Open Access Article
Open Access ArticlepH-responsive molecular assemblies of pyridylbutadiene derivative with cucurbit[7]uril†
Anuji K.
Vasu‡
a,
Raman
Khurana‡
bc,
Jyotirmayee
Mohanty
 *bc and
Sriram
Kanvah
*bc and
Sriram
Kanvah
 *a
*a
aDepartment of Chemistry, Indian Institute of Technology Gandhinagar, Palaj, Gandhinagar 382 355, India. E-mail: kanvah@gatech.edu
bRadiation & Photochemistry Division, Bhabha Atomic Research Centre, Mumbai, 400085, India. E-mail: jyotim@barc.gov.in
cHomi Bhabha National Institute, Training School Complex, Anushaktinagar, Mumbai, 400094, India
First published on 8th May 2018
Abstract
The interaction between red-emitting pyridinium derivative [4-((1E,3E)-4-(4-(dimethylamino)phenyl)buta-1,3-dien-1-yl)-1-methylpyridin-1-ium] (DABP) and cucurbit[7]uril was investigated using optical absorption, steady-state and time-resolved fluorescence and anisotropy measurements. The changes in the UV/Vis absorption, fluorescence spectral characteristics and lifetime, with a change in pH, reveal efficient binding of CB7 to the dye molecule. The cucurbit[7]uril encapsulated DABP brings ∼1.5 units upward pKa shift. The formation of supramolecular assemblies with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest stoichiometries with CB7 at different pH conditions have been verified from 1H NMR, isothermal titration calorimetric studies and geometry optimization calculations. CB7-dye complexation and the ensuing morphological changes were revealed by SEM, AFM and optical microscopy images. This pH-responsive supramolecular assemblies of red-emitting DABP dye can find potential applications in biological imaging, optical pH-sensor and the construction of building blocks for the supramolecular architectures.
1 host–guest stoichiometries with CB7 at different pH conditions have been verified from 1H NMR, isothermal titration calorimetric studies and geometry optimization calculations. CB7-dye complexation and the ensuing morphological changes were revealed by SEM, AFM and optical microscopy images. This pH-responsive supramolecular assemblies of red-emitting DABP dye can find potential applications in biological imaging, optical pH-sensor and the construction of building blocks for the supramolecular architectures.
Introduction
Water-soluble organic fluorophores have attracted much attention because of their potential applications as fluorescent sensors, probes, imaging agents in aqueous media. In particular, fluorophores absorbing and emitting in the lower energy region (>700 nm) of the spectrum are highly attractive.1,2 In many cases, the optical and electronic properties of these molecules strongly depend on the structural features and therefore can be fine-tuned by incorporating suitable electron donating or electron accepting groups or through extended π-conjugation.3–5 Importantly, these properties can further be modulated by the useful noncovalent host–guest interaction involving supramolecular receptors.6,7Cucurbit[n]urils (CBn) are a family of macrocyclic receptors consisting of an O-bonded C, N framework, with the rigid barrel-shaped hydrophobic cavity and highly electronegative portals containing carbonyl groups. Owing to the unique structural characteristics, CBs function as remarkable hosts for binding several organic fluorophores (guests) with greater affinity and selectivity through hydrophobic interactions involving the rigid cavities or ion–dipole interactions with the carbonyl groups.8,9 The inertness of the cavity and the absence of absorption or fluorescence in the visible region also impart CBs as a great host for supramolecular interaction studies.8,9 In particular, CBs are known to interact strongly with cationic fluorophores.10,11 Encapsulation of the guest in the CB cavity causes the guest to experience rigid or a lower polar environment and results in distinct photophysical and photochemical behavior.7,12 Among all the known CBs, cucurbit[7]uril (CB7) and cucurbit[8]uril (CB8) were extensively used for these investigations because of their water solubility and suitability of their cavity size for encapsulating different guest molecules.6,13–17 Cucurbiturils have been utilized as a template for stereoselective photodimerization,18 supramolecular catalysis,19 inhibition and disintegration of amyloid fibrils for therapeutics,20 radionuclide separation,21 construction of supramolecular assemblies,22,23 and to modulate photophysical properties10,24 for several cationic pyridinium or viologen derivatives.6,10,18,24–29 Inspired from these earlier reports, we describe optical absorption and emission properties of a cationic pyridinium derivative [4-((1E,3E)-4-(4-(dimethylamino)phenyl)buta-1,3-dien-1-yl)-1-methylpyridin-1-ium (DABP)], in water in the absence and presence of CB7. The dye (DABP) offers an advantageous emission in near IR region, ∼715 nm in water and exhibits strong solvatochromic emission owing to the push–pull effect from the presence of an electron withdrawing group (pyridinium cation) and electron donating (dimethylamine) group. The change in pH of the solutions allows tuning of the emission properties of the fluorophore. The results were detailed in the following sections.
Experimental
Materials and instruments
All the reagents utilized for this study were obtained from Sigma, Alfa Aesar, TCI or SD Fine Chemicals. The synthesized compound DABP was characterized using 1H and 13C NMR (Bruker Avance III-500 MHz) in D2O and DMSO-d6. UV-visible absorption spectra were recorded using UV-visible spectrophotometer (Analytik Jena, Specord plus 210 or Jasco V-650, Tokyo, Japan) and steady-state fluorescence studies were conducted using FS5 spectrofluorometer (Edinburgh Instruments, UK). Each set of samples were excited at the isosbestic point observed in the absorption spectra. Fluorescence quantum yields of the compound were estimated using Hoechst-33258 (Φ = 0.4 at pH 4.5)30 as a reference standard in acidic medium and Nile red (Φ = 0.38 in methanol)31 in the basic medium by comparison of integrated areas under the corrected emission spectrum curve of dye and that of the standard. Nanopure water (Millipore Gradient A10 system; conductivity of 0.06 μS cm−1) was used to prepare the sample solutions. HCl, as well as NaOH used for pH adjustment, was obtained from Merck Ltd., India. Solution pH was measured by a pH meter model PC2700 from Eutech Instruments, India. Time-resolved fluorescence measurements were carried out using a time-correlated single-photon-counting (TCSPC) spectrometer (Horiba Scientific, UK). In the present work, 374 nm and 445 nm diode lasers (100 ps, 1 MHz repetition rate) were used for the excitation of protonated and neutral form of the dye, respectively. A reconvolution procedure was used to analyze decay by using a proper instrument response function (IRF) obtained by using suspension of TiO2 particles in water as the light scatterer. The fluorescence decays [I(t)] were analyzed in general as a sum of exponentials (eqn.(1)):32,33I(t)=∑Bi![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−t/τi) exp(−t/τi) | (1) |
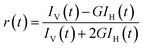 | (2) |
The structures of the complexes were optimized at the PM3/MM level by an energy minimization method using Gaussian 92.34
Isothermal titration calorimetric (ITC) experiments were carried out using a Microcal iTC 200 from Malvern, UK. 50 μM of dye solution in the sample cell was titrated by adding consecutively 19 injections of 2 μl of 5 mM of CB7 at both pH 3.0 and 9.5 conditions at 25 °C. The first data point was removed from the data set before curve fitting with Origin 7.0 software. Thermodynamical parameters of the complex formation were also evaluated using the estimated binding constant value and molar reaction enthalpy (ΔH).
Scanning electron microscopy (SEM) analysis was carried out using field emission SEM (JSM 7600F JEOL). For this purpose, one drop of the sample (∼10−5 M solution) was deposited on a Si-wafer mounted on an aluminium stub with the help of a double-sided adhesive carbon tape. The samples were heat-dried at 35 °C for 12 h and vacuum dried for 30 min to ensure complete removal of any residual water (and also for AFM measurements) and coated with platinum before being analyzed. The observed morphological changes are post evaporation processes optical microscopy analysis was carried out using Olympus BX51 polarizing microscope with image analyzer software. For this purpose, one drop of the sample (∼10−5 M solution) was deposited on a glass plate. Similar to SEM, the samples were heat-dried at 35 °C for 12 h and vacuum dried for 30 min to ensure removal of any residual water. Atomic force microscopy (AFM) analysis is done in peak force tunneling AFM module of scanning probe microscope (Nano Scope Multimode 8.0, Bruker) under ambient conditions, where variations in surface morphology were measured by featuring a closed loop X–Y head, Nanoscope 5 controller with Nanoscope 8.15 software. For this purpose, one drop of the sample [∼10−5 M] was spin coated on a high-grade AFM mica disc of 10 mm diameter.
Synthesis
The desired compound [4-((1E,3E)-4-(4-(dimethylamino)phenyl)buta-1,3-dien-1-yl)-1-methylpyridin-1-ium], was synthesized by a condensation reaction between methylpyridinium iodide and dimethylamino cinnamaldehyde (Scheme 1).35Results and discussions
Absorption and emission of DABP: effect of pH
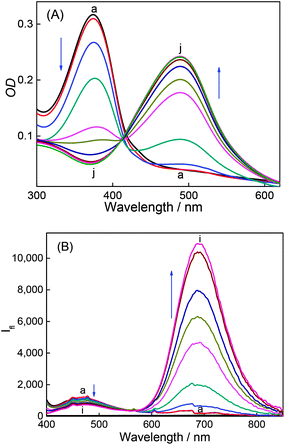
DABP absorbs at 448 nm (λa) in water and shows significant (∼32 nm) blue shift compared to organic solvents (∼480 nm).35 The blue-shift is a result of charge displacement of the unshared pair of electrons on the nitrogen atom of dimethylamine moiety through H-bonding obstructing the charge delocalization and increasing the transition energy.36,37 In strongly acidic media, the dye absorbs at 366 nm with a blue shift of ∼82 nm from its neutral counterpart [Fig. 1A]. At pH ∼ 4.5 the dye shows dual absorption bands at 448 nm and 367 nm. With increased acidic pH strength, the band at 448 nm decreases and subsequently disappears (Fig. 1A) yielding a major absorption band at ∼367 nm with the observance of an isosbestic point at 390 nm. These changes are attributed to the protonation of dimethylamine group and subsequent loss of its electron donating ability and intramolecular charge transfer (ICT) ability (Fig. S1, ESI†). The pKa measured from the absorbance data at different pH conditions is found to be 4.4 (Fig. 2).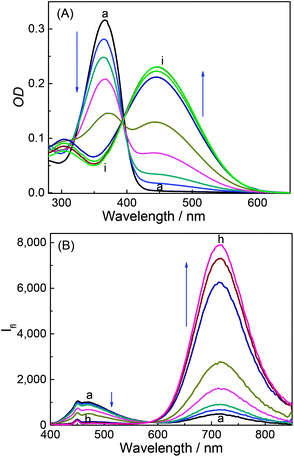 | ||
| Fig. 1 Absorption spectra (A) and emission spectra (B) of DABP at different pH solutions. pH: (a) 1.9, (b) 3.1, (c) 3.5, (d) 4.0, (e) 4.5, (f) 5.7, (g) 6.2, (h) 7.2 and (i) 8.4. | ||
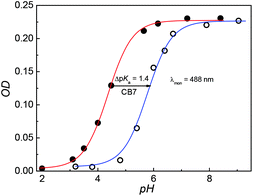 | ||
| Fig. 2 pH titration curves of DABP (20 μM) in the absence (Red) and the presence of 1 mM CB7 (Blue). The absorbance was measured at 488 nm. | ||
DABP shows strong solvatochromic emission shifts with an increase in solvent polarity culminating at ∼715 nm emission in water.35,38 These polarity dependent shifts are due to the formation of intramolecular charge transfer (ICT) excited state owing to the presence of strong donor (dimethylamine) and acceptor (pyridinium) groups. At pH ∼ 1.9, DABP emits at ∼476 nm. As the acidity decreases (with increased addition of a base), an increase in the emission intensity at ∼715 nm is noted along with a decrease in intensity of the 473 nm emission band (Fig. 1B). These emission peak shifts are attributed to the conversion of protonated dimethylamino moiety to the neutral form. The changes can also be distinguishable with the naked eye (Fig. S1, ESI†). The protonated dye (DABPH+) is colorless, and the neutral form (DABP) has a strong color. Increased alkalinity (pH 11.0) does not show any observable intensity or color changes.
Interaction of dye with CB7: pKa shift and binding constants
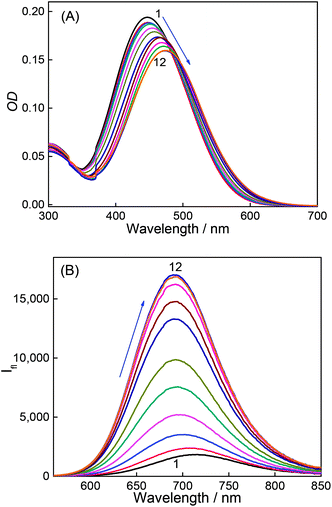
Fig. 3A shows absorption spectra of aqueous solutions of DABP at different pH in the presence of 1 mM CB7. Without CB7, at neutral and alkaline pH (pH ∼ 7.2 till pH 9.0), DABP shows only one absorption band at ∼448 nm. Interaction with CB7 in acidic or basic media results in +7 nm and +42 nm bathochromic absorption shifts respectively with an isosbestic point at 413 nm. As the acidic strength decreases, the absorbance also decreases. At pH ∼ 5.1, a peak at ∼490 nm was observed along with absorption band at 373 nm. Under pH neutral and basic conditions, only the peak at ∼490 nm is seen.In the presence of CB7, the absorption profile of DABP shows remarkable changes in both neutral and basic media. Calculation of pKa of supramolecular inclusion complex reveals a value of 5.9 (Fig. 2). It is known that the interaction with cucurbituril host increases pKa of aromatic amines.39–42 From the pKa curves, it is anticipated that DABP encapsulated in CB7 have higher propensity to remain in the protonated form than DABP in aqueous solution. Such differential binding affinity of the dye with the host molecules have been utilized for several chemical and biological applications.6,19,40,43–45
Similar to the absorption, the emission of DABP also shows variations post interaction with CB7. In the presence of 1 mM CB7, at neutral pH and basic pH, the dye shows a hypsochromic shift (−25 nm) in emission with maxima at ∼690 nm (Fig. 3B).
The titration of CB7 with the dye were performed and the absorption and emission spectral variation were monitored. Incremental addition of CB7 (0 μM to 362 μM) in 2.5 μM DABP yields absorption peak shifts (+10 nm) from 367 nm to 377 nm at ∼pH 3 (Fig. S2a†). Incremental addition of CB7 (0 μM to 83 μM) at ∼pH 9.5, also resulted in a +27 nm shift of absorption band from 448 nm to 475 nm (Fig. 4A). These absorption spectral changes indicate significant interaction of the dye with CB7 at both acidic and basic conditions. Similarly, the emission is also affected by the addition of CB7. At pH ∼ 3.0, an increase in intensity was observed for ∼470 nm along with a drop in intensity of the long wavelength band (∼715 nm) (Fig S2b†). At ∼pH 9.5, a blue shifted (−23 nm) emission at 691 nm was observed with enhanced intensity (Fig. 4B). Calculation of quantum yield of the (CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DABP) complex in acidic and basic media (pH 3 and 9.5) reveals ∼2.1 fold and ∼9 fold increment respectively in comparison to free dye (Table 1). This emission response also supports better host–guest interaction under alkaline conditions.
DABP) complex in acidic and basic media (pH 3 and 9.5) reveals ∼2.1 fold and ∼9 fold increment respectively in comparison to free dye (Table 1). This emission response also supports better host–guest interaction under alkaline conditions.
| Systems | λ abs (nm) | λ em (nm) | Φ f | τ f (ns) | τ r (ps) | pKa |
|---|---|---|---|---|---|---|
| a The lifetime of the fast decay component has kept fixed (∼40 ps, which is beyond the time resolution of our TCSPC instrument) during analysis. For all the decay fitting, χ2 values were obtained within 1.0–1.2. | ||||||
| Dye (pH 3) | 367 | 470 | 0.01 | — | — | 4.4 |
| Dye (pH 9.5) | 448 | 714 | 0.01 | — | — | |
Dye![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CB7 (pH 3) CB7 (pH 3) |
376 | 470 | 0.02 | <0.04a (60%), 5.1 (40%) | — | 5.9 |
Dye![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CB7 (pH 9.5) CB7 (pH 9.5) |
488 | 691 | 0.09 | 0.93 | 397 ± 50 | |
To calculate the association constant of CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DABP complex, the changes in the absorbance and fluorescence intensity were monitored at a particular wavelength (λabs at 448 nm and λem at 713 nm for pH 9.5, and λabs at 367 nm and λem at 480 nm for pH 3). At pH 3.0, the overall binding isotherms obtained from the absorption and fluorescence titrations displayed significant deviation from the usual 1
DABP complex, the changes in the absorbance and fluorescence intensity were monitored at a particular wavelength (λabs at 448 nm and λem at 713 nm for pH 9.5, and λabs at 367 nm and λem at 480 nm for pH 3). At pH 3.0, the overall binding isotherms obtained from the absorption and fluorescence titrations displayed significant deviation from the usual 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model, pointing to multiple equilibria present in the systems. A closer look at the initial part of the binding curve at pH 3.0 reveals that the increase in the fluorescence intensity apparently attains a quasi-saturation at a lower concentration of CB7 (∼20 μM), which corresponds to a 1
1 binding model, pointing to multiple equilibria present in the systems. A closer look at the initial part of the binding curve at pH 3.0 reveals that the increase in the fluorescence intensity apparently attains a quasi-saturation at a lower concentration of CB7 (∼20 μM), which corresponds to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding interaction. Beyond this quasi-saturation, the emission intensity slowly increased with further increase in host concentration (Fig. S3†) indicating the formation of higher-order complexes. Since the dye contains two binding sites at both the ends, we expect that the initial quasi-saturation point is mainly due to the 1
1 binding interaction. Beyond this quasi-saturation, the emission intensity slowly increased with further increase in host concentration (Fig. S3†) indicating the formation of higher-order complexes. Since the dye contains two binding sites at both the ends, we expect that the initial quasi-saturation point is mainly due to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex between CB7 and DABP and at a later stage, with a higher concentration of CB7, represents domination of the 2
1 complex between CB7 and DABP and at a later stage, with a higher concentration of CB7, represents domination of the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (CB7
1 (CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DABP) stoichiometric complex. Binding curves were fitted by non-linear regression method using the modified Benesi–Hildebrand equation for 2
DABP) stoichiometric complex. Binding curves were fitted by non-linear regression method using the modified Benesi–Hildebrand equation for 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex model (Method M1, ESI). At pH 3.0, K1 and K2 values were estimated to be (2.25 ± 0.5) × 106 M−1 and (5.15 ± 0.6) × 103 M−1 from the absorption titration and (1.8 ± 0.6) × 105 M−1 and (1.2 ± 0.8) × 103 M−1 from the fluorescence titration studies. The overall binding constant (K = K1 × K2) was found to be 1.15 × 1010 M−2 and 2.2 × 108 M−2 from absorption (Fig S3a†) and fluorescence (Fig S3b†) studies, respectively, for a 2
1 complex model (Method M1, ESI). At pH 3.0, K1 and K2 values were estimated to be (2.25 ± 0.5) × 106 M−1 and (5.15 ± 0.6) × 103 M−1 from the absorption titration and (1.8 ± 0.6) × 105 M−1 and (1.2 ± 0.8) × 103 M−1 from the fluorescence titration studies. The overall binding constant (K = K1 × K2) was found to be 1.15 × 1010 M−2 and 2.2 × 108 M−2 from absorption (Fig S3a†) and fluorescence (Fig S3b†) studies, respectively, for a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CB7
1 CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DABP stoichiometry. Similarly, at pH 9.5, the K1 and K2 values were estimated to be (4.9 ± 1.0) × 105 M−1 and (4.3 ± 0.7) × 104 M−1 from the absorption and (1.4 ± 0.7) × 105 M−1 and (1.6 ± 0.7) × 105 M−1 from the fluorescence measurements. The overall binding constant of dye with CB7 at pH 9.5 reveals 2.2 × 1010 M−2 with 2
DABP stoichiometry. Similarly, at pH 9.5, the K1 and K2 values were estimated to be (4.9 ± 1.0) × 105 M−1 and (4.3 ± 0.7) × 104 M−1 from the absorption and (1.4 ± 0.7) × 105 M−1 and (1.6 ± 0.7) × 105 M−1 from the fluorescence measurements. The overall binding constant of dye with CB7 at pH 9.5 reveals 2.2 × 1010 M−2 with 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CB7
1 CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DABP complexation stoichiometry (Fig. S4a and b†).
DABP complexation stoichiometry (Fig. S4a and b†).
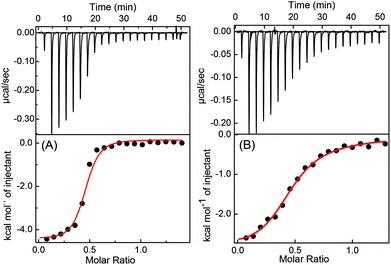
To get more insight into the binding interactions and thermodynamic parameters of the complexes at both the pHs, isothermal titration calorimetric (ITC) measurements were carried out. From the ITC data (Fig. 5) a sequential 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 binding model gave a satisfactory fit, similar to 2
2 binding model gave a satisfactory fit, similar to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (CB7
1 (CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DABP) model fitting for the absorption (Fig. S3A and S4A, ESI†) and fluorescence (Fig. S3B and S4B, ESI†) titration data. During the addition of CB7, first, there is a formation of 1
DABP) model fitting for the absorption (Fig. S3A and S4A, ESI†) and fluorescence (Fig. S3B and S4B, ESI†) titration data. During the addition of CB7, first, there is a formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex (K1) followed by 2
1 complex (K1) followed by 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (CB7
1 (CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DABP) complex formation (K2) at both the pHs. Hence, two binding constant values are obtained for each complex system (K1 = 6.1 × 103 M−1 and K2 = 6.3 × 106 M−1 for CB7
DABP) complex formation (K2) at both the pHs. Hence, two binding constant values are obtained for each complex system (K1 = 6.1 × 103 M−1 and K2 = 6.3 × 106 M−1 for CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DABP system at pH 3 and K1 = 5.6 × 104 M−1 and K2 = 7.3 × 104 M−1 for CB7
DABP system at pH 3 and K1 = 5.6 × 104 M−1 and K2 = 7.3 × 104 M−1 for CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DABP system at pH 9.5). The overall binding constant values (K = 3.8 × 1010 M−2 at pH 3 and 4.1 × 109 M−2 at pH 9.5) match well with the binding constant values obtained from the absorption and fluorescence titration data. The free energy changes for these complex formations at both the pHs were evaluated and are found to be energetically favorable. The details are provided in Note S1, ESI.†
DABP system at pH 9.5). The overall binding constant values (K = 3.8 × 1010 M−2 at pH 3 and 4.1 × 109 M−2 at pH 9.5) match well with the binding constant values obtained from the absorption and fluorescence titration data. The free energy changes for these complex formations at both the pHs were evaluated and are found to be energetically favorable. The details are provided in Note S1, ESI.†
Effect of CB7 complexation on the fluorescence lifetime and anisotropy of the dye
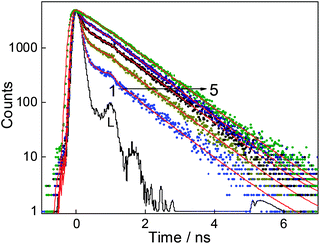
Fluorescence lifetime measurements are carried out to see the effect of CB7 on the excited state relaxation processes of DABP. The fluorescence lifetime of DABP at both the pH (3 and 9.5) is very short (<40 ps, beyond the time resolution of the used TCSPC instrument) because of the highly efficient nonradiative processes such as photoisomerization and intramolecular charge transfer from dimethylaniline group to methylpyridinium group. The modulation of the radiative properties of DABP on interaction with CB7 at both acidic and basic pH was evident from the significant increase in its excited-state lifetimes as shown in the fluorescence decay profiles presented in Fig. 6 and S5.† At pH 9.5, the decay became measurable upon addition of CB7 as the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex showed a biexponential decay having a significant contribution from faster decay components (<40 ps (68%)) along with a slower decay component of lifetime ∼930 ps (32%). In the presence of ∼12 μM CB7, the decay kinetics became single exponential (∼930 ps) which indicates severe retardation of the excited-state nonradiative relaxation channels, hence increasing the lifetime (Table S1†). At pH ∼ 3.0, as there is not much enhancement in the fluorescence intensity, the fluorescence lifetime was measured at the saturation concentration of CB7 (∼350 μM). The complex displayed biexponential decay with major contributions from the faster decay component [<40 ps (60%)] along with a slower decay component of a lifetime ∼5.1 ns (40%). The increased lifetime of DABP in the presence of CB7 at both the pH conditions could be attributed to the rigid environment of the dye in the CB7 cavity which restricts the nonradiative relaxation processes.
1 complex showed a biexponential decay having a significant contribution from faster decay components (<40 ps (68%)) along with a slower decay component of lifetime ∼930 ps (32%). In the presence of ∼12 μM CB7, the decay kinetics became single exponential (∼930 ps) which indicates severe retardation of the excited-state nonradiative relaxation channels, hence increasing the lifetime (Table S1†). At pH ∼ 3.0, as there is not much enhancement in the fluorescence intensity, the fluorescence lifetime was measured at the saturation concentration of CB7 (∼350 μM). The complex displayed biexponential decay with major contributions from the faster decay component [<40 ps (60%)] along with a slower decay component of a lifetime ∼5.1 ns (40%). The increased lifetime of DABP in the presence of CB7 at both the pH conditions could be attributed to the rigid environment of the dye in the CB7 cavity which restricts the nonradiative relaxation processes.
Time-resolved fluorescence anisotropy measurements give more insight into the hydrodynamic volume of the fluorophore which is linked to the size and structural arrangement of the complexed dye. It is expected that 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex formation between CB7 and DABP will be reflected in the increase in the rotational correlation time (τr) of the complex as compared to the free dye. It should be mentioned that the anisotropy decay of the free dye could not be measured using our TCSPC setup due to the very short lifetime of DABP at both pH conditions. However, the enhanced excited-state lifetime of DABP in the presence of CB7 at pH 9.5, allows the anisotropy to be recorded on the nanosecond time scale (Fig. S6†) and the τr value obtained from the single exponential fitting of the anisotropy decay is 396 ± 50 ps. This decrease in the rotational depolarization process indicates the increased rigidity and large molecular volume of the dye due to the complex formation. Since the fluorescence lifetime study shows that the major contribution is from the faster decay component at pH 3.0, the anisotropy measurement could not be carried out for the complex at this pH.
1 complex formation between CB7 and DABP will be reflected in the increase in the rotational correlation time (τr) of the complex as compared to the free dye. It should be mentioned that the anisotropy decay of the free dye could not be measured using our TCSPC setup due to the very short lifetime of DABP at both pH conditions. However, the enhanced excited-state lifetime of DABP in the presence of CB7 at pH 9.5, allows the anisotropy to be recorded on the nanosecond time scale (Fig. S6†) and the τr value obtained from the single exponential fitting of the anisotropy decay is 396 ± 50 ps. This decrease in the rotational depolarization process indicates the increased rigidity and large molecular volume of the dye due to the complex formation. Since the fluorescence lifetime study shows that the major contribution is from the faster decay component at pH 3.0, the anisotropy measurement could not be carried out for the complex at this pH.
1H NMR measurements
To understand the binding interactions, the NMR titrations were performed at pH 3 and 9.5. It is well known that encapsulation of guest molecules with CBs results in an upfield chemical shift of those protons that are within the hydrophobic cavity and a downfield shift was observed for protons located near to the carbonyl groups.46 NMR titrations at pH 3 solution were performed in deuterated hydrochloride (DCl). As shown in Fig. S7A,†1H NMR spectra of DABP in the presence of CB7 shows upfield shift of 0.03 ppm for N,N-dimethyl protons. The methyl group attached to the nitrogen of pyridinium moiety also shows an upfield shift. However, since this overlaps with protons of CB7, this upfield shift could not be quantified. Considerable upfield shifts (0.94–0.83 ppm) were observed for aromatic protons on the pyridinium ring and one proton of the conjugated double bond. Other aromatic protons and three protons of the double bonds show a downfield shift of 0.01 ppm to 0.36 ppm. NMR titrations at pH 9.5 were performed in NaOD. As shown in Fig. S7B,†1H NMR spectra of DABP in the presence of CB7 shows an upfield shift of 0.54 ppm and 0.49 ppm for aromatic protons those are near to N,N-dimethyl group, and a double bond. A 0.31 ppm upfield shift exhibited by aromatic protons and other two protons on the double bond. The pyridinium methyl group and methyl protons of N,N-dimethylamine group also shows an upfield shift of 0.08 ppm and 0.07 ppm. At pH 9.5, the appearance of proton resonances post complexation with CB7 is broad suggesting the presence of free and bound guest protons. From the NMR shifts, it is therefore confirmed that both protonated and neutral form of DABP shows 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CB7
1 CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dye binding.
dye binding.
Geometry optimization of the complexes
Computational calculations following semi-empirical method were performed to achieve better insight into the geometry and the nature of the interaction of both protonated and neutral forms of DABP with CB7. For this, we optimized the ground-state geometries of both protonated and neutral forms of DABP and CB7 and their complexes, namely, CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DABP (1
DABP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and (2
1) and (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for both the forms at PM3 level incorporating molecular mechanics (MM) correction using the Gaussian 92 suite of the package.35 In all the cases, the full geometry optimization was carried out without any symmetry constraint and in the absence of a solvent. The structure of 1
1) for both the forms at PM3 level incorporating molecular mechanics (MM) correction using the Gaussian 92 suite of the package.35 In all the cases, the full geometry optimization was carried out without any symmetry constraint and in the absence of a solvent. The structure of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes of the protonated form of DABP with CB7 was optimized by various input geometries, placing CB7 along the DABP molecule. In case of 1
1 complexes of the protonated form of DABP with CB7 was optimized by various input geometries, placing CB7 along the DABP molecule. In case of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex, the most stable structure was the one in which the butadiene moiety is placed inside the hydrophobic CB7 cavity (Fig. 7A) and the lowest ΔHf obtained for this arrangement is −68 kcal mol−1. To visualize the most stable geometry for the 2
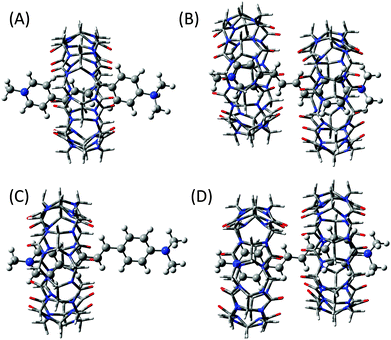
1 complex, the most stable structure was the one in which the butadiene moiety is placed inside the hydrophobic CB7 cavity (Fig. 7A) and the lowest ΔHf obtained for this arrangement is −68 kcal mol−1. To visualize the most stable geometry for the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex, geometry optimization was carried out by introducing two CB7 moieties from either end of the DABP molecule and the optimized structure is given in Fig. 7B. From the optimized parameters, the ΔHf value for the 2
1 complex, geometry optimization was carried out by introducing two CB7 moieties from either end of the DABP molecule and the optimized structure is given in Fig. 7B. From the optimized parameters, the ΔHf value for the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexation is estimated to be −90 kcal mol−1. In this structure, the stabilizing forces are the hydrogen bonding between the oxygen atoms of CB7 and the hydrogen atoms of butadiene moiety with bond distance about 2.5 Å and the ion–dipole interaction between the charged nitrogen atoms of N-methyl pyridinium and dimethyl anilinium groups of DABP and the carbonyl portals of CB7.
1 complexation is estimated to be −90 kcal mol−1. In this structure, the stabilizing forces are the hydrogen bonding between the oxygen atoms of CB7 and the hydrogen atoms of butadiene moiety with bond distance about 2.5 Å and the ion–dipole interaction between the charged nitrogen atoms of N-methyl pyridinium and dimethyl anilinium groups of DABP and the carbonyl portals of CB7.
Similarly, geometry optimization was attempted for the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes of the neutral form of DABP. Unlike protonated form, the positioning of pyridinium group of DABP inside the CB7 cavity was the most stable structure (Fig. 7C) due to the ion–dipole interaction between the N-methyl pyridinium group and the carbonyl portal of CB7. The lowest ΔHf value is estimated as −40 kcal mol−1. For the 2
1 complexes of the neutral form of DABP. Unlike protonated form, the positioning of pyridinium group of DABP inside the CB7 cavity was the most stable structure (Fig. 7C) due to the ion–dipole interaction between the N-methyl pyridinium group and the carbonyl portal of CB7. The lowest ΔHf value is estimated as −40 kcal mol−1. For the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex, a similar procedure was adopted to find out the most stable geometry. Hydrogen bonding between the oxygen atoms of CB7 and the hydrogen atoms of butadiene moiety and the ion–dipole interaction between the charged nitrogen atoms of N-methyl pyridinium group of DABP and the carbonyl portals of CB7 are the stabilizing forces for the optimized structure as shown in Fig. 7D. From the optimized parameter, the ΔHf value for the 2
1 complex, a similar procedure was adopted to find out the most stable geometry. Hydrogen bonding between the oxygen atoms of CB7 and the hydrogen atoms of butadiene moiety and the ion–dipole interaction between the charged nitrogen atoms of N-methyl pyridinium group of DABP and the carbonyl portals of CB7 are the stabilizing forces for the optimized structure as shown in Fig. 7D. From the optimized parameter, the ΔHf value for the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexation is calculated as −18 kcal mol−1. Though the energetics favor a 1
1 complexation is calculated as −18 kcal mol−1. Though the energetics favor a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex, however, at higher concentration of CB7 the equilibrium shifts toward the 2
1 complex, however, at higher concentration of CB7 the equilibrium shifts toward the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry.
1 stoichiometry.
Complexation induced morphological changes
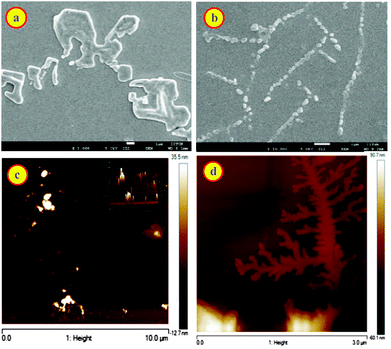
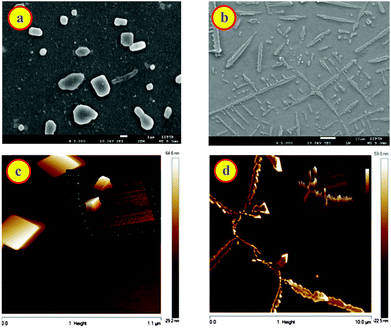
Macrocycle assisted assemblies are very attractive because of the potential applications as smart materials.22,24,47–49 To investigate the formation of such molecular assemblies, we examined the morphology of dye, CB7, and inclusion complex of the CB7–dye complex at different pH solutions using SEM, AFM, and optical microscopic techniques. Fig. S8a† shows square/cylindrical shaped structures for CB7 at pH 3. This confined structure of CB7 was confirmed from optical microscopy images as well (Fig. S8c, ESI†). The SEM image of DABP at pH 3 reveals small-rod like morphology (Fig. S8b, ESI†) having a dimension of ∼610 nm. Optical microscopy image (Fig. S8d†) supports the results observed from SEM. In the presence of CB7 (10 μM), SEM image of DABP shows nano-rods with a layered shape where particles are immersed inside (Fig. 8a) that is different to the SEM of the CB7 or the dye alone (Fig. S10a and b, ESI†). These morphological differences show the formation of macrocycle induced complexes. In the presence of higher concentrations of CB7 (200 μM), the nano-rods come closer and reveal wire like symmetry (Fig. 8b), but closer examination reveals that the rod-like structures do not connect with each other. AFM images (Fig. 8c, d and S12†) confirm the formation of such structures in a continuous manner.The experiments were also performed for CB7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dye complex at pH 9.5. Fig. 9a, b and S11a† demonstrates the morphological changes. At lower CB7 concentration, SEM images show distinct macrostructures with an average length of 1.58 μm that is greater than free dye and CB7 (Fig. 9a, S10 and S11, ESI†). In the presence of 200 μM concentration of CB7, SEM image of DABP shows rod shape geometry teeth like projections (Fig. 9b). These rods are not in uniform in size, and because of the prodigious interaction of DABP with CB7 at higher concentration (200 μM) these complexes come together and form branched structures. Reasonable doubt can appear that these structures could be due to the CB7 self-organization.
dye complex at pH 9.5. Fig. 9a, b and S11a† demonstrates the morphological changes. At lower CB7 concentration, SEM images show distinct macrostructures with an average length of 1.58 μm that is greater than free dye and CB7 (Fig. 9a, S10 and S11, ESI†). In the presence of 200 μM concentration of CB7, SEM image of DABP shows rod shape geometry teeth like projections (Fig. 9b). These rods are not in uniform in size, and because of the prodigious interaction of DABP with CB7 at higher concentration (200 μM) these complexes come together and form branched structures. Reasonable doubt can appear that these structures could be due to the CB7 self-organization.
To check this, we recorded SEM image of CB7 (200 μM) at pH 9.5, and that showed agglomeration of CB7 molecules (Fig. S10a, ESI†). AFM images (Fig. 9d) display a continuous structure in the form of ribbons or rods at higher CB7 concentrations, and at low concentrations of CB7 these continuous structures break down to square type structures. These insights from AFM confirm the observations from SEM, optical microscopy, and 1H NMR. Because of the formation of higher order insoluble complex, actual fluorescence enhancement was not measurable. Nevertheless, the higher order complexes were elucidated through SEM and AFM images.
Conclusions
The results obtained from steady-state absorption, fluorescence, NMR and isothermal titration calorimetric measurements indicate that dye produces different supramolecular assemblies with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest stoichiometries with CB7 at different pH conditions. The formation of these assemblies at different pH conditions have been further verified by geometry optimization calculation. The increase in fluorescence intensity of dye upon complexation is attributed to the restrictions imposed on the intramolecular motion of dye by the hydrophobic and rigid CB7 cavity. Changes in pH reveal tunable absorption and emission properties in the presence of CB7. SEM, AFM and optical microscopic images reveal changes in morphology of the encapsulated dye. These pH-responsive luminescent properties of DABP–CB7 assemblies can find applications in bio-imaging, organic electronics, light-emitting devices, optical-pH sensor, etc.
1 host–guest stoichiometries with CB7 at different pH conditions. The formation of these assemblies at different pH conditions have been further verified by geometry optimization calculation. The increase in fluorescence intensity of dye upon complexation is attributed to the restrictions imposed on the intramolecular motion of dye by the hydrophobic and rigid CB7 cavity. Changes in pH reveal tunable absorption and emission properties in the presence of CB7. SEM, AFM and optical microscopic images reveal changes in morphology of the encapsulated dye. These pH-responsive luminescent properties of DABP–CB7 assemblies can find applications in bio-imaging, organic electronics, light-emitting devices, optical-pH sensor, etc.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors acknowledge a financial grant from BRNS (Board of Research in Nuclear Sciences) India (37(2)/14/05/2016), Bhabha Atomic Research Center (BARC) and IIT Gandhinagar for overall support. General help from Mr Palash Jana, IIT Gandhinagar, is acknowledged. Mr R. Khurana acknowledges BARC for providing him a research fellowship. We also thank Dr A. C. Bhasikuttan and Dr N. Barooah, Radiation & Photochemistry Division, BARC, India for their support during the experiments and fruitful discussion.Notes and references
- J. O. Escobedo, O. Rusin, S. Lim and R. M. Strongin, Curr. Opin. Chem. Biol., 2010, 14, 64–70 CrossRef CAS.
- G. Hong, A. L. Antaris and H. Dai, Nat. Biomed. Eng., 2017, 1, 0010 CrossRef.
- W. T. Mason, Fluorescent and luminescent probes for biological activity: a practical guide to technology for quantitative real-time analysis, Academic Press, 1999 Search PubMed.
- L. Yuan, W. Lin, K. Zheng, L. He and W. Huang, Chem. Soc. Rev., 2013, 42, 622–661 RSC.
- Q. Yang, Z. Ma, H. Wang, B. Zhou, S. Zhu, Y. Zhong, J. Wang, H. Wan, A. Antaris, R. Ma, X. Zhang, J. Yang, X. Zhang, H. Sun, W. Liu, Y. Liang and H. Dai, Adv. Mater., 2017, 29, 1605497 CrossRef.
- J. W. Lee, S. Samal, N. Selvapalam, H.-J. Kim and K. Kim, Acc. Chem. Res., 2003, 36, 621–630 CrossRef CAS.
- A. C. Bhasikuttan, J. Mohanty, W. M. Nau and H. Pal, Angew. Chem., Int. Ed., 2007, 119, 4198–4200 CrossRef.
- G. Ghale and W. M. Nau, Acc. Chem. Res., 2014, 47, 2150–2159 CrossRef CAS.
- A. C. Bhasikuttan, H. Pal and J. Mohanty, Chem. Commun., 2011, 47, 9959–9971 RSC.
- D. A. Ivanov, N. K. Petrov, E. A. Nikitina, M. V. Basilevsky, A. I. Vedernikov, S. P. Gromov and M. V. Alfimov, J. Phys. Chem. A, 2011, 115, 4505–4510 CrossRef CAS.
- R. Khurana, N. Barooah, A. C. Bhasikuttan and J. Mohanty, Org. Biomol. Chem., 2017, 15, 8448–8457 RSC.
- M. E. Aliaga, L. Garcia-Rio, M. Pessego, R. Montecinos, D. Fuentealba, I. Uribe, M. Martin-Pastor and O. Garcia-Beltran, New J. Chem., 2015, 39, 3084–3092 RSC.
- J. Lagona, P. Mukhopadhyay, S. Chakrabarti and L. Isaacs, Angew. Chem., Int. Ed., 2005, 44, 4844–4870 CrossRef CAS.
- K. Kim, N. Selvapalam, Y. H. Ko, K. M. Park, D. Kim and J. Kim, Chem. Soc. Rev., 2007, 36, 267–279 RSC.
- L. Isaacs, Chem. Commun., 2009, 619–629 RSC.
- R. N. Dsouza, U. Pischel and W. M. Nau, Chem. Rev., 2011, 111, 7941–7980 CrossRef CAS.
- H. Dube, M. R. Ams and J. Rebek, J. Am. Chem. Soc., 2010, 132, 9984–9985 CrossRef CAS.
- M. V. S. N. Maddipatla, M. Pattabiraman, A. Natarajan, K. Srivastav, J. T. Mague and V. Ramamurthy, Org. Biomol. Chem., 2012, 10, 9219–9222 RSC.
- L. Zheng, S. Sonzini, M. Ambarwati, E. Rosta, O. A. Scherman and A. Herrmann, Angew. Chem., Int. Ed., 2015, 54, 13007–13011 CrossRef CAS.
- A. C. Bhasikuttan and J. Mohanty, Chem. Commun., 2017, 53, 2789–2809 RSC.
- T. Goel, N. Barooah, M. B. Mallia, A. C. Bhasikuttan and J. Mohanty, Chem. Commun., 2016, 52, 7306–7309 RSC.
- F. Benyettou, K. Nchimi-Nono, M. Jouiad, Y. Lalatonne, I. Milosevic, L. Motte, J.-C. Olsen, N. i. Saleh and A. Trabolsi, Chem.–Eur. J., 2015, 21, 4607–4613 CrossRef CAS.
- N. Barooah, A. Kunwar, R. Khurana, A. C. Bhasikuttan and J. Mohanty, Chem.–Asian J., 2017, 12, 122–129 CrossRef CAS.
- P. Wang, Y. Lin, M. Smith, S. Feng, B. Song, S. Yang and J. Hu, Chem. Commun., 2014, 50, 11950–11953 RSC.
- Y. Mao, K. Liu, G. Lv, Y. Wen, X. Zhu, H. Lan and T. Yi, Chem. Commun., 2015, 51, 6667–6670 RSC.
- E. Masson, X. Ling, R. Joseph, L. Kyeremeh-Mensah and X. Lu, RSC Adv., 2012, 2, 1213–1247 RSC.
- L. Yuan, R. Wang and D. H. Macartney, J. Org. Chem., 2007, 72, 4539–4542 CrossRef CAS.
- M. S. A. Khan, D. Heger, M. Necas and V. Sindelar, J. Phys. Chem. B, 2009, 113, 11054–11057 CrossRef CAS.
- V. Kolman, M. S. A. Khan, M. Babinský, R. Marek and V. Sindelar, Org. Lett., 2011, 13, 6148–6151 CrossRef CAS.
- N. Barooah, J. Mohanty, H. Pal, S. K. Sarkar, T. Mukherjee and A. C. Bhasikuttan, Photochem. Photobiol. Sci., 2011, 10, 35–41 RSC.
- J. Jose and K. Burgess, J. Org. Chem., 2006, 71, 7835–7839 CrossRef CAS.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, 2006 Search PubMed.
- D. V. O'Connor and D. Phillips, Time Correlated Single Photon Counting, Academic Press, 1984 Search PubMed.
- M. J. Frisch, G. W. Trucks, M. Head-Gorden, P. M. W. Gill, M. W. Wong, J. B. Foresman, B. G. Johnson, H. B. Schlegel, M. A. Robb, E. S. Replogle, R. Gomperts, J. L. Andres, K. Rahavachari, J. S. Binkley, C. Gonzalez, R. Martin, L. D. J. Fox, D. J. Defrees, J. Baker, J. J. P. Stewart and J. A. Pople, Gaussian 92, Gaussian Inc., Pittsburgh, PA, 1992 Search PubMed.
- A. K. Vasu and S. Kanvah, Dyes Pigm., 2017, 142, 230–236 CrossRef CAS.
- J. Figueras, J. Am. Chem. Soc., 1971, 93, 3255–3263 CrossRef CAS.
- B. Carlotti, G. Consiglio, F. Elisei, C. G. Fortuna, U. Mazzucato and A. Spalletti, J. Phys. Chem. A, 2014, 118, 7782–7787 CrossRef CAS PubMed.
- H. Agnihotri, A. K. Vasu, V. Palakollu and S. Kanvah, Photochem. Photobiol. Sci., 2015, 14, 2159–2167 RSC.
- N. Barooah, J. Mohanty, H. Pal and A. C. Bhasikuttan, J. Phys. Chem. B, 2012, 116, 3683–3689 CrossRef CAS.
- M. Shaikh, J. Mohanty, P. K. Singh, W. M. Nau and H. Pal, Photochem. Photobiol. Sci., 2008, 7, 408–414 RSC.
- A. L. Koner and W. M. Nau, Supramol. Chem., 2007, 19, 55–66 CrossRef CAS.
- A. Praetorius, D. M. Bailey, T. Schwarzlose and W. M. Nau, Org. Lett., 2008, 10, 4089–4092 CrossRef CAS.
- M. D. Pluth, R. G. Bergman and K. N. Raymond, Science, 2007, 316, 85–88 CrossRef CAS.
- F. Ortega-Caballero, C. Rousseau, B. Christensen, T. E. Petersen and M. Bols, J. Am. Chem. Soc., 2005, 127, 3238–3239 CrossRef CAS.
- D. H. Leung, R. G. Bergman and K. N. Raymond, J. Am. Chem. Soc., 2007, 129, 2746–2747 CrossRef CAS.
- V. Sindelar, K. Moon and A. E. Kaifer, Org. Lett., 2004, 6, 2665–2668 CrossRef CAS.
- X. Lu and E. Masson, Langmuir, 2011, 27, 3051–3058 CrossRef CAS.
- H. Sawai, T. Matsuura, H. Kakiuchi, T. Ohgi, Y. Shiraishi and N. Toshima, Chem. Lett., 2012, 41, 1160–1162 CrossRef CAS.
- J. Pennakalathil, E. Jahja, E. S. Özdemir, Ö. Konu and D. Tuncel, Biomacromolecules, 2014, 15, 3366–3374 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional supporting graphs and method M1. See DOI: 10.1039/c8ra03355b |
| ‡ Authors A. K. V. and R. K. contributed equally. |
| This journal is © The Royal Society of Chemistry 2018 |