Supramolecular hydrogels from unprotected dipeptides: a comparative study on stereoisomers and structural isomers†
Ottavia
Bellotto
 a,
Slavko
Kralj
a,
Slavko
Kralj
 b,
Rita
De Zorzi
b,
Rita
De Zorzi
 a,
Silvano
Geremia
a,
Silvano
Geremia
 a and
Silvia
Marchesan
a and
Silvia
Marchesan
 *a
*a
aUniversity of Trieste, Chem. Pharm. Sc. Dept., Via Giorgieri 1, 34127 Trieste, Italy. E-mail: smarchesan@units.it
bJožef Stefan Institute, Materials Synthesis Dept., Jamova 39, Ljubljana, Slovenia
First published on 3rd September 2020
Abstract
Amino acid stereoconfiguration has been shown to play a key role in the self-assembly of unprotected tripeptides into hydrogels under physiological conditions. Dramatic changes were noted for hydrophobic sequences based on the diphenylalanine motif from the formation of amorphous aggregates in the case of homochiral peptides to nanostructured and stable hydrogels in the case of heterochiral stereoisomers. Herein, we report that by further shortening the sequence to a dipeptide, the overall differences between isomers are less marked, with both homo- and hetero-chiral dipeptides forming gels, although with different stability over time. The soft materials are studied by a number of spectroscopic and microcopic techniques, and single-crystal X-ray diffraction to unveil the supramolecular interactions of these hydrogel building blocks.
Introduction
Short peptides and their analogues have been attracting great interest in recent years as building blocks for soft matter.1–7 They offer several advantages over other molecular classes, such as ease of preparation and scale-up, low cost, inherent biocompatibility and biodegradability, and the possibility to encode biological messages.8 In particular, it has been shown that a sequence as short as a tripeptide possesses, on average, the ideal number of non-carbon atoms required to maximise ligand–receptor interactions to develop drug-like molecules.9 There is a plethora of endogenous tripeptide sequences that are well-known for their bioactivity, e.g., RGD for cell adhesion, and the topic was recently reviewed.10What is less-known is that also dipeptides can exert some biological effect, and they are very relevant for instance to the food industry.11 They could find application to improve taste in drug formulations, and they have been long studied as food supplements.12–15 New biological activities continue to emerge, e.g., Phe–Leu was shown to act as antidepressant,16 anxiolytic,17 and, to a major extent of its structural isomer Leu–Phe, to be an angiotensin I-converting enzyme (ACE) inhibitor.18 Short repeats of Leu and Phe can activate neutrophils,19 and the activity is maintained, or even boosted, when the sequence alternates D- and L-amino acids,20,21 giving scope to study heterochiral short peptides.
Dipeptides containing Leu and Phe thus appeared as interesting candidates for self-assembly into soft matter. To the best of our knowledge, Ile–Phe is the only unprotected dipeptide reported to form a stable hydrogel,22 while Phe–Phe hydrogels were reported to be metastable,23,24 unless the dipeptide was cyclised to the corresponding 2,5-diketopiperazine,24 or further modified, e.g., by adding a p-nitro substitution on the Phe benzene ring.24 The removal of just one methylene unit from the gelling Ile–Phe to give Val–Phe was sufficient to disrupt self-assembling ability in water,22 and the same applied to Phe–Val that was too hydrophilic to gel unless it was cyclised to the corresponding 2,5-piperazinedione.24 Indeed, the design of linear unprotected dipeptide gelators is a challenging aim, whilst their cyclic derivatives have been more widely applied, for instance for enzyme mimicry, drug release, and photo-responsive systems.25–28
In addition, amino acid chirality is an interesting tool to fine-tune self-assembly, as it was reported to have dramatic effects in the case of unprotected tripeptide stereoisomers. For instance, in the case of Leu–Phe–Phe and Phe–Leu–Phe, at high concentration the homochiral L-peptides precipitated into amorphous aggregates, while heterochiral isomers self-organised into stable, nanostructured hydrogels.29–32 However, the effects of combining D- and L-amino acids on dipeptides is still unknown, hence for this work the ability to form supramolecular hydrogels was tested for the unprotected dipeptides reported in Table 1. The mirror-images D-Leu–D-Phe, L-Leu–D-Phe, D-Phe–D-Leu, and L-Phe–D-Leu were not included, because enantiomers display the same supramolecular behaviour in an achiral environment. Thus, the current investigation aims to cover the self-organisation ability of all possible sequence and stereoconfiguration combinations of Leu and Phe in an unprotected dipeptide.
| Dipeptide | Gel | Time (min) | Stable? | mgc (mM) | HPLC Rt (min) |
|---|---|---|---|---|---|
| L-Leu–L-Phe | Yes | 24 | YES | 40 | 11.7 |
| D-Leu–L-Phe | Yes | 12 | YES | 40 | 13.0 |
| L-Phe–L-Leu | No | n.a. | n.a. | n.a. | 11.6 |
| D-Phe–L-Leu | Yes | <1 | NO | 20 | 13.1 |
Results and discussion
All dipeptides were synthesised according to standard protocols on solid-phase and purified by reverse-phase HPLC.29 Their purity and identity were confirmed by 1H-NMR, 13C-NMR, and ESI-MS (see ESI†). They were dissolved in phosphate-buffered saline (PBS) with the aid of sonication and heating, then they were left to cool down to room temperature. Hydrogelation was first probed by the inversion tube test, next it was confirmed by oscillatory rheology measurements. As can be seen from Table 1, all dipeptides, except for L-Phe–L-Leu, gelled. Therefore, the effects of changing amino acid chirality were less dramatic on dipeptides than what reported for unprotected tripeptides.The viscoelastic properties of each sample were probed by oscillatory rheometry, starting with time sweeps (Fig. 1). In the case of Leu–Phe, gelation time doubled going from the D,L-heterochiral to the L-homochiral sequence (Fig. 1A and B). Higher peptide concentrations led to faster kinetics and higher moduli (see ESI†). In the case of Phe–Leu, the D,L-heterochiral peptide formed a metastable hydrogel, while the L-homochiral did not gel at all (Fig. 1C and D). Stress sweeps (Fig. 2) revealed no significant differences between Leu–Phe stereoisomers (Fig. 2A and B), while the metastable hydrogel formed by D-Phe–L-Leu disassembled during the test (Fig. 2D), thus not allowing for an accurate analysis. The same issue affected the frequency sweeps, while in the case of Leu–Phe stereoisomers, both the elastic modulus G′ and the viscous modulus G′′ were independent from the applied frequency, with G′ > G′′, as expected for stable hydrogels (see ESI†).
 | ||
| Fig. 2 Oscillatory rheology stress sweeps for the four dipeptides. Red stars mark gel-to-sol transitions. | ||
Overall, from the rheological analysis, we inferred that heterochirality promoted hydrogelation, since in the case of Leu–Phe, it reduced gelation time, while in the case of Phe–Leu, it yielded a gel in contrast to the non-gelling L-isomer. The reasons behind this phenomenon could lie in the hydrophobicity increase, as supported by HPLC retention times (Rt),33 which were higher for heterochiral than homochiral isomers (Table 1). It was recently demonstrated on hydrophobic tripeptides that heterochirality oriented the side chains on the same side of the peptide backbone, contrarily to the L-isomers. As a result, an amphipathic conformation arose only for heterochiral tripeptides, with net segregation between hydrophilic and hydrophobic regions that allowed for the successful self-organization into stable superstructures.29,32
Transmission electron microscopy (TEM) micrographs (Fig. 3) confirmed a network of anisotropic structures for L-Leu–L-Phe, D-Leu–L-Phe, and D-Phe–L-Leu. The amorphous aggregates formed by L-Phe–L-Leu did not have nanoscale features that could be seen by TEM. Rigid fibers with heterogeneous size were noted for L-Leu–L-Phe (Fig. 3A), which arose from the association of thinner fibrils (Fig. 3B) that were difficult to distinguish individually. Similar was the case of D-Leu–L-Phe (Fig. 3C), although in this case 12 ± 2 nm-wide (n = 100) individual fibrils were clearly visible (Fig. 3D). Finally, for the metastable hydrogel formed by D-Phe–L-Leu, instances of crystal nucleation and clusters of short fibrils were seen (Fig. 3E and F), in agreement with its transient nature confirmed by the rheological analysis. Hydrogels are often the kinetic product of peptide self-assembly, while crystals are the thermodynamic product.34 Indeed, within an hour, single crystals arose from the disassembly of the metastable gel, giving the opportunity for XRD investigation. Although a crystal and a gel are clearly two different phases, it was recently shown for a similar Phe-derived gelator that they share key intermolecular interactions,35 while differing mainly in the long-range order and hydration level.
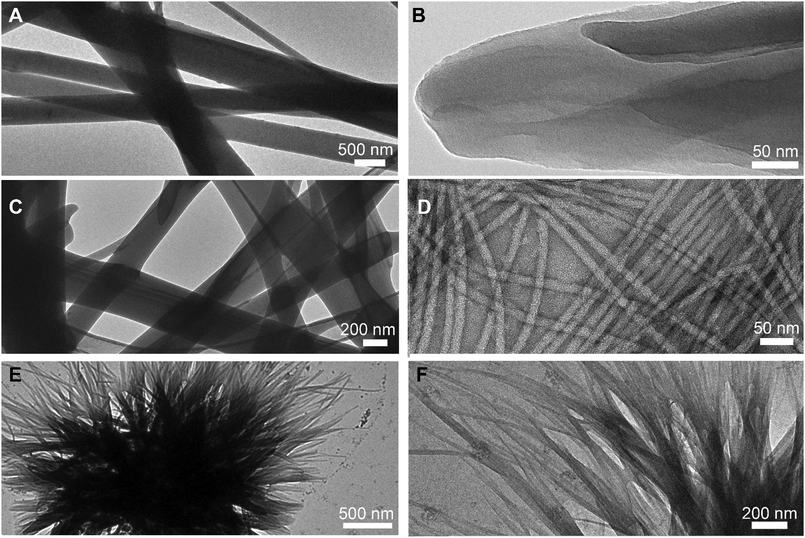 | ||
| Fig. 3 TEM micrographs of self-organised dipeptides at lower (left) and higher (right) magnification. (A) and (B) L-Leu–L-Phe; (C) and (D) D-Leu–L-Phe; (E) and (F) D-Phe–L-Leu. | ||
The crystal structures of homochiral L-Leu–L-Phe and L-Phe–L-Leu were reported by Görbitz36 as part of a series of studies on hydrophobic dipeptides (Fig. 4A and B).37,38 Both compounds displayed a remarkably similar packing into hydrophilic nanotubes, thanks to their amphipathic conformation. The inner, water-filled, cavity featured the amide-rich peptide backbone, while the hydrophobic side-chains were displayed on the outer surface, where they faced those of the other channels, and, only in the case of gelling L-Leu–L-Phe, they interdigitated with each other into zippers (see ESI,† Fig. S27). On the contrary, D-Phe–L-Leu – reported in the present work for the first time – assembled into alternating hydrophilic and hydrophobic layers devoid of interactions between the latter (Fig. 4C). Interestingly, the interdigitation of the aromatic side chains of Phe into dry zippers that exclude solvent is a common feature of the unprotected dipeptides that form stable hydrogels, i.e., Ile–Phe22,39 and Leu–Phe36 (see Fig. S27 in the ESI†), while it is absent in the non-gelling Phe–Leu,36 Val–Phe,40 and Phe–Val.41 This feature is very common for amyloid structures42 and may play a role in providing stability to the hydrogels.30 In terms of hydrogen bonding pattern, surprisingly, gelling L-Leu–L-Phe and non-gelling L-Phe–L-Leu displayed analogous head-to-tail extended interactions. It is possible, though, that the latter successfully established such network of interactions only in the crystal phase, while gelation may have been hampered by its inability to effectively establish Phe zippers. By contrast, D-Phe–L-Leu featured water molecules bridging between N-termini and engaging in hydrogen bonding also with the amide carbonyl moiety; the amide N–H atoms were hydrogen-bound to the C-termini, which also interacted through hydrogen-bonding through water as a bridging element (Fig. 4D). We inferred that the presence of localised interactions, as opposed to extended networks of hydrogen bonding, and the absence of Phe zippers, may be key to the metastability of the hydrogel formed by D-Phe–L-Leu.
 | ||
| Fig. 4 Single-crystal XRD structures of (A) L-Leu–L-Phe,36 (B) L-Phe–L-Leu,36 and (C) and (D) D-Phe–L-Leu (CCDC 2012848),† highlighting the alternation of hydrophobic and hydrophilic layers (C) and the hydrogen bonding pattern (D), hydrogen bonding involving only peptide molecules as black dashes, hydrogen bonding involving both peptide and water molecules in blue dashes. | ||
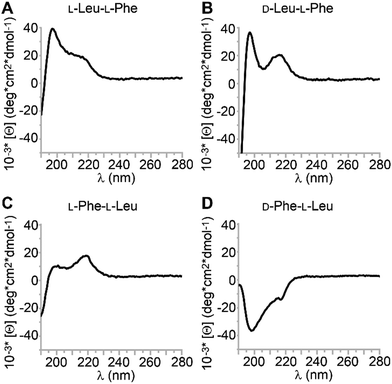
Circular dichroism (CD) is a useful technique to probe the spatial arrangement of chiral molecules. In the case of longer sequences, it is widely applied to determine conformation thanks to the vast literature on well-established signatures that can be ascribed to α-helices, β-sheets, and so on. What is less known is that also single amino acids display CD spectra, which are positive in the 200–250 nm UV region for the L-stereoconfiguration.43 While their D-mirror images are expected to display negative mirror-image spectra, the case of heterochiral sequences is more complex and difficult to predict. It has been reported that the stereoconfiguration of the N-terminal,44 central,45 or C-terminal46,47 amino acid dictates the sign of the CD spectrum, but clearly other factors come into play, and the observed trends appear to be sequence-specific.48,49 CD spectra of the four dipeptides are reported in Fig. 5. In this study, there was only one CD spectrum that was negative in the 200–230 nm region for D-Phe–L-Leu (Fig. 5D), with two minima at 200 and 219 nm. A similar case, but opposite in sign, was displayed by the other two gelling peptides L-Leu–L-Phe and D-Leu–L-Phe (Fig. 5A and B). The CD signature was very similar to what reported for gelling D-Phe–L-Phe–L-Leu,50 and for L-Phe–D-Leu–L-Phe, for which a combination of experimental and molecular dynamics revealed it corresponded to a population of conformations in the non-assembled state, whereby the most visited displayed dihedral angles typical of β-structures (sheets and turns).29 The non-assembling L-Phe–L-Leu peptide was the only one displaying the maximum at 219 nm of higher intensity than that at 200 nm (Fig. 5C). We inferred that the distribution of conformations populated by this non-gelling sequence was different relative to the gelators. CD spectra were also acquired in the hydrogel state, however, due to the presence of salts and high peptide concentration, it was not possible to acquire meaningful data in the far UV range. Interestingly, the CD spectra above 220 nm were all positive, including D-Phe–L-Leu, for which a sign inversion occurred with assembly (see ESI†).
Peptide conformation was also investigated by Attenuated Total Reflectance (ATR) FT-IR on gel samples. Fig. 6 shows the amide region for the three dipeptides in the gel state (Fig. 6A, B and D), and for the non-gelling L-Phe–L-Leu (Fig. 6C) in the precipitate obtained at the same concentration. While it would be rather controversial to assign typical peptide conformations to a dipeptide, the canonical signatures can provide a useful reference for the hydrogen bonding pattern that arises in the supramolecular assemblies. In all cases, a signal was clearly visible in the 1670–1680 cm−1 range, where β-turns are located for longer peptides.51 A second peak in the amide I region was clearly visible in all samples and it occurred in the β-structure region (1600–1625 cm−1). The metastable gel formed by D-Phe–L-Leu was the only sample to display an amide signal in the disordered region at 1641 cm−1, which could explain its instability.
 | ||
| Fig. 6 Amide region of the ATR-IR spectra of the four dipeptides in the gel (A) (B) and (D) or precipitate (C) form. | ||
In the amide II region, all gelling samples displayed two maxima (≈1525 and 1560 cm−1), which were both downshifted of 5–10 cm−1 for the non-gelling peptide. For comparison, the reported L-Ile–L-Phe gelator IR signal at 1570 cm−1 arose from strong association between the carboxylate and ammonium charged termini in the supramolecular state, and was absent for non-assembling L-Val–L-Phe.22 In addition, the lack of such extended interactions resulted in a signal for the carboxylate group to 1598 cm−1 for the non-assembling L-Val–L-Phe, as noticed in this work for the metastable D-Phe–L-Leu.22 This is in agreement with the interactions noticed in the crystal structure (Fig. 4D). By contrast, the other heterochiral dipeptide, the gelling D-Leu–L-Phe, was the only sample to display a signal at 1717 cm−1 that is typically ascribed to carboxylic functionalities that are strongly engaged in hydrogen bonding in the protonated form.52 Overall, we inferred from the ATR-IR and the XRD analyses that the stable gelators had two distinctive features; (1) they engaged in Phe zippers; (2) they displayed an extended network of hydrogen bonds between N- and C-termini, while the metastable gel displayed more disorder and only localised hydrogen-bond networks.
Conclusions
In this work, the four dipeptides L-Leu–L-Phe, D-Leu–L-Phe, L-Phe–L-Leu, and D-Phe–L-Leu were investigated for self-assembly into hydrogels in phosphate buffer. They represented all possible combinations of Leu and Phe in unprotected dipeptides, since their mirror-images D-Leu–D-Phe, L-Leu–D-Phe, D-Phe–D-Leu, and L-Phe–D-Leu display analogous supramolecular behaviour in an achiral environment. Except for L-Phe–L-Leu, all the other compounds gelled, albeit D-Phe–L-Leu metastable hydrogel converted into crystals within an hour. The hydrogels arose from a network of fibrils, often bundling into rigid fibers of heterogeneous size, as shown by TEM.Overall, the presence of Phe at the C-terminus was associated with better gelling ability, and heterochirality increased dipeptide hydrophobicity, and promoted hydrogelation. CD analysis suggested a different distribution of conformations in solution for the non-gelling L-Phe–L-Leu, relative to the other dipeptides, whose spectra were similar to D-Phe–L-Phe–L-Leu50 and L-Phe–D-Leu–L-Phe.29 Spectroscopic and single-crystal X-ray diffraction analyses suggested a very similar, and extended, hydrogen bonding network between N- and C-termini, together with Phe zippers, as distinctive features of stable gelators, in agreement with the literature.22
We can conclude that there was no single parameter that was crucial for the determination of a dipeptide gelling ability, which appeared to be the result of a fine equilibrium between different properties. In any case, while gelling or non-gelling compounds clearly displayed a rather diverse set of features in terms of ability to form nanotubes, only stable gelators featured extended networks of hydrogen bonds and Phe zippers.30,53 It is possible that this latter feature, which is well-established for amyloids,42,54 promotes stability of this kind of hydrogels that are driven by the hydrophobic collapse in water. Further studies on other sequences will be needed to verify this hypothesis and clearly identify key features for the future design of supramolecular systems based on unprotected dipeptides.
Experimental section
Materials and general methods
2-Chlorotrityl resin, O-benzotriazole-N,N,N,N′-tetramethyl-uronium-hexafluoro-phosphate (HBTU), and Fmoc protected amino acids were purchased from GL Biochem (Shanghai) Ltd. All solvents were purchased from Merck, at analytical grade. Piperidine, trifluoroacetic acid (TFA), N,N-diisopropyl ethylamine (DIPEA), triisopropyl silane (TIPS) were from Acros. Sodium dihydrogen phosphate and disodium hydrogen phosphate were from BDH AnalaR. High purity Milli-Q-water with a resistivity greater than 18 M Ω cm was obtained from an in-line Millipore RiOs/Origin system. 1H-NMR and 13C-NMR spectra were recorded at 400 MHz and 100 MHz on a Varian Innova Instrument with chemical shift reported as ppm (in DMSO with tetramethylsilane as internal standard). ESI-MS spectra were recorded on an Agilent 6120 single quadrupole LC-MS system.Dipeptide synthesis, purification, and self-assembly
Each dipeptide was synthesised by standard solid-phase methods and purified by reversed-phase HPLC as described previously.29 Each dipeptide was dissolved in phosphate-buffered saline solution at the desired concentration (40 mM, unless otherwise indicated) with the aid of an ultrasound bath (Branson 500) for a few seconds, followed by vial immersion in an oil bath at 100 °C until a clear solution was obtained. Upon cooling to room temperature, hydrogels formed as described in the text, except for L-Phe–L-Leu, which precipitated into amorphous aggregates.Oscillatory rheology
Dynamic time sweep rheological analysis was conducted on a Malvern Kinexus Ultra Plus Rheometer (Alfatest, Milan, Italy) with a 20 mm stainless steel parallel plate geometry. The temperature was maintained at 25 °C using a Peltier temperature controller. Samples were prepared and immediately analysed with a gap of 1 mm. Time sweeps were recorded using a frequency of 1 Hz and a controlled stress of 1.00 Pa or 0.50 Pa for the metastable hydrogel. Frequency sweeps were recorded using a controlled stress of 1.00 Pa, except for the metastable gel for which a value of 0.50 Pa was set, and then stress sweeps were recorded using a frequency of 1 Hz for all compounds.Transmission electron microscopy (TEM) imaging
The peptide self-organization was visualized with a transmission electron microscope (TEM). Briefly, 5 μL of the peptide samples were poured on a copper-lacey carbon-coated 300 mesh grids, while a TEM grid was exposed for 6 min. under ultraviolet (UV) ozone cleaner just before material deposition. After 1 min of adsorption, the excess material was drawn off, and 5 μL of a 2% aqueous potassium phosphotungstate at pH 7.2 was poured on the grids. Grids were air-dried until needed and TEM images were acquired using Jeol JEM 2100 instrument at 100 kV.Single-crystal XRD
Single crystals of D-Phe–L-Leu (CCDC 2012848) were obtained after approximately 1 hour of hydrogel preparation as described above.† A rectangular-shaped single crystal of the peptide was collected with a loop, cryoprotected by dipping the crystal in glycerol, and stored frozen in liquid nitrogen. The crystal was mounted on the diffractometer at the synchrotron Elettra, Trieste (Italy), beamline XRD1, using the robot present at the facility. Temperature was kept at 100 K. Diffraction data were collected by the rotating crystal method using synchrotron radiation, wavelength 0.70 Å. Reflections were indexed and integrated in the C2 space group, unit cell parameters 16.870 Å, 5.770 Å, 19.890 Å, 105.90°. The structure was solved by direct methods and refined anisotropically. Further details on structure determination and cell unit parameters are provided in the ESI.†Circular dichroism spectroscopy
A 0.1 mm quartz cell was used on a Jasco J815 Spectropolarimeter, with 1 s integrations, 1 accumulation and a step size of 1 nm with a bandwidth of 1 nm at 25 °C. Samples were prepared at a peptide concentration of 1 mM in milliQ water to analyse the non-assembled state, or at 40 mM in PBS to analyse the assembled form. Shown spectra are the average of at least 5 measurements.Attenuated total reflectance (ATR) infrared spectroscopy
The infrared (IR) spectra were recorded with a Jasco 4700 FT-IR, equipped with an ATR Pro One. A drop of the hydrogel was placed on a silicon wafer, and then dried under vacuum overnight. Spectra were acquired with 132 accumulations and 2 cm−1 resolution.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the Italian Ministry of Education and Research MIUR through the SIR program, “HOT-SPOT” project, personal research grant no. RBSI14A7PL to S. M. and by the Slovenian Research Agency (ARRS) through the core funding No. P2-0089 and project No. J1-7302. The authors acknowledge Elettra Sincrotrone Trieste for providing access to its synchrotron radiation facilities and Maurizio Polentarutti, Nicola Demitri and Giorgio Bais for assistance in using beamline XRD1. The authors also acknowledge the CENN Nanocenter (Slovenia) for the access to electron microscopy, and Mr Marco Tedesco (University of Trieste) for his kind technical assistance with ATR-IR measurements.Notes and references
- S. Mondal, S. Das and A. K. Nandi, Soft Matter, 2020, 16, 1404–1454 RSC.
- V. Castelletto, J. Seitsonen, K. M. Tewari, A. Hasan, R. M. Edkins, J. Ruokolainen, L. M. Pandey, I. W. Hamley and K. H. A. Lau, ACS Macro Lett., 2020, 9, 494–499 CrossRef CAS.
- P. Chakraborty, Y. Tang, T. Yamamoto, Y. Yao, T. Guterman, S. Zilberzwige-Tal, N. Adadi, W. Ji, T. Dvir, A. Ramamoorthy, G. Wei and E. Gazit, Adv. Mater., 2020, 32, 1906043 CrossRef CAS.
- C. G. Pappas, N. Wijerathne, J. K. Sahoo, A. Jain, D. Kroiss, I. R. Sasselli, A. S. Pina, A. Lampel and R. V. Ulijn, ChemSystemsChem, 2020, 2, e2000013 CrossRef.
- A. D. Martin, J. P. Wojciechowski, H. Warren, M. in het Panhuis and P. Thordarson, Soft Matter, 2016, 12, 2700–2707 RSC.
- S. Motamed, M. P. Del Borgo, K. Kulkarni, N. Habila, K. Zhou, P. Perlmutter, J. S. Forsythe and M. I. Aguilar, Soft Matter, 2016, 12, 2243–2246 RSC.
- A. D. Ariawan, B. Sun, J. P. Wojciechowski, I. Lin, E. Y. Du, S. C. Goodchild, C. G. Cranfield, L. M. Ittner, P. Thordarson and A. D. Martin, Soft Matter, 2020, 16, 4800–4805 RSC.
- J. Li, R. Xing, S. Bai and X. Yan, Soft Matter, 2019, 15, 1704–1715 RSC.
- P. Ung and D. A. Winkler, J. Med. Chem., 2011, 54, 1111–1125 CrossRef CAS.
- I. W. Hamley, Chem. Rev., 2017, 117, 14015–14041 CrossRef CAS.
- M. Gallego, L. Mora and F. Toldrá, Food Prod., Process. Nutr., 2019, 1, 2 CrossRef.
- S. Chakrabarti, S. Guha and K. Majumder, Nutrients, 2018, 10, 1738 CrossRef.
- J. L. Zambonino Infante, C. L. Cahu and A. Peres, J. Nutr., 1997, 127, 608–614 CrossRef CAS.
- T. Verri, A. Barca, P. Pisani, B. Piccinni, C. Storelli and A. Romano, J. Comp. Physiol., B, 2017, 187, 395–462 CrossRef CAS.
- J. M. Rouanet, J. L. Zambonino Infante, B. Caporiccio and C. Pejoan, Ann. Nutr. Metab., 1990, 34, 175–182 CrossRef CAS.
- T. Mizushige, T. Uchida and K. Ohinata, Sci. Rep., 2020, 10, 2257 CrossRef CAS.
- T. Mizushige, N. Kanegawa, A. Yamada, A. Ota, R. Kanamoto and K. Ohinata, Neurosci. Lett., 2013, 543, 126–129 CrossRef CAS.
- S. Ono, M. Hosokawa, K. Miyashita and K. Takahashi, Int. J. Food Sci. Technol., 2006, 41, 383–386 CrossRef CAS.
- A. Dalpiaz, A. Scatturin, G. Vertuani, R. Pecoraro, P. A. Borea, K. Varani, S. Traniello and S. Spisani, Eur. J. Pharmacol., 2001, 411, 327–333 CrossRef CAS.
- A. Dalpiaz, M. E. Ferretti, R. Pecoraro, E. Fabbri, S. Traniello, A. Scatturin and S. Spisani, Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol., 1999, 1432, 27–39 CrossRef CAS.
- J. D. Higgins, G. J. Bridger, C. K. Derian, M. J. Beblavy, P. E. Hernandez, F. E. Gaul, M. J. Abrams, M. C. Pike and H. F. Solomon, J. Med. Chem., 1996, 39, 1013–1015 CrossRef CAS.
- N. S. de Groot, T. Parella, F. X. Aviles, J. Vendrell and S. Ventura, Biophys. J., 2007, 92, 1732–1741 CrossRef.
- M. P. Conte, N. Singh, I. R. Sasselli, B. Escuder and R. V. Ulijn, Chem. Commun., 2016, 52, 13889–13892 RSC.
- M. Kurbasic, S. Semeraro, A. M. Garcia, S. Kralj, E. Parisi, C. Deganutti, R. De Zorzi and S. Marchesan, Synthesis, 2019, 2829–2838 CAS.
- C. Balachandra and T. Govindaraju, J. Org. Chem., 2020, 85, 1525–1536 CrossRef CAS.
- J. Karcher and Z. L. Pianowski, Chem. – Eur. J., 2018, 24, 11605–11610 CrossRef CAS.
- A. J. Kleinsmann and B. J. Nachtsheim, Org. Biomol. Chem., 2020, 18, 102–107 RSC.
- A. J. Kleinsmann and B. J. Nachtsheim, Chem. Commun., 2013, 49, 7818–7820 RSC.
- A. M. Garcia, D. Iglesias, E. Parisi, K. E. Styan, L. J. Waddington, C. Deganutti, R. De Zorzi, M. Grassi, M. Melchionna, A. V. Vargiu and S. Marchesan, Chem, 2018, 4, 1862–1876 CAS.
- S. Marchesan, L. Waddington, C. D. Easton, D. A. Winkler, L. Goodall, J. Forsythe and P. G. Hartley, Nanoscale, 2012, 4, 6752–6760 RSC.
- D. Iglesias, M. Melle-Franco, M. Kurbasic, M. Melchionna, M. Abrami, M. Grassi, M. Prato and S. Marchesan, ACS Nano, 2018, 12, 5530–5538 CrossRef CAS.
- A. V. Vargiu, D. Iglesias, K. Styan, L. Waddington, C. Easton and S. Marchesan, Chem. Commun., 2016, 52, 5912–5915 RSC.
- H. Bolt, C. Williams, R. Brooks, R. Zuckermann, S. Cobb and E. Bromley, Pept. Sci., 2017, 108, e23014 CrossRef.
- J. Wang, K. Liu, R. Xing and X. Yan, Chem. Soc. Rev., 2016, 45, 5589–5604 RSC.
- A. M. Garcia, R. Lavendomme, S. Kralj, M. Kurbasic, O. Bellotto, M. C. Cringoli, S. Semeraro, A. Bandiera, R. De Zorzi and S. Marchesan, Chem. – Eur. J., 2020, 26, 1880–1886 CrossRef CAS.
- C. H. Görbitz, Chem. – Eur. J., 2001, 7, 5153–5159 CrossRef.
- C. H. Görbitz, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2018, 74, 311–318 CrossRef.
- C. H. Görbitz, Chem. – Eur. J., 2007, 13, 1022–1031 CrossRef.
- C. Görbitz, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2004, 60, o371–o373 CrossRef.
- C. Görbitz, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2002, 58, 512–518 CrossRef.
- C. Görbitz, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2000, 56, 1496–2498 CrossRef.
- M. R. Sawaya, S. Sambashivan, R. Nelson, M. I. Ivanova, S. A. Sievers, M. I. Apostol, M. J. Thompson, M. Balbirnie, J. J. Wiltzius, H. T. McFarlane, A. O. Madsen, C. Riekel and D. Eisenberg, Nature, 2007, 447, 453–457 CrossRef CAS.
- N. Amdursky and M. M. Stevens, ChemPhysChem, 2015, 16, 2768–2774 CrossRef CAS.
- U. Orcel, M. De Poli, M. De Zotti and J. Clayden, Chem. – Eur. J., 2013, 19, 16357–16365 CrossRef CAS.
- S. Marchesan, K. E. Styan, C. D. Easton, L. Waddington and A. V. Vargiu, J. Mater. Chem. B, 2015, 3, 8123–8132 RSC.
- Y. Fu, B. Li, Z. Huang, Y. Li and Y. Yang, Langmuir, 2013, 29, 6013–6017 CrossRef CAS.
- M. Wang, P. Zhou, J. Wang, Y. Zhao, H. Ma, J. R. Lu and H. Xu, J. Am. Chem. Soc., 2017, 139, 4185–4194 CrossRef CAS.
- S. Lin, Y. Li, B. Li and Y. Yang, Langmuir, 2016, 32, 7420–7426 CrossRef CAS.
- Q. Xing, J. Zhang, Y. Xie, Y. Wang, W. Qi, H. Rao, R. Su and Z. He, ACS Nano, 2018, 12, 12305–12314 CrossRef CAS.
- M. C. Cringoli, C. Romano, E. Parisi, L. J. Waddington, M. Melchionna, S. Semeraro, R. De Zorzi, M. Grönholm and S. Marchesan, Chem. Commun., 2020, 56, 3015–3018 RSC.
- E. Goormaghtigh, J.-M. Ruysschaert and V. Raussens, Biophys. J., 2006, 90, 2946–2957 CrossRef CAS.
- Z. Gu, R. Zambrano and A. McDermott, J. Am. Chem. Soc., 1994, 116, 6368–6372 CrossRef CAS.
- S. Mondal, L. Adler-Abramovich, A. Lampel, Y. Bram, S. Lipstman and E. Gazit, Nat. Commun., 2015, 6, 8615 CrossRef CAS.
- L. K. Chang, J. H. Zhao, H. L. Liu, J. W. Wu, C. K. Chuang, K. T. Liu, J. T. Chen, W. B. Tsai and Y. Ho, J. Biomol. Struct. Dyn., 2010, 28, 39–50 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Spectroscopy, rheology, single-crystal XRD data. CCDC 2012848. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d0sm01191f |
| This journal is © The Royal Society of Chemistry 2020 |