†). For this purpose, each segment was normalized by the sum of the three related segments (aliphatic, saccharide, and amide) and correlated with the analyzed organic compounds. The best correlation was for saccharides (R2 = 0.608), where the saccharide segment (segment 3) of 1H-NMR analysis was correlated with the total saccharide concentration (Fig. 2). This relationship indicates that 1H-NMR can be successfully used for semi-quantitative analysis of saccharide content in bioaerosols. No significant correlation (R2 = −0.002) was detected between amino acids and the amide segment (segment 7), which could be due to weak 1H-NMR signals of amide groups,70 a limited number of quantified amino acids, and overlap with signals from other functional groups (i.e., aromatic H in segment 7, Table 2). A negative correlation (R2 = −0.442) was found between individual fatty acid analysis and the aliphatic proton region (segment 1). This may be attributed to other compounds (i.e., peptides)102 that contribute to strong aliphatic signals.4 Conclusion
The chemical composition of selected bioaerosols (pollen, fungi, algae, bacteria) was characterized using qualitative (1H-NMR) and quantitative (GC-MS, UPLC-MS, UV-Vis-NIR) analysis. 1H-NMR analysis revealed that protons from aliphatic, saccharide, and OH groups had the biggest contributions to the overall proton distribution among all bioaerosols. Bacillus and Pedobacter bacteria, western gall rust fungus, spirulina algae, and rabbitbrush pollen had the largest contribution from aliphatic protons (31.3–73.8%), while lodgepole pine pollen had the largest contribution from saccharide protons (35.7–36.1%). Amide signals were some of the weakest, ranging from 0.3% in western gall rust fungus to 3.4% in Pedobacter bacteria.The chemical composition varies significantly between bioaerosol types and species. The quantitative analysis of selected organics (saccharides, amino acids, and fatty acids) showed that the saccharide glucose, the amino acids proline, leucine, isoleucine, alanine, and phenylalanine, and the fatty acids palmitic, oleic, linoleic, linolenic, and stearic (except in Bacillus bacteria) are common among analyzed bioaerosol aqueous extracts. The major organic species were found to be saccharides (up to 18.4% of total dry mass), followed by amino acids (up to 2.1% of total dry mass), and fatty acids (up to 2.6% total dry mass). The high saccharide concentrations may due to the high water solubility of most monosaccharides.66 Both pollen species (lodgepole pine and rabbitbrush) were found to have the highest concentrations of saccharides (Fig. 2), which can be explained by the tendency of pollen grains to easily rupture in the presence of water.61 Differences in saccharide concentrations of July 2020, June 2021, and May 2022 lodgepole pine samples may be explained by seasonal variations in pollen saccharides,94 where the highest sugar concentrations are found earlier in the season (May 2022). This is the first study, to our knowledge, that provided a chemical analysis of western gall rust fungus, which is known to affect pine trees around the world.37,40,103 The correlation analysis between qualitative and quantitative results showed a good correlation (R2 = 0.608) between the saccharide 1H-NMR segment and the total saccharide concentration, which suggests that the 1H-NMR qualitative data reflects the quantitative analysis of saccharides. For Bacillus and spirulina, the highest contribution of analyzed organic species came from amino acids, while the major fraction of organics for pollen was saccharides. Based on our chemical analysis of these various bioaerosol species, it can be concluded that proper tracer analysis of bioaerosols in ambient samples should use fatty acids in addition to amino acids and saccharides, as was suggested by Axelrod et al. (2021).53
Several constraints of the study include the limited number of organic species (saccharides, amino acids, fatty acids, etc.) and bioaerosol species analyzed. Future bioaerosol studies may benefit from extracting bioaerosols in solvents of different polarity (i.e., acetone, dichloromethane, hexane) and applying other analytical methods for further functional group characterization. For example, Fourier-Transform Infrared Spectroscopy (FTIR) can provide information regarding the polarity of the bioaerosols and higher-resolution NMR (>500 MHz 1H-NMR) can yield a more detailed analysis of the protons present. An extensive chemical analysis should be conducted and the contribution of other chemical species to the bioaerosols can be explored, such as the contribution of phosphorous-containing compounds in bioaerosols.104 Further, pollen samples may be collected and analyzed at different times over the course of one season to observe a more defined seasonal variation in chemical composition. Subsequent studies should also consider analysis of ambient samples for identification of bioaerosols, using the chemical compositions provided in this study as tracers.
The results of the present study provide valuable insight of bioaerosol contribution to atmospheric chemistry and essential data for future bioaerosol research. It has been highlighted in a recent review101 that the role bioaerosols play in atmospheric processes have been greatly overlooked and largely understudied mainly due to the complexity of bioaerosols and diversity of their sources. Therefore, as Kumari and Yadav (2024)101 pointed out, there are large uncertainties in assessing bioaerosol regional and global climate effects, especially in source apportionment and climate models. As such, our detailed chemical analysis of bioaerosols can help to understand the impact of bioaerosols on atmospheric organic carbon, as well as their role in cloud formation processes such as CCN and INP. Moreover, the chemical constituents can be utilized for better identification and classification of bioaerosols in atmospheric aerosol samples, as suggested in Samburova et al. (2013).6 Our fundamental data may be relevant in future research regarding increases in concentrations of pollen and algae due to climate change.30,105–107 For a broader impact, these results can be used for studies in other fields, such as medicine, nutrition, biofuels, toxicology, agriculture, among others.42,44,58,108 For example, all the analyzed bioaerosols are known to have significant effects on human and animal health, as mentioned previously, and understanding their chemical composition in a medical context is significant and necessary. Likewise, the organic and functional group composition of spirulina is relevant from a nutrition and biofuel perspective.46,88,109
Data availability
The data used in this article is available in the ESI.†Author contributions
Palina Bahdanovich – conceptualization, methodology, formal analysis, data curation, writing – original draft, writing – review and editing, visualization. Kevin Axelrod – validation, writing – review and editing. Andrey Y. Khlystov – validation, writing – review and editing, funding acquisition. Vera Samburova – conceptualization, methodology, validation, writing – review and editing, supervision, funding acquisition.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This study was funded by the National Science Foundation, grant AGS-1760328, the Nevada DRIVE Scholarship (University of Nevada, Reno, USA), and the DRI Graduate Student Fellowship (Desert Research Institute, Reno, NV, USA). The authors would like to thank Dr Alison Murray (Desert Research Institute, Reno, NV, USA) for the donation of cultured bacteria species for our bioaerosol study. The authors would also like to thank Dr Yeongkwon Son (Desert Research Institute, NV, USA) for their analytical expertise in UPLC-MS method development and Dr Alexander Eletsky (University of Georgia, GA, USA) for their 1H-NMR expertise.References
- J. Fröhlich-Nowoisky, C. J. Kampf, B. Weber, J. A. Huffman, C. Pöhlker, M. O. Andreae, N. Lang-Yona, S. M. Burrows, S. S. Gunthe, W. Elbert, H. Su, P. Hoor, E. Thines, T. Hoffmann, V. R. Després and U. Pöschl, Atmos. Res., 2016, 182, 346–376 CrossRef.
- R. M. Bowers, A. P. Sullivan, E. K. Costello, J. L. Collett, R. Knight and N. Fierer, Appl. Environ. Microbiol., 2011, 77, 6350–6356 CrossRef CAS PubMed.
- G. Mainelis, Aerosol Sci. Technol., 2020, 54, 496–519 CrossRef CAS PubMed.
- K. H. Kim, E. Kabir and S. A. Jahan, J. Environ. Sci., 2018, 67, 23–35 CrossRef CAS PubMed.
- M. Lazaridis, Atmosphere, 2019, 10 DOI:10.3390/atmos10120786.
- V. Samburova, A. Gannet Hallar, L. R. Mazzoleni, P. Saranjampour, D. Lowenthal, S. D. Kohl and B. Zielinska, Environ. Chem., 2013, 10, 370–380 CrossRef CAS.
- J. A. Huffman, A. J. Prenni, P. J. Demott, C. Pöhlker, R. H. Mason, N. H. Robinson, J. Fröhlich-Nowoisky, Y. Tobo, V. R. Després, E. Garcia, D. J. Gochis, E. Harris, I. Müller-Germann, C. Ruzene, B. Schmer, B. Sinha, D. A. Day, M. O. Andreae, J. L. Jimenez, M. Gallagher, S. M. Kreidenweis, A. K. Bertram and U. Pöschl, Atmos. Chem. Phys., 2013, 13, 6151–6164 CrossRef.
- C. J. Schumacher, C. Pöhlker, P. Aalto, V. Hiltunen, T. Petäjä, M. Kulmala, U. Pöschl and J. A. Huffman, Atmos. Chem. Phys., 2013, 13, 11987–12001 CrossRef.
- V. R. Després, J. A. Huffman, S. M. Burrows, C. Hoose, A. S. Safatov, G. Buryak, J. Fröhlich-Nowoisky, W. Elbert, M. O. Andreae, U. Pöschl and R. Jaenicke, Tellus B, 2012, 64 DOI:10.3402/tellusb.v64i0.15598.
- R. Jaenicke, Science, 2005, 308, 73 CrossRef CAS PubMed.
- P. Hyde and A. Mahalov, J. Air Waste Manage. Assoc., 2020, 70, 71–77 CrossRef CAS PubMed.
- J. M. Schiffer, L. E. Mael, K. A. Prather, R. E. Amaro and V. H. Grassian, ACS Cent. Sci., 2018, 4, 1617–1623 CrossRef CAS PubMed.
- J. Burkart, J. Gratzl, T. M. Seifried, P. Bieber and H. Grothe, Biogeosciences, 2021, 18, 5751–5765 CrossRef.
- F. D. Pope, Environ. Res. Lett., 2010, 5 DOI:10.1088/1748-9326/5/4/044015.
- O. Möhler, P. J. Demott, G. Vali and Z. Levin, Biogeosciences, 2007, 4, 1059–1071 CrossRef.
- D. I. Haga, S. M. Burrows, R. Iannone, M. J. Wheeler, R. H. Mason, J. Chen, E. A. Polishchuk, U. Pöschl and A. K. Bertram, Atmos. Chem. Phys., 2014, 14, 8611–8630 CrossRef.
- J. Sun and P. A. Ariya, Atmos. Environ., 2006, 40, 795–820 CrossRef CAS.
- H. Bauer, H. Giebl, R. Hitzenberger, A. Kasper-Giebl, G. Reischl, F. Zibuschka and H. Puxbaum, J. Geophys. Res.: Atmos., 2003, 108 DOI:10.1029/2003jd003545.
- G. C. Cornwell, C. S. McCluskey, T. C. J Hill, E. T. Levin, N. E. Rothfuss, S.-L. Tai, M. D. Petters, P. J. DeMott, S. Kreidenweis, K. A. Prather and S. M. Burrows, Sci. Adv., 2023, 9 DOI:10.1126/sciadv.adg3715.
- P. DasSarma and S. DasSarma, Curr. Opin. Microbiol., 2018, 43, 24–30 CrossRef CAS PubMed.
- G. Gartner, M. Stoyneva-Gartner and B. Uzunov, Toxins, 2021, 13 DOI:10.3390/toxins13050322.
- D. J. Smith, D. W. Griffin, R. D. McPeters, P. D. Ward and A. C. Schuerger, Aerobiologia, 2011, 27, 319–332 CrossRef.
- S. D. Warren and L. L. St. Clair, AIMS Environ. Sci., 2021, 8, 498–516 Search PubMed.
- S. V. M. Tesson, C. A. Skjøth, T. Šantl-Temkiv and J. Löndahl, Appl. Environ. Microbiol., 2016, 82, 1978–1991 CrossRef CAS PubMed.
- H. Sénéchal, N. Visez, D. Charpin, Y. Shahali, G. Peltre, J. P. Biolley, F. Lhuissier, R. Couderc, O. Yamada, A. Malrat-Domenge, N. Pham-Thi, P. Poncet and J. P. Sutra, Sci. World J., 2015, 2015 DOI:10.1155/2015/940243.
- M. Kampa and E. Castanas, Environ. Pollut., 2008, 151, 362–367 CrossRef CAS PubMed.
- G. D’Amato, C. E. Baena-Cagnani, L. Cecchi, I. Annesi-Maesano, C. Nunes, I. Ansotegui, M. D’Amato, G. Liccardi, M. Sofia and W. G. Canonica, Multidiscip. Respir. Med., 2013, 8 DOI:10.1186/2049-6958-8-12.
- P. E. Taylor, K. W. Jacobson, J. M. House and M. M. Glovsky, Int. Arch. Allergy Immunol., 2007, 144, 162–170 CrossRef PubMed.
- S. Mayer, V. Curtui, E. Usleber and M. Gareis, Mycotoxin Res., 2007, 23, 94–100 CrossRef CAS PubMed.
- N. W. May, N. E. Olson, M. Panas, J. L. Axson, P. S. Tirella, R. M. Kirpes, R. L. Craig, M. J. Gunsch, S. China, A. Laskin, A. P. Ault and K. A. Pratt, Environ. Sci. Technol., 2018, 52, 397–405 CrossRef CAS PubMed.
- W. R. L. Anderegg, J. T. Abatzoglou, L. D. L. Anderegg, L. Bielory, P. L. Kinney and L. Ziska, Proc. Natl. Acad. Sci. U. S. A., 2021, 118 DOI:10.1073/pnas.2013284118.
- W. W. Carmichael and G. L. Boyer, Harmful Algae, 2016, 54, 194–212 CrossRef PubMed.
- N. Center for Health Statistics, Table A-2. Selected Respiratory Diseases Among Adults Aged 18 and over, by Selected Characteristics, United States, 2018, 2018 Search PubMed.
- B. V. Chileen, K. K. McLauchlan, P. E. Higuera, M. Parish and B. N. Shuman, Holocene, 2020, 30, 1493–1503 CrossRef.
- T. M. Faske, A. C. Agneray, J. P. Jahner, L. M. Sheta, E. A. Leger and T. L. Parchman, Evol. Appl., 2021, 14, 2881–2900 CrossRef PubMed.
- M. Jędryczka, B. Żuraw, P. Zagórski, J. Rodzik, K. Mędrek, I. A. Pidek, W. Haratym, J. Kaczmarek and M. Sadyś, Pol. Polar Res., 2023, 44, 313–338 Search PubMed.
- K. M. Old, W. J. Libby and J. H. Russell, Silvae Genet., 1986, 35, 145–149 Search PubMed.
- J. M. Powell and Y. Hiratsuka, Can. Plant Dis. Surv., 1973, 53, 67–71 Search PubMed.
- J. Hoffman and S. Hagle, Western Gall Rust Management, 2011 Search PubMed.
- T. D. Ramsfield, D. J. Kriticos, D. R. Vogler and B. W. Geils, N. Z. J. For. Sci., 2007, 37, 143–152 Search PubMed.
- K. M. Hughes, D. Price, A. A. J. Torriero, M. R. E. Symonds and C. Suphioglu, Int. J. Mol. Sci., 2022, 23 DOI:10.3390/ijms23084313.
- W. Tan, J. Wang, W. Bai, J. Qi and W. Chen, Sci. Rep., 2020, 10, 6012 CrossRef CAS PubMed.
- J. Bjerketorp, J. J. Levenfors, C. Nord, B. Guss, B. Öberg and A. Broberg, Front. Microbiol., 2021, 12 DOI:10.3389/fmicb.2021.642829.
- A. Hashem, B. Tabassum and E. F. Abd-Allah, Saudi J. Biol. Sci., 2019, 26, 1291–1297 CrossRef CAS PubMed.
- L. Merrill, J. Dunbar, J. Richardson and C. R. Kuske, J. Forensic Sci., 2006, 51, 559–565 CrossRef CAS PubMed.
- A. M. Delort, M. Vaïtilingom, P. Amato, M. Sancelme, M. Parazols, G. Mailhot, P. Laj and L. Deguillaume, Atmos. Res., 2010, 98, 249–260 CrossRef CAS.
- T. S. Vo, D. H. Ngo and S. K. Kim, in Handbook of Marine Microalgae: Biotechnology Advances, Academic Press, 2015, pp. 299–308 Search PubMed.
- N. Sahu and A. D. Tangutur, Aerobiologia, 2015, 31, 89–97 CrossRef.
- R. M. B. O. Duarte and A. C. Duarte, Magn. Reson. Chem., 2015, 53, 658–666 CrossRef CAS PubMed.
- M. C. G. Chalbot, G. Gamboa da Costa and I. G. Kavouras, Appl. Magn. Reson., 2013, 44, 1347–1358 CrossRef CAS.
- Š. Horník, J. Sýkora, J. Schwarz and V. Ždímal, ACS Omega, 2020, 5, 22750–22758 CrossRef PubMed.
- Y. Suzuki, M. Kawakami and K. Akasaka, Environ. Sci. Technol., 2001, 35, 2656–2664 CrossRef CAS PubMed.
- K. Axelrod, V. Samburova and A. Y. Khlystov, Sci. Total Environ., 2021, 799 DOI:10.1016/j.scitotenv.2021.149254.
- M. Kuznetsova, C. Lee and J. Aller, Mar. Chem., 2005, 96, 359–377 CrossRef CAS.
- W. Elbert, P. E. Taylor, M. O. Andreae and U. Pöschl, Atmos. Chem. Phys., 2007, 7, 4569–4588 CrossRef CAS.
- P. Bahdanovich, K. Axelrod, A. Y. Khlystov and V. Samburova, Analytica, 2022, 3, 394–405 CrossRef.
- A. D. Estillore, J. V Trueblood and V. H. Grassian, Chem. Sci., 2016, 7, 6604–6616 RSC.
- R. Kumar, V. Bansal, M. B. Patel, A. S. Sarpal and J. Algal, Biomass Util., 2014, 5, 36–45 Search PubMed.
- E. Pacini, M. Guarnieri and M. Nepi, Protoplasma, 2006, 228, 73–77 CrossRef CAS PubMed.
- D. D. Hughes, C. B. A. Mampage, L. M. Jones, Z. Liu and E. A. Stone, Environ. Sci. Technol. Lett., 2020, 7, 409–414 CrossRef CAS.
- C. B. A. Mampage, D. D. Hughes, L. M. Jones, N. Metwali, P. S. Thorne and E. A. Stone, Atmos. Environ. X, 2022, 15 DOI:10.1016/j.aeaoa.2022.100177.
- E. Gute, R. O. David, Z. A. Kanji and J. P. D. Abbatt, ACS Earth Space Chem., 2020, 4, 2312–2319 CrossRef CAS.
- W. G. Lindsley, B. J. Green, F. M. Blachere, S. B. Martin, B. F. Law, P. A. Jensen and M. P. Schafer, NIOSH Manual of Analytical Methods (NMAM), Sampling and Characterization of Bioaerosols, 5th edn, 2017 Search PubMed.
- S. Boonpo, S. Kungwankunakorn and J. Adv, Agric. Technol., 2017, 4, 345–349 Search PubMed.
- VnmrJ 4.0 (Revision A), Agilent Technologies, Inc., Santa Clara, CA, 2013 Search PubMed.
- M. Jr. Jones and S. A. Fleming, in Organic Chemistry, ed. E. Fahlgren, C. L. Talmadge and R. Cotton, W.W. Norton & Company, Inc., New York, NY 10110, 5th edn, 2014 Search PubMed.
- MestReNova 14.2, Mastrelab Research S.L., Escondido, CA, 2020 Search PubMed.
- G. V. D. Tiers, J. Phys. Chem., 1958, 62, 1151–1152 CrossRef CAS.
- R. Ellson, R. Stearns, M. Mutz, C. Brown, B. Browning, D. Harris, S. Qureshi, J. Shieh and D. Wold, Comb. Chem. High Throughput Screening, 2005, 8, 489–498 CrossRef CAS PubMed.
- E. Pretsch, P. Bühlmann and M. Badertscher, Structure Determination of Organic Compounds: Tables of Spectral Data, Springer, Berlin, Germany, 4th edn, 2009 Search PubMed.
- 1H-NMR Chemical Shifts, https://www.chemistryconnected.com/courses/NMR/index.html, accessed August 2024 Search PubMed.
- NMR Solvent Data Chart, https://isotope.com/literature-nuclear-magnetic-resonance-nmr-solvents, accessed August 2024 Search PubMed.
- C. Schummer, O. Delhomme, B. M. R. Appenzeller, R. Wennig and M. Millet, Talanta, 2009, 77, 1473–1482 CrossRef CAS PubMed.
- F. Muth, P. R. Breslow, P. Masek and A. S. Leonard, Behav. Ecol., 2018, 29, 1371–1379 Search PubMed.
- R. Manning, Bee World, 2001, 82, 60–75 CrossRef.
- V. Samburova, M. S. Lemos, S. Hiibel, S. Kent Hoekman, J. C. Cushman and B. Zielinska, J. Am. Oil Chem. Soc., 2013, 90, 53–64 CrossRef CAS.
- Y. Maltsev and K. Maltseva, Rev. Environ. Sci. Biotechnol., 2021, 20, 515–547 CrossRef CAS.
- J. Salimon, B. M. Abdullah and N. Salih, Chem. Cent. J., 2011, 5 DOI:10.1186/1752-153X-5-67.
- K.-S. Liu, J. Am. Oil Chem. Soc., 1994, 71, 1179–1187 CrossRef CAS.
- A. M. Burja, R. E. Armenta, H. Radianingtyas and C. J. Barrow, J. Agric. Food Chem., 2007, 55, 4795–4801 CrossRef CAS PubMed.
- C. Reichhardt and L. Cegelski, Mol. Phys., 2014, 112, 887–894 CrossRef CAS PubMed.
- J. A. H. Romaniuk and L. Cegelski, Philos. Trans. R. Soc., B, 2015, 370 DOI:10.1098/rstb.2015.0024.
- S. C. Pemmaraju, D. Sharma, N. Singh, R. Panwar, S. S. Cameotra and V. Pruthi, Appl. Biochem. Biotechnol., 2012, 167, 1119–1131 CrossRef CAS PubMed.
- S. Mohapatra, B. Sarkar, D. P. Samantaray, A. Daware, S. Maity, S. Pattnaik and S. Bhattacharjee, Environ. Tech., 2017, 38, 3201–3208 CrossRef CAS PubMed.
- T. Beltrani, S. Chiavarini, D. O. Cicero, M. Grimaldi, C. Ruggeri, E. Tamburini and C. Cremisini, Int. J. Biol. Macromol., 2015, 72, 1090–1096 CrossRef CAS PubMed.
- P. Rajasekar, S. Palanisamy, R. Anjali, M. Vinosha, M. Elakkiya, T. Marudhupandi, M. Tabarsa, S. G. You and N. M. Prabhu, Int. J. Biol. Macromol., 2019, 141, 809–821 CrossRef CAS PubMed.
- M. Kavisri, M. Abraham, G. Prabakaran, M. Elangovan and M. Moovendhan, Biomass Convers. Biorefin., 2021, 13, 10147–10154 CrossRef.
- A. S. Sarpal, B. K. Sharma, J. Scott, R. kumar, V. Sugmaran, A. Chopra, V. Bansal and N. K. Rajagopalan, J. Anal. Bioanal. Tech., 2016, 1, 17–41 Search PubMed.
- M. C. G. Chalbot and I. G. Kavouras, Nat. Prod. Commun., 2019, 14 DOI:10.1177/1934578X19849972.
- A. M. Otify, A. M. El-Sayed, C. G. Michel and M. A. Farag, Metabolomics, 2019, 15 DOI:10.1007/s11306-019-1581-7.
- M. R. Brown, J. Exp. Mar. Biol. Ecol., 1991, 145, 79–99 CrossRef CAS.
- K. Madhavi, S. L. V Venkataraman and P. V Salimath, Phytochem., 1987, 26, 2267–2269 CrossRef.
- R. Chaiklahan, N. Chirasuwan, P. Triratana, V. Loha, S. Tia and B. Bunnag, Int. J. Biol. Macromol., 2013, 58, 73–78 CrossRef CAS PubMed.
- P. Fu, K. Kawamura, M. Kobayashi and B. R. T. Simoneit, Atmos. Environ., 2012, 55, 234–239 CrossRef CAS.
- R. G. Vendruscolo, M. M. X. Facchi, M. M. Maroneze, M. B. Fagundes, A. J. Cichoski, L. Q. Zepka, J. S. Barin, E. Jacob-Lopes and R. Wagner, Food Res. Int., 2018, 109, 204–212 CrossRef CAS PubMed.
- S. Bashir, M. K. Sharif, M. S. Butt and M. Shahid, Pak. J. Sci. Ind. Res., Ser. B, 2016, 59, 12–19 CAS.
- A. Andreeva, E. Budenkova, O. Babich, S. Sukhikh, E. Ulrikh, S. Ivanova, A. Prosekov and V. Dolganyuk, Molecules, 2021, 26, 2767 CrossRef CAS PubMed.
- H. Ozler, S. Pehlivan and F. Bayrak, Asian J. Plant Sci., 2009, 8 DOI:10.3923/ajps.2009.308.312.
- S. A. Erdyneeva, V. G. Shiretorova, Z. A. Tykheev and L. D. Radnaeva, Chem. Nat. Compd., 2021, 57, 741–742 CrossRef CAS.
- T. Kaneda, Bacteriol. Rev., 1977, 41, 391–418 CrossRef CAS PubMed.
- K. Kumari and S. Yadav, Int. J. Environ. Res., 2024, 18 DOI:10.1007/s41742-024-00633-2.
- M. F. Mesleh, W. A. Shirley, C. E. Heise, N. Ling, R. A. Maki and R. P. Laura, J. Biol. Chem., 2007, 282, 6338–6346 CrossRef CAS PubMed.
- K. M. Old, Aust. For., 1981, 44, 178–184 CrossRef.
- K. Violaki, A. Nenes, M. Tsagkaraki, M. Paglione, S. Jacquet, R. Sempéré and C. Panagiotopoulos, npj Clim. Atmos. Sci., 2021, 4 DOI:10.1038/s41612-021-00215-5.
- N. W. May, N. E. Olson, M. Panas, J. L. Axson, P. S. Tirella, R. M. Kirpes, R. L. Craig, M. J. Gunsch, S. China, A. Laskin, A. P. Ault and K. A. Pratt, Environ. Sci. Technol., 2018, 52, 397–405 CrossRef CAS PubMed.
- Y. Zhang and A. L. Steiner, Nat. Commun., 2022, 13, 1–10 Search PubMed.
- V. Zingales, M. Taroncher, P. A. Martino, M. J. Ruiz and F. Caloni, Toxins, 2022, 14 DOI:10.3390/toxins14070445.
- B. D. Wahlen, M. R. Morgan, A. T. McCurdy, R. M. Willis, M. D. Morgan, D. J. Dye, B. Bugbee, B. D. Wood and L. C. Seefeldt, Energy Fuels, 2013, 27, 220–228 CrossRef CAS.
- M. Hannon, J. Gimpel, M. Tran, B. Rasala and S. Mayfield, Biofuels, 2010, 1, 763–784 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ea00083h |
| This journal is © The Royal Society of Chemistry 2024 |
