A nano perspective behind the COVID-19 pandemic
Camila Pedroso
Silveira†
a,
Andressa da Cruz
Schneid†
a,
Iris Renata Sousa
Ribeiro
ab,
Flávia Elisa
Galdino
ab and
Mateus Borba
Cardoso
 *ab
*ab
aBrazilian Synchrotron Light Laboratory (LNLS), Brazilian Center for Research in Energy and Materials (CNPEM), Postal Code 13083-970, Campinas, Brazil. E-mail: cardosomb@lnls.br
bInstitute of Chemistry (IQ), University of Campinas (UNICAMP), Post Office Box 6154, Postal Code 13083-970, Campinas, SP, Brazil
First published on 4th August 2021
Abstract
The global pandemic scenario has definitely pushed the scientific community to develop COVID-19 vaccines at unprecedented speed. Nevertheless, a worldwide vaccination campaign is still far from being achieved, making the usual precautionary measures as necessary as at the beginning of the outbreak. Many aspects of the SARS-CoV-2 infectious potential and disease severity do not solely rely on interactions at the molecular level but also on physical–chemical parameters that often involve nanoscale effects. Here the SARS-CoV-2 journey to infect a susceptible host is reviewed, focusing on the nanoscale aspects that play a role in the viral infectivity and disease progression. These nanoscale-driven interactions are essential to establish mitigation-related strategies.
Introduction
While new and more transmissible strains of the severe acute respiratory coronavirus 2 (SARS-CoV-2) threaten the world, the scientific community is incessantly trying to unravel molecular aspects that are key to the coronavirus disease 2019 (COVID-19) pathogenesis.1 Molecular findings on how the severe acute respiratory coronavirus 2 (SARS-CoV-2) interacts with the cellular machinery were essential to define decision-making elements to prevent transmission.2,3 For instance, the early finding that the virus enters the cell through the specific binding between its spike protein and the angiotensin-converting enzyme 2 (ACE2) cellular receptor – especially abundant on cells of the nasal mucosa – was key for world health authorities to recommend the use of facemasks.4 Nevertheless, many aspects of viral transmission and infection are not merely related to molecular interactions between viral protein domains and their specific cellular receptors but instead ruled by physical–chemical aspects that often fall in the nanoscale range.Briefly, the SARS-CoV-2 journey to infect a susceptible host starts when the virus is released to the environment within water-based droplets from contaminated respiratory fluids.5 At this stage, transmission is governed by micro-scale aerodynamic effects. Whether viral-bearing droplets will stay suspended in air (as airborne droplets) or follow a ballistic pathway depends on their aerodynamic size, which also plays a role in their deposition pattern in the respiratory tract after they are inhaled.6 At first, most of the viral-bearing droplets settle in the nose, where the virus has to overcome intermolecular and hydrophobic interactions with the mucus layer. The mucus acts as a first physical defence barrier and filtrates foreign particulate matter by affinity and size through pores that favour the passage of nanoscale entities of up to ca. 200 nm.7 This gives the SARS-CoV-2 (approximately 100 nm diameter size) an advantage to cross the hydrogel net and infect epithelial cells – rich in ACE2 receptors.3 Evidently, after infection, the virus initiates a constant cycle of replication and release of new viral particles, which can either be expelled to the environment or infect other cells along the respiratory tract. Reaching the alveoli, the deposition of nanoscale particles is favoured by the small and branched alveolar architecture.8 As the viral infection route moves to the deep lungs, micro-scale elements give place to nano-interactions that become gradually more relevant and pronounced. Deep lung infection down-regulates the secretion of pulmonary surfactant and leads to alveolar collapse since this surfactant is responsible for regulating the lung surface tension to allow expansion and compressing during breathing.9 The SARS-CoV-2 infection in the alveoli induces an increase of tissue permeability, leading to the infiltration of inflammatory macrophages that transit between the lungs and the blood.9 Together with the alveolar collapse, this panorama allows the virus to translocate to endothelial cells and eventually reach the bloodstream,9,10 where nano-parameters rule the viral journey.
The COVID-19 multidisciplinary characteristic opens space for nanomaterial scientists to step in and share a leading role in the pandemic.11 SARS-CoV-2 can be seen as an airborne nanoparticle that finds its target cells through inhalation and travels from the air to the bloodstream with high efficiency.12 In the past decade, the consolidation of bio–nano sciences leveraged the scientific knowledge of how specific nanoparticle physical–chemical characteristics may affect the biological setting.13 Thus, nanoscience can provide the community with cues from the interaction of nanomaterials with the biological machinery, and the behaviour of nanoparticles in the respiratory tract and blood can be paralleled to the SARS-CoV-2 infection route. Herein, we propose to turn the spotlight to physical–chemical and nano-structural aspects of the COVID-19 pandemic for the benefit of better understanding the viral behaviour and the established preventive measures. We describe the viral journey from the early transmission stages, passing through the upper and lower respiratory tracts, finally reaching the blood, and translating nanoparticle studies to the pandemic panorama, focusing on aspects that favour the viral tropism.
Transmission and lung deposition
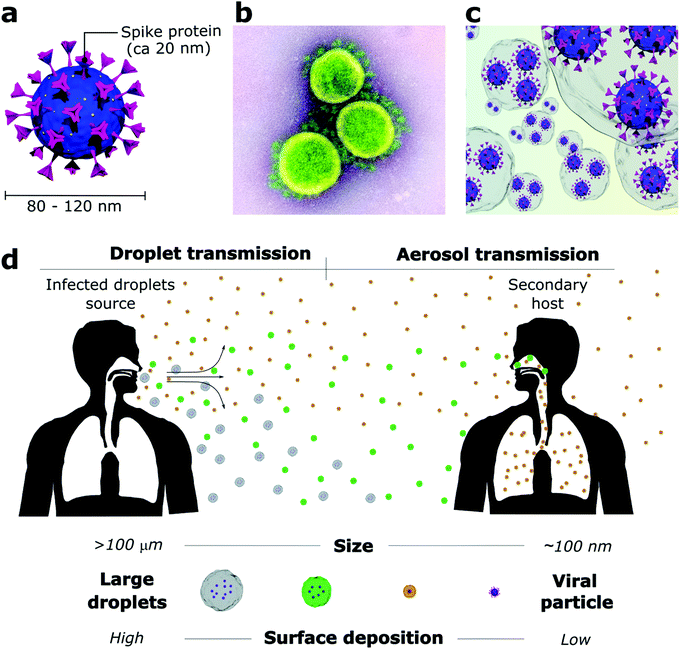
The outbreak of the COVID-19 pandemic has required scientists to quickly figure which viral transmission routes were at stake in an urgent need to find measurements that could rapidly mitigate the SARS-CoV-2 spread. While SARS-CoV-2 particles are only about 80–120 nm2 (Fig. 1a and b), they do not travel through the air as individualized bare entities but rather within large water-based droplets that carry a viral load (Fig. 1c) which are transmitted from person to person through saliva and respiratory fluids generated during coughing, sneezing, talking and even breathing.14–17 These mechanisms of viral shedding produce viral-containing droplets that are conveniently categorized as larger droplets (>10 μm) and smaller droplets or aerosols (<10 μm). Conventionally, air transmission routes are divided according to the droplet size due to their aerodynamic characteristics (Fig. 1d). Specifically for SARS-CoV-2, global health authorities have not clear guidance on the extent of aerosol contribution, and other transmission routes are still considered meaningful. However, mounting evidence suggests that aerosol transmission is responsible for the major part of infections, despite difficulties in sampling and quantifying virus-containing aerosol particles in real-world environments.18Droplets larger than 100 μm (called ballistic droplets) settle rapidly and can directly infect a person within their ballistic pathway or indirectly through contaminated surfaces where they land.16,19 These droplets are the major source of surface contamination as they quickly fall from the emitted air-jet due to gravitational forces and reach the surface before their complete evaporation.19 Droplets from 10 to 100 μm stay longer in the air jet but usually dry before landing on the surface. They are the primary source of direct person-to-person contamination, for example, when someone sneezes or coughs near others.16 Droplets smaller than 10 μm evaporate fast after being released and, as part of the bioaerosol (aerosol enclosed by mucus and saliva residue), remain suspended for prolonged periods and travel much greater distances.17,20 A decrease from 100 μm to 80 μm in droplet diameter increases its reach from 1 to 3 m before landing on a surface.15 Non-ballistic droplets are poorly affected by gravity and are more susceptible to the influence of heat zones, relative humidity, and air currents.17,19 Thus, providing good ventilation to closed environments is mandatory to dilute the droplet and aerosol concentration and decrease transmission rates. Otherwise, the virus-containing aerosol can linger in the air for hours or even be dragged through the whole building by the air-conditioning system.17,21
The controversial debate between scientists and global health authorities became evident when the World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC) launched the first recommendations to minimize the SARS-CoV-2 spread based on the concept that ballistic droplets are the primary source of viral transmission, an idea promoted by Turner in 1941.22 Ballistic droplets land no farther than 2 m from their source, therefore, the 2 m social distancing rule fits appropriately to prevent person-to-person transmission. However, Turner's study overlooked the aerosol contribution to a viral transmission.22,23 In the past few decades, several reports have shown the high aerosol generation by respiratory events and their ability to spread beyond 2 m. Aerosolised viral-loaded droplets can reach people several meters away from the emitter,23,24 evidencing the importance of facemask wearing. Initially, mask-wearing was intended to block person-to-person transmission, and it was recommended only for health professionals. As the COVID-19 outbreak escalated, masks were made mandatory for the general population to prevent transmission in public environments.25 Fortunately, proper mask-wearing (covering nose and mouth without any gaps) is also efficient to protect individuals from spreading or inhaling infected aerosol – contradicting the common belief that the mask fibres would not be able to filtrate small droplets.26,27 Studies have proven that even cloth masks can efficiently block 80% of droplets smaller than 300 nm and more than 90% of the larger ones. The capture of ultrasmall droplets (from 100 nm to 1 μm) happens mainly by mechanical interception. Thus, the droplet interacts with the fibre surface by Brownian motion diffusion when the droplets collide randomly with the fibre.28,29
The droplets act as viral carriers and protect the virus against environmental factors during their release from the host. Mainly composed of water (>95%), droplets start to evaporate as soon as the air jet is exhaled.6,30 Naturally, evaporation is directly affected by relative humidity and temperature, happening faster under high temperatures and low relative humidity conditions.15,30 Therefore, it is expected that large droplets evaporate to become aerosols that remain airborne for more extended periods.20 The viral load in each droplet is proportional to its size when emitted.6 Hence, dehydrated droplets that were initially large can carry high amounts of virions and result in a more efficient viral infection. This is one of the reasons why airborne transmission is so alarming. Moreover, aerosols are more likely to deposit directly in the deep lungs,31 where they avoid the defence mechanisms of the upper respiratory tract and may result in more severe infections. A study that measured the total viral load released by influenza-infected patients showed that 65% of the entire released viral content was concentrated in droplets smaller than 4 μm.32 Another work showed that the dose required for an influenza infection is 100–1000 fold lower for aerosol inoculation than intranasal inoculation.33
In the past few months, researchers have been collecting evidence to prove the major contribution of aerosol transmission in the COVID-19 pandemic. Several cases reported the contamination of individuals without having direct contact with a primary host.34 One of them took place in China, where SARS-CoV-2 infected three families at an air-conditioned restaurant. Video recordings showed no interaction between the families, indicating that the airflow had an essential role in the virus transmission.35 Likewise, SARS-CoV-2 RNA has been detected in air samples from hospital hallways and surroundings as well as in rooms used by COVID-19-positive patients. These findings support the potential for airborne transmission, although the viral viability is rarely checked.36–38 Furthermore, a study that evaluated the impact of mitigations measures on the pandemic outcomes showed that the transmission rates in China, Italy, and New York (USA) were barely affected by social distancing but highly affected by face covering, which corroborates that SARS-CoV-2 might be transmitted predominantly through the airborne route.25,34
It is known that environmental factors (humidity, temperature, and sunlight) can interfere in the viral lifetime. Non-enveloped viruses, in general, are more likely to remain active under high relative humidity levels,39,40 so they preserve structural water molecules that are essential to avoid denaturation of surface proteins. Enveloped viruses (e.g., influenza and coronaviruses), in turn, are more viable in low relative humidity levels (20–30%).39,41 In this condition, the lipidic envelope is more stable and, therefore, can provide greater protection against inactivating agents.40 Temperature can also impact the integrity of viral proteins and genetic material, thus, enveloped viruses tend to be more viable at low temperatures (7–8 °C) compared to high (>30 °C).39 Other environmental factors also affect viral viability, such as UV radiation and air chemical composition.40 UV radiation is widely known for its antimicrobial activity, and it is commonly used to sterilize laboratory settings. However, in natural environments, the UV wavelength window represents just a small portion of the sunlight. It results in a poor effect in viral inactivation, which is also true for artificial white lights.40 Air pollutants interact with the virus in different ways depending on their nature. It has been reported that sulphur and nitrogen dioxides lead to virus deactivation while ozone molecules might act as a virus incubator, maintaining its viability and participating in its transport.40,42
After inhalation, the place where the virus deposits in the lung highly depends on the droplet aerodynamic size, density, inhalation airflow, and airway architecture. Droplets larger than 10 μm are caught early in the nose and throat and are prone to accumulate non-homogeneously. Their deposition is governed by inertial impaction through the upper airways, where the flow is turbulent and has high speed.43,44 Under strong air streamlines, the particle inertia rises and increases the particle-wall impact, resulting in more significant particle entrapment.43,45 Droplets between 2 to 10 μm are mainly governed by gravitational sedimentation and reach the tracheobronchial tree, where the flow is slower and the particle residence time is longer. Conversely, smaller droplets (<1 μm) are driven by Brownian diffusion. Thus, they deposit throughout the entire respiratory tract extension and reach the alveolar region, where the airspeed is near zero.43,44,46 Little is known about the SARS-CoV-2 deposition pattern. However, recent evidence shows that most of the viral load concentrates in the nose and upper respiratory tract at the beginning of infection.47,48 From there, the virus replicates and reaches other respiratory tract sites in a constant host re-infection loop.47 The lung anatomy also plays an essential role in the deposition pattern. Infants’ airways have relatively smaller diameters, directly impacting the particle access to the lungs, i.e., its size naturally selects smaller particles.49,50 This may contribute to the lower COVID-19 severity and infection rates observed in children. Similarly, the initial viral load received by the host is decisive on the disease evolution. A recent study evaluated the infective dose necessary to identify detectable RNA viral units in the lungs after aerosol exposure. The authors show that rats exposed to 25 minutes of the aerosol-containing virus at 36 TCID50 per min (median tissue culture infectious dose per minute) were required to cause COVID-19 infection in mice expressing human ACE2.51 It is pertinent to pinpoint that 1 min of loud talking, for instance, generates at least 1000 virus-containing droplets.38,52
It is relevant to reiterate that alveolar deposition favours smaller particles.8 Therefore, pathogen particles that travel along the respiratory tract within nanoscale droplets or in their individualized form are more likely to infect alveolar pneumocytes. Studies performed with nanoparticles allowed Geiser and Kreyling (2010) to estimate the alveoli deposition rate.8 On average, the number of nanoparticles deposited per cell can go from 6 per day with one single inhalation to 120 per hour with multiple inhalations over time, considering the nanoparticle aerosol concentration at the maximum value (approx. 106 cm−3) possible due to colloidal limitations.8 We can use these calculations to emphasize that time is crucial to SARS-CoV-2 transmission and disease development. The longer a person stays in a contaminated environment, especially indoors, the greater will be his/her initial viral load.
Upper airways
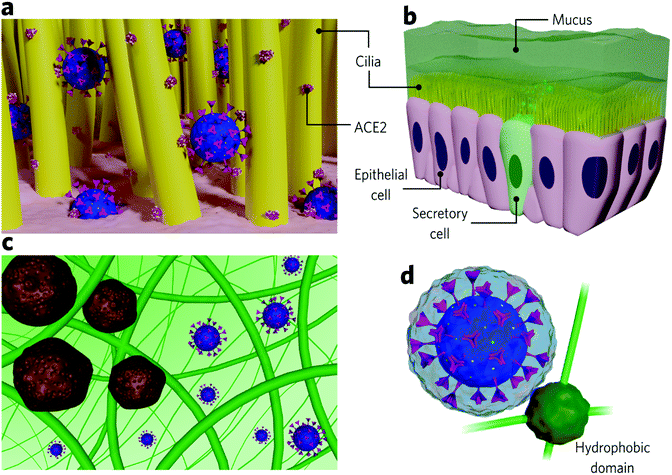
The nose is the most susceptible site for SARS-CoV-2 infection, and, indeed, recent findings show that it is where the virus predominantly accumulates at the initial stages of the disease.27 Virus-containing droplets predominantly deposit in the nose, where ACE2 receptors are more abundant than on cells from the lower airways.3,53 The epithelium lining presents motile cilia responsible for protecting the cells from particulate matter by mucociliary clearance. Some studies show that ACE2 is also highly expressed along the cilia, impairing pathogen clearance during SARS-CoV-2 infection (Fig. 2a).54 ACE2 expression varies among populations, which influences the susceptibility to SARS-CoV-2 and disease severity. Smokers are associated with higher ACE2 expression along the epithelial airway,55 which may be one of the reasons why this group has proven to be more susceptible to COVID-19 complications. Conversely, children are associated with lower ACE2 expression,56,57 and, indeed, they are at a lower risk of infection.Inhaled virions first come upon mucus before reaching the epithelial cells. Mucus covers many of the epithelial surfaces (Fig. 2b) and, in the airways, lines from the nose to the terminal bronchioles58 act as a defence barrier against pathogens. It consists of a heterogeneous and dynamic cross-linked hydrogel that filtrates foreign pathogens and particulates by size exclusion and intermolecular and electrostatic interactions (Fig. 2c).59,60 It is negatively charged due to the high sialic acid and sulfate content and presents small hydrophobic regions composed of highly hydrophobic proteins.7,60 Produced by secretory cells located between epithelial cells, mucus maintains the local hydrated since it comprises about 95% water. The remaining 5% contains mucins and other proteins, DNA, lipids, salts, cells, and cellular debris.59,61 Mucins are glycoproteins of 10–40 MDa, responsible for forming the mucous cross-linked net and conferring it with high viscosity (3–4 orders of magnitude higher than water).59,62,63 The airway mucus is 5–100 μm thick and is constantly renewed (each 10 min for nasal mucus, for example).7,59
Airway epithelial ciliated cells present an apical layer of motile cilia (Fig. 2b) that beat co-ordinately to allow mucus renewal and, consequently, pathogen elimination, which is key to preventing pathogens from getting in contact with underlying epithelial cells.59,64 The mucous hydrogel can protect epithelial cells as long as the pathogen diffusion is slower than the renewal period.7 Pathogen diffusion depends on intermolecular interactions with the hydrogel as well as on its hydrodynamic size compared to the diameters of the pores. Studies that quantitatively determine viral diffusivity in mucus are few. However, it is possible to establish a credible analogy from nanoparticle studies. A given particle experiences high diffusivity in mucus if it is small enough to pass through its pores and if no strong interaction is established (Fig. 2c).60 In this case, the viscosity perceived by the particle is close to the water. The barrier properties are considered more efficient against large viruses due to expected higher viscosities perceived.65,66 The mucus pores size distribution ranges from 20 to 1800 nm, depending on the region. It presents a medium-size around 150 nm in the nose, slowing down the diffusion or immobilizing viruses bigger than this.60 Viruses able to diffuse in the airway mucus and reach epithelial cells are typically about 30–200 nm in size (Rhinoviruses: 15–30 nm; adenovirus: 60–90 nm; SARS-CoV-2:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80–120 nm; influenza virus: 84–170 nm).7,67,68 In general, non-enveloped viruses present higher diffusion rates than enveloped ones mainly due to their smaller sizes.7
80–120 nm; influenza virus: 84–170 nm).7,67,68 In general, non-enveloped viruses present higher diffusion rates than enveloped ones mainly due to their smaller sizes.7
Nanoparticle studies show that those with net neutral surface charges are the most mucus-penetrating, while the mucus efficiently traps positive nanoparticles through strong electrostatic interaction with mucins. Similarly, hydrophobic nanoparticles diffuse slower than hydrophilic ones due to interactions with hydrophobic mucin cores.59,63 Generally, infectious viruses present a negative net charge (isoelectric points approximately between 2 and 9)69 and, therefore, are expected to diffuse in mucus due to electrostatic repulsion with negatively charged mucins. Enveloped viruses (covered by an additional protein-containing lipid bilayer), in most cases, present slower diffusion rates than capsid viruses which are also due to the higher probability of interaction with the hydrophobic mucus domains.7 SARS-CoV-2 is an enveloped virus. Therefore slower diffusion rates would be expected due to possible interactions of its lipid bilayer with mucous hydrophobic domains. Although diffusion rates for SARS-CoV-2 have not been investigated, we should also note that the protein anchored in its lipid bilayer (spike protein) is reasonably glycosylated, which increases its hydrophilicity.70 Moreover, the spike protein is projected about 20 nm outwards the envelope,71,72 which may sterically hinder the interaction of the hydrophobic mucin regions with its lipid bilayer (Fig. 2d).
Once SARS-CoV-2 reaches and infects the mucous-underlying epithelium, it induces several cell dysfunctions, including cilia alterations that result in mucociliary transport impairment.73,74 Mucus overproduction has been observed in COVID-19 patients, and its accumulation in the airways can also impair ciliary movement, which facilitates viral internalisation.64,75 Slower ciliary beat frequency and longer nasal mucociliary clearance times are naturally observed in the elderly, who shows higher infectivity and severity rates of COVID-19.76 The loss of smell reported by some COVID-19 patients may come from epithelial ciliary damage since olfactory receptors accumulate on sensory cilia, which also express the specific receptors for SARS-CoV-2 entry.77,78 Recent findings revealed that SARS-CoV-2 entries into, and is released from, epithelial cells through the apical domain,79 similar to SARS-CoV and other respiratory viruses.80 This means that, after replication, new viral particles are released back to the lumen, from where they may be thrown to the environment through coughing, speaking, or breathing – which is the major reason why facemasks are the most effective way to prevent SARS-CoV-2 transmission. From the lumen, virions can also infect other epithelial cells or reach the alveoli, where they may, in rare severe cases, translocate to the bloodstream and attack other organs.80
Lower airways
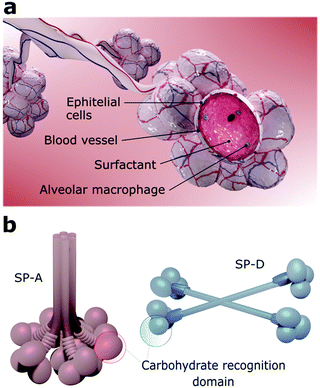
The viruses that can reach the lower airways find a favourable environment to infect alveolar cells since they also express ACE2 receptors.53,81 The alveoli are constituted by a layer of pulmonary surfactants (PS) film at the air–liquid interface, mobile cells (e.g., alveolar macrophages), a highly-differentiated epithelium, and a subepithelial connective tissue that contains blood and lymphatic vessels (Fig. 3a).8 Together, PS, alveolar macrophages, and epithelial cells are defence barriers against SARS-CoV-2 and other pathogens by preventing them from reaching the bloodstream.82 If the virus overcomes all these barriers, it may reach the bloodstream since the distance from the air in the alveolar lumen to the capillary blood flow is less than 400 nm.81PS is a membrane-based lipid–protein system essential for breathing since it regulates the surface tension of the lungs and therefore stabilizes the whole respiratory surface.83,84 It protects the lungs against inhaled particles and microbes, avoiding injuries and infections.85 Approximately 10% of this monolayer consists of proteins and 90% of phospholipids, including the amphiphilic dipalmitoylphosphatidylcholine (DPPC) and neutral lipids like cholesterol.84 Four surfactant proteins (SP) constitute the PS, designated as SP-A, SP-B, SP-C, and SP-D.86,87 SP-A and SP-D are large hydrophilic proteins responsible for the PS metabolism and immune regulation.86,87 Conversely, SP-B and SP-C are small and highly hydrophobic proteins that form and maintain stable surface-active domains at the interface and, therefore, are responsible for keeping the surfactant surface tension.88 Particularly, proteins SP-A and SP-D, also known as pulmonary collectins, bear significant roles in the host immune response. Collectins are characterised by conserved carbohydrate-recognition domains formed by trimeric clusters (Fig. 3b) that present low affinity for single monosaccharides but high affinity for clustered oligosaccharides, usually present in viruses other microorganisms.86,87 Both SP-A and SP-D, for instance, bind to mannose and glucose and poorly to galactose, while SP-A binds preferentially to N-acetylmannosamine and fucose-type glycans. SP-A and SP-D specifically recognise pathogens by using various binding motifs within the carbohydrate-recognition domain. Several studies have shown that these proteins interact with specific cellular receptors to mediate cellular apoptosis to enhance pathogen phagocytosis and activate receptors that induce inflammatory responses.87–89 Recognition of pathogens is, in most cases, mediated by the Ca2+-dependent binding of the C-type lectin domains of SP-A or SP-D to carbohydrate structures on the surface of the microorganisms, which leads to neutralization, agglutination, and enhanced phagocytosis.88,90 A recent analysis of the site-specific glycans of SARS-CoV-2 spike protein revealed an abundance of oligomannose-type glycans, hybrid afucosylated and fucosylated glycans, and complex-type glycans,91 suggesting that SP-A and SP-D might have key roles on SARS-CoV-2 neutralization.
The use of soap and water is the easiest and one of the most cost-effective preventive measures against SARS-CoV-2 infection by contact with contaminated surfaces. Soaps are chemically composed of sodium and potassium salts of saturated or unsaturated amphiphilic long-chain fatty acids that function as surfactant molecules. These molecules penetrate and disrupt the envelope lipid bilayer, releasing the viral content and therefore inactivating the virus.92,93 PS can also inhibit virions that arrive in the alveoli. However, its efficacy depends on the initial viral load. SARS-CoV-2 virions that pass the surfactant layer suppress the production of PS by infecting type II pneumocytes,94–96 which are responsible for the PS synthesis and secretion.97 The drop in PS production makes it difficult for the lungs to keep the alveoli open, leading to their collapse.94 This process comes with a generalised dysfunction of the alveolar cells, including the alveolar macrophages. These cells are a vital part of the innate immune response and express ACE2 receptors. Recent findings show that alveolar macrophages are depleted upon SARS-CoV-2 infection, and inflammatory macrophages massively permeate the damaged tissue.9 This unbalanced cascade is believed to induce a hyperinflammatory state, referred to as cytokine storm, characterised by a hyperproduction of cytokines that lead to significant cytotoxic effects.98,99 It is worth mentioning that advanced age naturally decreases surfactant production, resulting in high probabilities of severe COVID-19 cases in the elderly.100 Thus, exogenous surfactant substitution has proven a valid therapeutic option.101,102 Works focused on SARS-CoV-2 have shown that the early administration of natural lung surfactant decreased extracorporeal membrane oxygenation therapy and ventilation time in patients with severe acute respiratory distress syndrome.92,94,100 Additionally, surfactant therapy can also prevent viral spread. The use of surfactant-based gargle or mouthwash was shown to interfere with the formation of the viral envelope, leading to the inactivation of the virus transmitted via saliva, for example.103
The behaviour of SARS-CoV-2 in the lungs can be better understood when compared with studies performed with lab-synthesized nanoparticles. Recently, a simulation study evaluated the rates of nanoparticle deposition and adsorption in a microfluidic lung-on-a-chip device under several breathing patterns.104 The authors found that physical exercising increases NP lung deposition due to changes in the airflow volume and high breathing frequency. Similarly, breath-holding while smoking also increases nanoparticle deposition.104 The higher deposition rates upon smoking, together with other physiological aspects, may be one of the reasons why smokers are at higher risk of developing severe SARS-CoV-2 infection. Likewise, the return to the gyms should be analysed with extra care since physical activities could lead to the inhalation and deposition of higher viral loads in case of a contaminated environment, especially without proper ventilation.
The interaction between nanoparticles and PS has been considered an alternative strategy to engineer new nanoparticle-based pulmonary drug delivery systems and assess nanotoxicity profiles.105 Model nanoparticles have been extensively applied to understand how they penetrate the pulmonary surfactant layer to reach the underlying target cells.106 In this sense, hydrophobic interactions between the nanoparticle and the pulmonary surfactant proteins and lipids play a decisive role. Hydrophilic nanoparticles are expected to penetrate the monolayer quickly. At the same time, hydrophobic NPs are unlikely to translocate through it due to their interaction with the hydrophobic domains, which often results in their entrapment. Moreover, the strong interaction between hydrophobic NPs and PS may lead to aggregate protrusions, disrupting the lining structure.107,108
Naturally, nanoparticles in contact with PS readily interact with their proteins and lipids to form an adsorbed biomolecular layer referred to as surfactant corona, which dresses the nanostructure with a biological identity and modulates nanoparticle-cell interactions. The surfactant corona formation depends on the nanomaterial size,105 charges,109 chemical functionalization,110 and hydrophobicity.108 The presence of nanoparticles in the alveoli may negatively impact its lining fluid, either by changing its physiological viscosity111 or by depletion of proteins and lipids that come out of the surfactant to adsorb on the nanoparticle surface.109,112 A reduction in hydrophobic proteins, for instance, impairs the PS ability to regulate the alveolar surface tension, which is crucial to reduce the mechanical stress in breathing and thus avoid alveolar collapse.94 The surfactant corona composition also plays a significant role in nanoparticle clearance to prevent them from reaching the microvascular endothelium.110 Specifically, SP-A-enriched corona was shown to enhance cellular uptake of nanoparticles by alveolar macrophages and other phagocytic cells.113 In relation to SARS-CoV-2, studies evaluating the virus interaction with proteins and lipids from the PS could be valuable to understand if any specific biomolecule plays a role in viral neutralization or cellular uptake, which, in turn, could corroborate the understanding of potential therapeutical strategies as well as about the viral pathophysiology.
Reaching the blood and other organs
Once deposited in the alveoli, foreign nanostructures can translocate to the bloodstream.114 The mechanisms by which this process occurs are not completely elucidated. Still, viral or synthetic nanoparticles find a suitable environment to reach the bloodstream after lung deposition, mainly when inflammation is triggered.81,115 For SARS-CoV-2, the immune response begins early after inhalation, when they encounter airway epithelial cells and mucosal immune cells.116 The innate immune system (our first line of defence) discerns self from non-self through evolutionarily-conserved molecular patches that exist on pathogens, called pathogen-associated molecular patterns (PAMPs).117 PAMPs are broadly shared by microbes and include lipopolysaccharides (LPS), glycoproteins, and nucleic acid motifs. Our cells have evolved to encode cellular receptors that recognize PAMPs by pattern recognition receptors (PRRs), which, upon recognition, initiate the upregulation of proinflammatory factors.117 The inflammatory response induces an increase in vascular permeability, and immune cells can translocate to the site of infection. This, in turn, may also accelerate the translocation of viral nanoparticles to the bloodstream.Upon recognition and binding the SARS-CoV-2 spike protein to the ACE2 cellular receptor, its viral RNA – a PAMP – is recognized by PRRs, activating antiviral signalling pathways. Severe cases of SARS-CoV-2 are associated with high levels of proinflammatory cytokines and chemokines that lead to an uncontrolled and generalized inflammatory response (the aforementioned cytokine storm).98,99 This event, together with the alveolar disruption caused by SARS-CoV-2, leads to higher vascular permeability and tissue leakage, favouring the passage of the virus to endothelial cells, which also present ACE2 receptors. The hyperinflammatory state developed in some patients can cause microvascular damage and endothelial dysfunction, which, in rare severe cases, can evolve to systemic hypercoagulability and to thromboembolic complications.118 Once in the blood, SARS-CoV-2 can potentially reach all organs and, considering the ACE2 density in several tissues, it can become a systemic disease and evolve to a cardiovascular, renal, or neurologic condition.
The increase in the endothelial permeability due to inflammation has meaningful consequences for patients with pre-existing respiratory conditions, like asthma,119 which is a risk factor for COVID-19. Asthmatic patients usually present higher inflammatory responses to pathogens, inducing more tissue damage and, consequently, alveolar-blood particle translocation. This has been confirmed in nanoparticle models, in which asthmatic mice presented considerably higher translocation rates after gold nanoparticle inhalation.120 In healthy patients, most inhaled nanoparticles are retained in the lungs, and only a small percentage can reach the bloodstream (0.5–3%).121,122 However, the prevalence of inflammation before nanoparticle inhalation affects pulmonary retention and translocation rates. In a study performed with mice, healthy animals retained more than 80% of inhaled nanoparticles in the lungs while LPS-primed animals retained only 25%. Moreover, the number of nanoparticles found in the spleen of these primed animals was 5–6 fold higher if compared to healthy animals.123 As with synthetic nanoparticles, SARS-CoV-2 translocation to the blood takes place via degenerated lung tissue with acute inflammation98 and, therefore, the results from engineered nanoparticles portray a comparable scenario to SARS-CoV-2. The low particle translocation rates observed for synthetic nanomaterials are analogous to the low percentage of SARS-CoV-2-infected people that present blood-related complications. However, other physiological aspects are implicated in the illness severity.
Nanoparticles have been extensively used as models to boost immune responses.124 It is now broadly accepted that they can interact with receptors from the innate arm.125 Interestingly, nanoparticle interactions with the biological machinery are shape-dependent. Inspired by naturally occurring geometric patterns, nanomaterial scientists have devoted considerable attention to the role of nanoparticle morphology on specific biological responses. Particularly, spiky nanoparticles seem to engage in cellular pathways that trigger higher immune responses than their spherical counterparts, as they have shown to induce higher cytokine levels.126,127 A study comparing the biodistribution of the nanoparticles of distinct geometries revealed that spiky nanoparticles were the only ones to accumulate in the lungs.126 These branched nanoparticles often resemble viral structures. An intriguing aspect of viral nanoparticles is their defined geometric arrangements.12 SARS-CoV-2, for example, presents a spherical envelope anchored with spikes of about 20 nm71,72 (Fig. 1a). Nevertheless, what bio–nano sciences indicate is that morphology matters. However, whether viral geometry plays an evolutionary role in viral tropism is a topic that requires and merits further investigation.
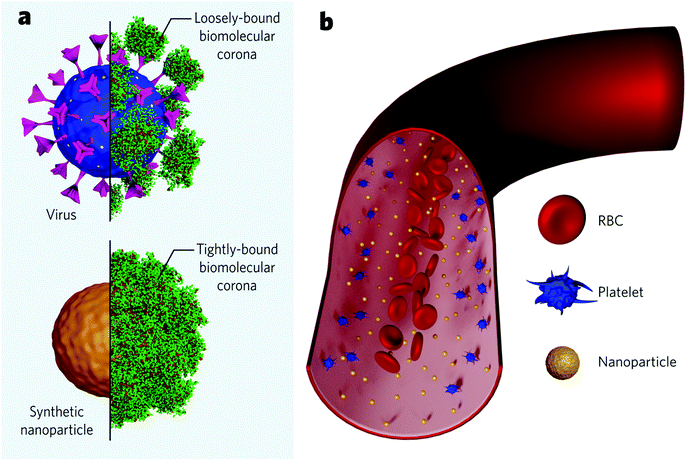
It has been inferred that the differences in the cellular response coming from shaped nanoparticles are due to variations in their protein adsorption profile. Upon contact with blood, synthetic nanoparticles instantly interact with biomolecules from the medium to form an adsorbed layer (so-called biomolecular corona) that confers the nanoparticle an endogenous identity (Fig. 4a). This biomolecular dressing becomes the interface of interaction between the particles and the cellular machinery and often hides the original particle functionalization. Specific proteins commonly present in the corona (e.g., complement factors and immunoglobulins) are believed to tag nanoparticles for clearance – in analogy to the body's immune response to microbial entities. Researchers recently started to investigate the formation of viral protein corona and its implications on viral pathogenesis. Current findings show that viruses behave like synthetic nanoparticles when placed in extracellular environments and associate with proteins from the medium to form a protein corona. Viruses were found to present bio-fluid-dependent protein profiles particular to each virus type, similar to artificial nanoparticles, although the total amount of associated proteins is considerably lower (Fig. 4a). Viral protein corona formed in human plasma was shown to present high amounts of complement proteins and immunoglobulins, accounting for more than 60% of the total protein composition.128 Noteworthily, the corona composition seems to affect viral infectivity. Coronae enriched with immunoglobulins and complement factors could impart infectivity, while coronae enriched with adhesion and anchoring proteins increased viral infectivity.129 These findings suggest that protein corona studies could be explored, for instance, to find cues of the SARS-CoV-2 behaviour in the blood of different patients and, possibly, to consolidate a rapid screening to predict the susceptibility to infection for each subject. To date, it is unknown if higher serum levels of specific proteins are associated with a higher likelihood of developing severe cases. However, some post-infection upregulated proteins have been suggested as biomarkers to predict the COVID-19 severity.130–132 On the other hand, it has been increasingly accepted that some proteins are associated with a lower risk of infection. This is the case for anti-A antibodies from the ABO blood group, which seem to block the interaction of the spike protein with the ACE2 receptor.133,134 In this case, protein corona experiments associated with cellular studies could indicate the nature of this interaction. It is worth mentioning, though, that viral protein corona studies should account for differences compared to those for synthetic nanomaterials. Viruses present considerably fewer proteins in their corona. They seem to be more loosely bound, probably due to the high glycan content on the viral surface (Fig. 4a upper panel), which would act as a shield and inhibit the interaction with specific proteins. Recent findings suggest that the viral targeting ability is kept, and receptor-mediated interactions are not hindered despite the biomolecular dressing.135 Evidently, more studies are needed to understand to what extent the protein corona influences the viral tropism.
Different from most synthetic nanoparticles, viruses count on a structure refined through evolution to guarantee its endurance in the host. Made of the same elemental structures as cells (proteins, lipids, and genetic material), viruses do not suffer, for example, from aggregation due to the high physiological ionic strength.128 Nevertheless, once in the bloodstream, both viruses and synthetic nanoparticles are subjected to flow dynamics. A given particle must first marginate to the endothelium wall to bind to a cellular receptor. In this regard, the margination mechanisms of microscale entities have been widely understood within the field of blood rheology. Red blood cells (RBCs), the major cellular component in the blood (45%), play a pivotal role. Briefly speaking, RBCs are highly deformable and flexible structures which movement dramatically depends on parameters like shear rate, haematocrit concentration, blood viscosity, etc.136 RBCs tend to move radially towards the vessel core, while stiffer particles – such as platelets – are pushed towards the wall in a mechanism called RBC-enhanced diffusivity137,138 (Fig. 4b). This mechanism favours the interaction of stiffer particles with endothelium cells. Nanoscale entities, however, are significantly smaller than RBCs (approximately 100-fold), which makes them less subjected to the RBC-enhanced diffusivity effect and more subjected to Brownian diffusion.139,140 Therefore, nanoparticles tend to uniformly spread across the vessel's entire cross-section (Fig. 4b). This highlights the significance of nanoparticle concentration on targeting rates, since the more nanoparticles populate the vessel, the more nanoparticles get near the walls to bind the receptors. While injected nano-formulations (with a finite number of particles) get diluted along with the bloodstream, viral replication grants the constant addition of new viral particles to the system, increasing the chances of successful binding events.
It has been shown that stiff particles (e.g., 3000 MPa) migrate more towards the vessel wall than soft particles (e.g., 3 MPa) and, therefore, associate more with endothelium cells.141 The Young's modulus of some of the ss-RNA enveloped viruses vary from 200 to 3300 MPa.142,143 This considerable stiffness helps the virus protect its payload – the genetic message – and is beneficial for targeting. Some deformability levels are desirable since more flattened surfaces present larger contact areas and favour multivalent interactions, which increases the endocytosis rate. In this regard, viruses demonstrate remarkable flexibility, and some capsids can deform up to 70% of their diameter.144 Moreover, lipidic structures – like cells and enveloped viruses like SARS-CoV-2 – present some degree of protein diffusivity, where ligands and receptors can move throughout the lipidic layer to maximize the number of bonds and, therefore, increase internalisation rates.145,146 These nano-structural features confer SARS-CoV-2 with incredible advantage to a successful targeting despite the hostile environment dictated by blood.
Conclusions
The combination of scientific areas that are not strictly biological highly benefits a complete overview of the COVID-19 pandemic. For instance, physical–chemical parameters dictate several stages of the transmission route. Thus, aerosol research has much contributed to our understanding of the potential SARS-CoV-2 airborne route. Yet, we lack a complete comprehension of how the virus travels and infects people through the air, which is essential to withhold the total number of cases, especially now with the appearance of more transmissible viral lineages.Biological processes happen at the nanoscale level. To what extent the viral dimension is an evolutionary advantage, we cannot say for sure. However, we can hardly argue against the viral efficiency – especially the SARS-CoV-2. It certainly makes an advantage for nanomaterial scientists, who can virtually design and build any model nanoparticle to interact with the biological machinery, including those inspired by viruses. Given their dimensions, viral nanoparticles and synthetic nanoparticles are governed by the same physical–chemical rules. This makes the knowledge interchange between nanoscience and virology extremely valuable. Some well-established concepts in bio–nano have recently started to be translated into virus research. For instance, the biomolecular corona formed around viruses seems to hold helpful cues regarding viral infectivity.
As the virus infiltrates the body to develop a systemic disease, nanometric aspects become increasingly meaningful – from the ability to trespass the upper and lower airway barriers to the nanoscale interactions with biomolecules and cells. Of course, physiological and molecular factors are also at stake. For instance, interactions with the host immune system and the cell internalisation mechanisms are not to be overlooked. SARS-CoV-2 spike protein targets the ACE2 receptor, which is commonly expressed in various tissues, giving the potential for the virus to infect several organs. Adding to that, as seen in other viruses, SARS-CoV-2 spike protein is projected the spherical viral structure outwards. It presents considerable conformational flexibility in the event of receptor-binding, which happens with astonishing avidity. Taken together, the COVID-19 pandemic can be perceived not only within the molecular frame but also considering nanoscale structural features that wind up favouring viral infectivity.
Author contributions
C. P. S., A. C. S., I. R. S. R., and F. E. G. contributed with writing – original draft and review & editing. M. B. C. contributed with the conceptualization, supervision, and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support of the Fundação de Amparo à Pesquisa do Estado de São Paulo (processes 2015/25406-5, 2017/21318-0, 2019/14007-3, 2018/00763-8 and 2018/09555-9). Mateus B. Cardoso acknowledges CNPq for a productivity fellowship.Notes and references
- Nanomedicine and the COVID-19 vaccines, Nat. Nanotechnol., 2020, 15, 963 Search PubMed.
- C. G. K. Ziegler, S. J. Allon, S. K. Nyquist, I. M. Mbano, V. N. Miao, C. N. Tzouanas, Y. Cao, A. S. Yousif, J. Bals, B. M. Hauser, J. Feldman, C. Muus, M. H. Wadsworth, S. W. Kazer, T. K. Hughes, B. Doran, G. J. Gatter, M. Vukovic, F. Taliaferro, B. E. Mead, Z. Guo, J. P. Wang, D. Gras, M. Plaisant, M. Ansari, I. Angelidis, H. Adler, J. M. S. Sucre, C. J. Taylor, B. Lin, A. Waghray, V. Mitsialis, D. F. Dwyer, K. M. Buchheit, J. A. Boyce, N. A. Barrett, T. M. Laidlaw, S. L. Carroll, L. Colonna, V. Tkachev, C. W. Peterson, A. Yu, H. B. Zheng, H. P. Gideon, C. G. Winchell, P. L. Lin, C. D. Bingle, S. B. Snapper, J. A. Kropski, F. J. Theis, H. B. Schiller, L. E. Zaragosi, P. Barbry, A. Leslie, H. P. Kiem, J. A. L. Flynn, S. M. Fortune, B. Berger, R. W. Finberg, L. S. Kean, M. Garber, A. G. Schmidt, D. Lingwood, A. K. Shalek, J. Ordovas-Montanes, N. Banovich, A. Brazma, T. Desai, T. E. Duong, O. Eickelberg, C. Falk, M. Farzan, I. Glass, M. Haniffa, P. Horvath, D. Hung, N. Kaminski, M. Krasnow, M. Kuhnemund, R. Lafyatis, H. Lee, S. Leroy, S. Linnarson, J. Lundeberg, K. Meyer, A. Misharin, M. Nawijn, M. Z. Nikolic, D. Pe’er, J. Powell, S. Quake, J. Rajagopal, P. R. Tata, E. L. Rawlins, A. Regev, P. A. Reyfman, M. Rojas, O. Rosen, K. Saeb-Parsy, C. Samakovlis, H. Schiller, J. L. Schultze, M. A. Seibold, D. Shepherd, J. Spence, A. Spira, X. Sun, S. Teichmann, F. Theis, A. Tsankov, M. van den Berge, M. von Papen, J. Whitsett, R. Xavier, Y. Xu and K. Zhang, Cell, 2020, 181, 1016–1035.e19 CrossRef CAS PubMed.
- W. Sungnak, N. Huang, C. Bécavin, M. Berg, R. Queen, M. Litvinukova, C. Talavera-López, H. Maatz, D. Reichart, F. Sampaziotis, K. B. Worlock, M. Yoshida, J. L. Barnes, N. E. Banovich, P. Barbry, A. Brazma, J. Collin, T. J. Desai, T. E. Duong, O. Eickelberg, C. Falk, M. Farzan, I. Glass, R. K. Gupta, M. Haniffa, P. Horvath, N. Hubner, D. Hung, N. Kaminski, M. Krasnow, J. A. Kropski, M. Kuhnemund, M. Lako, H. Lee, S. Leroy, S. Linnarson, J. Lundeberg, K. B. Meyer, Z. Miao, A. V. Misharin, M. C. Nawijn, M. Z. Nikolic, M. Noseda, J. Ordovas-Montanes, G. Y. Oudit, D. Pe’er, J. Powell, S. Quake, J. Rajagopal, P. R. Tata, E. L. Rawlins, A. Regev, P. A. Reyfman, O. Rozenblatt-Rosen, K. Saeb-Parsy, C. Samakovlis, H. B. Schiller, J. L. Schultze, M. A. Seibold, C. E. Seidman, J. G. Seidman, A. K. Shalek, D. Shepherd, J. Spence, A. Spira, X. Sun, S. A. Teichmann, F. J. Theis, A. M. Tsankov, L. Vallier, M. van den Berge, J. Whitsett, R. Xavier, Y. Xu, L. E. Zaragosi, D. Zerti, H. Zhang, K. Zhang, M. Rojas and F. Figueiredo, Nat. Med., 2020, 26, 681–687 CrossRef CAS PubMed.
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N. H. Wu, A. Nitsche, M. A. Müller, C. Drosten and S. Pöhlmann, Cell, 2020, 181, 271–280.e8 CrossRef CAS PubMed.
- M. Richard, A. Kok, D. de Meulder, T. M. Bestebroer, M. M. Lamers, N. M. A. Okba, M. Fentener van Vlissingen, B. Rockx, B. L. Haagmans, M. P. G. Koopmans, R. A. M. Fouchier and S. Herfst, Nat. Commun., 2020, 11, 3496 CrossRef CAS PubMed.
- V. Stadnytskyi, C. E. Bax, A. Bax and P. Anfinrud, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 11875–11877 CrossRef CAS PubMed.
- R. A. Cone, Adv. Drug Delivery Rev., 2009, 61, 75–85 CrossRef CAS PubMed.
- M. Geiser and W. G. Kreyling, Part. Fibre Toxicol., 2010, 7, 2 CrossRef PubMed.
- M. Merad and J. C. Martin, Nat. Rev. Immunol., 2020, 20, 355–362 CrossRef CAS PubMed.
- Z. Abassi, Y. Knaney, T. Karram and S. N. Heyman, Front. Immunol., 2020, 11, 1–5 CrossRef PubMed.
- P. A. S. Kinaret, G. del Giudice and D. Greco, Nano Today, 2020, 35, 100945 CrossRef CAS PubMed.
- I. R. S. Ribeiro, R. F. da Silva, C. P. Silveira, F. E. Galdino and M. B. Cardoso, Nano Today, 2021, 36, 101012 CrossRef CAS PubMed.
- I. Lynch and K. A. Dawson, Nano Today, 2008, 3, 40–47 CrossRef CAS.
- W. Chen, N. Zhang, J. Wei, H. L. Yen and Y. Li, Build. Environ., 2020, 176, 106859 CrossRef.
- X. Xie, Y. Li, A. T. Y. Chwang, P. L. Ho and W. H. Seto, Indoor Air, 2007, 17, 211–225 CrossRef CAS PubMed.
- T. P. Weber and N. I. Stilianakis, J. Infect., 2008, 57, 361–373 CrossRef PubMed.
- M. A. Kohanski, L. J. Lo and M. S. Waring, Int. Forum Allergy Rhinol., 2020, 10, 1173–1179 CrossRef PubMed.
- Y. Liu, Z. Ning, Y. Chen, M. Guo, Y. Liu, N. K. Gali, L. Sun, Y. Duan, J. Cai, D. Westerdahl, X. Liu, K. Xu, K. fai Ho, H. Kan, Q. Fu and K. Lan, Nature, 2020, 582, 557–560 CrossRef CAS PubMed.
- C. P. Cummins, O. J. Ajayi, F. V. Mehendale, R. Gabl and I. M. Viola, Phys. Fluids, 2020, 32, 083302 CrossRef CAS PubMed.
- L. Morawska, Indoor Air, 2006, 16, 335–347 CrossRef CAS PubMed.
- C. Chen and B. Zhao, Indoor Air, 2010, 20, 95–111 CrossRef CAS PubMed.
- C. E. Turner, M. W. Jennison and H. E. Edgerton, Am. J. Public Health Nation's Health, 1941, 31, 319–324 CrossRef CAS PubMed.
- N. R. Jones, Z. U. Qureshi, R. J. Temple, J. P. J. Larwood, T. Greenhalgh and L. Bourouiba, BMJ, 2020, 370, m3223 CrossRef PubMed.
- L. Setti, F. Passarini, G. De Gennaro, P. Barbieri, M. G. Perrone, M. Borelli, J. Palmisani, A. Di Gilio, P. Piscitelli and A. Miani, Int. J. Environ. Res. Public Health, 2020, 17, 2932 CrossRef CAS PubMed.
- R. Zhang, Y. Li, A. L. Zhang, Y. Wang and M. J. Molina, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 14857–14863 CrossRef CAS PubMed.
- M. van der Sande, P. Teunis and R. Sabel, PLoS One, 2008, 3, 3–8 CrossRef PubMed.
- K. A. Prather, C. C. Wang and R. T. Schooley, Science, 2020, 368, 1422–1424 CrossRef CAS PubMed.
- A. Konda, A. Prakash, G. A. Moss, M. Schmoldt, G. D. Grant and S. Guha, ACS Nano, 2020, 14, 6339–6347 CrossRef CAS PubMed.
- A. Tcharkhtchi, N. Abbasnezhad, M. Zarbini Seydani, N. Zirak, S. Farzaneh and M. Shirinbayan, Bioact. Mater., 2021, 6, 106–122 CrossRef CAS PubMed.
- Y. Drossinos and N. I. Stilianakis, Aerosol Sci. Technol., 2020, 54, 639–643 CrossRef CAS.
- N. H. L. Leung, D. K. W. Chu, E. Y. C. Shiu, K. H. Chan, J. J. McDevitt, B. J. P. Hau, H. L. Yen, Y. Li, D. K. M. Ip, J. S. M. Peiris, W. H. Seto, G. M. Leung, D. K. Milton and B. J. Cowling, Nat. Med., 2020, 26, 676–680 CrossRef CAS PubMed.
- W. G. Lindsley, F. M. Blachere, R. E. Thewlis, A. Vishnu, K. A. Davis, G. Cao, J. E. Palmer, K. E. Clark, M. A. Fisher, R. Khakoo and D. H. Beezhold, PLoS One, 2010, 5, e15100 CrossRef CAS PubMed.
- R. Tellier, Emerging Infect. Dis., 2006, 12, 1657–1662 CrossRef PubMed.
- S. Tang, Y. Mao, R. M. Jones, Q. Tan, J. S. Ji, N. Li, J. Shen, Y. Lv, L. Pan, P. Ding, X. Wang, Y. Wang, C. R. MacIntyre and X. Shi, Environ. Int., 2020, 144, 106039 CrossRef CAS PubMed.
- F. W. Moses, R. Gonzalez-Rothi and G. Schmidt, Emerging Infect. Dis., 2020, 26, 2298 CrossRef CAS PubMed.
- M. Coccia, Sci. Total Environ, 2020, 729, 138474 CrossRef CAS PubMed.
- X. Ou, Y. Liu, X. Lei, P. Li, D. Mi, L. Ren, L. Guo, R. Guo, T. Chen, J. Hu, Z. Xiang, Z. Mu, X. Chen, J. Chen, K. Hu, Q. Jin, J. Wang and Z. Qian, Nat. Commun., 2020, 11, 1620 CrossRef CAS PubMed.
- J. L. Santarpia, D. N. Rivera, V. L. Herrera, M. J. Morwitzer, H. M. Creager, G. W. Santarpia, K. K. Crown, D. M. Brett-Major, E. R. Schnaubelt, M. J. Broadhurst, J. V. Lawler, S. P. Reid and J. J. Lowe, Sci. Rep., 2020, 10, 12732 CrossRef CAS PubMed.
- J. W. Tang, J. R. Soc., Interface, 2009, 6, S737–S746 Search PubMed.
- S. A. Sattar and M. K. Ijaz, Crit. Rev. Environ. Control, 1987, 17, 89–131 CrossRef.
- J. Shaman and M. Kohn, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 3243–3248 CrossRef CAS PubMed.
- M. M. Menebo, Sci. Total Environ., 2020, 737, 139659 CrossRef CAS PubMed.
- C. Kleinstreuer and Z. Zhang, Annu. Rev. Fluid Mech., 2010, 42, 301–334 CrossRef.
- T. C. Carvalho, J. I. Peters and R. O. Williams III, Int. J. Pharm., 2011, 406, 1–10 CrossRef CAS PubMed.
- M. Rahimi-Gorji, O. Pourmehran, M. Gorji-Bandpy and T. B. Gorji, J. Mol. Liq., 2015, 209, 121–133 CrossRef CAS.
- G. D. Leikauf, S. H. Kim and A. S. Jang, Exp. Mol. Med., 2020, 52, 329–337 CrossRef CAS PubMed.
- B. G. Madas, P. Füri, Á. Farkas, A. Nagy, A. Czitrovszky, I. Balásházy, G. G. Schay and A. Horváth, Sci. Rep., 2020, 10, 22430 CrossRef CAS PubMed.
- R. Wölfel, V. M. Corman, W. Guggemos, M. Seilmaier, S. Zange, M. A. Müller, D. Niemeyer, T. C. Jones, P. Vollmar, C. Rothe, M. Hoelscher, T. Bleicker, S. Brünink, J. Schneider, R. Ehmann, K. Zwirglmaier, C. Drosten and C. Wendtner, Nature, 2020, 581, 465–469 CrossRef PubMed.
- I. Amirav, M. T. Newhouse, S. Minocchieri, J. A. Castro-Rodriguez and K. G. Schüepp, J. Allergy Clin. Immunol., 2010, 125, 1206–1211 CrossRef CAS PubMed.
- K. G. Schüepp, D. Straub, A. Möller and J. H. Wildhaber, J. Aerosol Med., 2004, 17, 153–156 CrossRef PubMed.
- L. Bao, H. Gao, W. Deng, Q. Lv, H. Yu, M. Liu, P. Yu, J. Liu, Y. Qu, S. Gong, K. Lin, F. Qi, Y. Xu, F. Li, C. Xiao, J. Xue, Z. Song, Z. Xiang, G. Wang, S. Wang, X. Liu, W. Zhao, Y. Han, Q. Wei and C. Qin, J. Infect. Dis., 2020, 222, 551–555 CrossRef CAS PubMed.
- J. Ma, X. Qi, H. Chen, X. Li, Z. Zhang, H. Wang, L. Sun, L. Zhang, J. Guo, L. Morawska, S. A. Grinshpun, P. Biswas, R. C. Flagan and M. Yao, Clin. Infect. Dis., 2021, 72, e652–e654 CrossRef CAS PubMed.
- Y. J. Hou, K. Okuda, C. E. Edwards, D. R. Martinez, T. Asakura, K. H. Dinnon, T. Kato, R. E. Lee, B. L. Yount, T. M. Mascenik, G. Chen, K. N. Olivier, A. Ghio, L. V. Tse, S. R. Leist, L. E. Gralinski, A. Schäfer, H. Dang, R. Gilmore, S. Nakano, L. Sun, M. L. Fulcher, A. Livraghi-Butrico, N. I. Nicely, M. Cameron, C. Cameron, D. J. Kelvin, A. de Silva, D. M. Margolis, A. Markmann, L. Bartelt, R. Zumwalt, F. J. Martinez, S. P. Salvatore, A. Borczuk, P. R. Tata, V. Sontake, A. Kimple, I. Jaspers, W. K. O’Neal, S. H. Randell, R. C. Boucher and R. S. Baric, Cell, 2020, 182, 429–446.e14 CrossRef CAS PubMed.
- I. T. Lee, T. Nakayama, C.-T. Wu, Y. Goltsev, S. Jiang, P. A. Gall, C.-K. Liao, L.-C. Shih, C. M. Schürch, D. R. McIlwain, P. Chu, N. A. Borchard, D. Zarabanda, S. S. Dholakia, A. Yang, D. Kim, H. Chen, T. Kanie, C.-D. Lin, M.-H. Tsai, K. M. Phillips, R. Kim, J. B. Overdevest, M. A. Tyler, C. H. Yan, C.-F. Lin, Y.-T. Lin, D.-T. Bau, G. J. Tsay, Z. M. Patel, Y.-A. Tsou, A. Tzankov, M. S. Matter, C.-J. Tai, T.-H. Yeh, P. H. Hwang, G. P. Nolan, J. V. Nayak and P. K. Jackson, Nat. Commun., 2020, 11, 5453 CrossRef CAS PubMed.
- M. Matusiak and C. M. Schürch, Respir. Res., 2020, 21, 252 CrossRef CAS PubMed.
- J. B. Steinman, F. M. Lum, P. P. K. Ho, N. Kaminski and L. Steinman, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 24620–24626 CrossRef CAS PubMed.
- Y. Yao, H. Wang and Z. Liu, Clin. Exp. Allergy, 2020, 50, 1313–1324 CrossRef CAS PubMed.
- N. Sanders, C. Rudolph, K. Braeckmans, S. C. De Smedt and J. Demeester, Adv. Drug Delivery Rev., 2009, 61, 115–127 CrossRef CAS PubMed.
- X. Murgia, B. Loretz, O. Hartwig, M. Hittinger and C.-M. Lehr, Adv. Drug Delivery Rev., 2018, 124, 82–97 CrossRef CAS PubMed.
- J. Leal, H. D. C. Smyth and D. Ghosh, Int. J. Pharm., 2017, 532, 555–572 CrossRef CAS PubMed.
- L. Serra, J. Doménech and N. A. Peppas, Eur. J. Pharm. Biopharm., 2009, 71, 519–528 CrossRef CAS PubMed.
- S. K. Lai, Y.-Y. Wang, D. Wirtz and J. Hanes, Adv. Drug Delivery Rev., 2009, 61, 86–100 CrossRef CAS PubMed.
- B. S. Schuster, J. S. Suk, G. F. Woodworth and J. Hanes, Biomaterials, 2013, 34, 3439–3446 CrossRef CAS PubMed.
- J. V. Fahy and B. F. Dickey, N. Engl. J. Med., 2010, 363, 2233–2247 CrossRef CAS PubMed.
- S. S. Olmsted, J. L. Padgett, A. I. Yudin, K. J. Whaley, T. R. Moench and R. A. Cone, Biophys. J., 2001, 81, 1930–1937 CrossRef CAS PubMed.
- J. T. Huckaby and S. K. Lai, Adv. Drug Delivery Rev., 2018, 124, 125–139 CrossRef CAS PubMed.
- N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang, J. Song, X. Zhao, B. Huang, W. Shi, R. Lu, P. Niu, F. Zhan, X. Ma, D. Wang, W. Xu, G. Wu, G. F. Gao and W. Tan, N. Engl. J. Med., 2020, 382, 727–733 CrossRef CAS PubMed.
- S. Dreschers and C. Adams, Encyclopedia of Virology, Elsevier, 2008, pp. 541–548 Search PubMed.
- B. Michen and T. Graule, J. Appl. Microbiol., 2010, 109, 388–397 CrossRef CAS PubMed.
- A. C. Walls, Y.-J. Park, M. A. Tortorici, A. Wall, A. T. McGuire and D. Veesler, Cell, 2020, 181, 281–292.e6 CrossRef CAS PubMed.
- H. Yao, Y. Song, Y. Chen, N. Wu, J. Xu, C. Sun, J. Zhang, T. Weng, Z. Zhang, Z. Wu, L. Cheng, D. Shi, X. Lu, J. Lei, M. Crispin, Y. Shi, L. Li and S. Li, Cell, 2020, 183, 730–738.e13 CrossRef CAS PubMed.
- S. Klein, M. Cortese, S. L. Winter, M. Wachsmuth-melm and C. J. Neufeldt, Nat. Commun., 2020, 1–8 Search PubMed.
- M. Hubert, G. D. De Melo, S. Levallois, F. Larrous, J. Fernandes, S. Gellenoncourt, O. Gorgette, C. Thouvenot, A. Mallet, S. Gobaa, P. Lledo, M. Lecuit, D. Duffy, V. Michel, O. Schwartz and L. A. Chakrabarti, bioRxiv, 2020 DOI:10.1101/2020.10.06.328369.
- N. Zhu, W. Wang, Z. Liu, C. Liang, W. Wang, F. Ye, B. Huang, L. Zhao, H. Wang, W. Zhou, Y. Deng, L. Mao, C. Su, G. Qiang, T. Jiang, J. Zhao, G. Wu, J. Song and W. Tan, Nat. Commun., 2020, 11, 3910 CrossRef CAS PubMed.
- J. He, S. Cai, H. Feng, B. Cai, L. Lin, Y. Mai, Y. Fan, A. Zhu, H. Huang, J. Shi, D. Li, Y. Wei, Y. Li, Y. Zhao, Y. Pan, H. Liu, X. Mo, X. He, S. Cao, F. Hu, J. Zhao, J. Wang, N. Zhong, X. Chen, X. Deng and J. Chen, Protein Cell, 2020, 11, 680–687 CrossRef CAS PubMed.
- J. C. Ho, K. N. Chan, W. H. Hu, W. K. Lam, L. Zheng, G. L. Tipoe, J. Sun, R. Leung and K. W. Tsang, Am. J. Respir. Crit. Care Med., 2001, 163, 983–988 CrossRef CAS PubMed.
- W. Li, M. Li and G. Ou, FEBS J., 2020, 287, 3672–3676 CrossRef CAS PubMed.
- K. Bilinska, P. Jakubowska, C. S. Von Bartheld and R. Butowt, ACS Chem. Neurosci., 2020, 11, 1555–1562 CrossRef CAS PubMed.
- A. Milewska, A. Kula-Pacurar, J. Wadas, A. Suder, A. Szczepanski, A. Dabrowska, K. Owczarek, A. Marcello, M. Ochman, T. Stacel, Z. Rajfur, M. Sanak, P. Labaj, W. Branicki and K. Pyrc, J. Virol., 2020, 94, e00957 CrossRef CAS PubMed.
- C.-T. K. Tseng, J. Tseng, L. Perrone, M. Worthy, V. Popov and C. J. Peters, J. Virol., 2005, 79, 9470–9479 CrossRef CAS PubMed.
- W. Yang, J. I. Peters and R. O. Williams, Int. J. Pharm., 2008, 356, 239–247 CrossRef CAS PubMed.
- C. Mühlfeld, B. Rothen-Rutishauser, F. Blank, D. Vanhecke, M. Ochs and P. Gehr, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2008, 294, L817–L829 CrossRef PubMed.
- A. Hidalgo, A. Cruz and J. Pérez-Gil, Biochim. Biophys. Acta, Biomembr., 2017, 1859, 1740–1748 CrossRef CAS PubMed.
- K. Yue, X. Sun, J. Tang, Y. Wei and X. Zhang, Int. J. Mol. Sci., 2019, 20, 3281 CrossRef CAS PubMed.
- R. K. Harishchandra, M. Saleem and H.-J. Galla, J. R. Soc., Interface, 2010, 7, S15–S26 CrossRef CAS PubMed.
- U. Kishore, T. J. Greenhough, P. Waters, A. K. Shrive, R. Ghai, M. F. Kamran, A. L. Bernal, K. B. M. Reid, T. Madan and T. Chakraborty, Mol. Immunol., 2006, 43, 1293–1315 CrossRef CAS PubMed.
- J. R. Wright, Nat. Rev. Immunol., 2005, 5, 58–68 CrossRef CAS PubMed.
- A. Haczku, J. Allergy Clin. Immunol., 2008, 122, 861–879 CrossRef CAS PubMed.
- A. Watson, M. J. S. Phipps, H. W. Clark, C.-K. Skylaris and J. Madsen, J. Innate Immun., 2019, 11, 13–28 CrossRef CAS PubMed.
- K. L. Hartshorn, E. C. Crouch, M. R. White, P. Eggleton, A. I. Tauber, D. Chang and K. Sastry, J. Clin. Invest., 1994, 94, 311–319 CrossRef CAS PubMed.
- Y. Watanabe, J. D. Allen, D. Wrapp, J. S. McLellan and M. Crispin, Science, 2020, 369, eabb9983 CrossRef PubMed.
- H. Takano, Med. Hypotheses, 2020, 144, 110020 CrossRef CAS PubMed.
- N. K. Chaudhary, N. Chaudhary, M. Dahal, B. Guragain, S. Rai, R. Chaudhary, K. Sachin, R. Lamichhane-Khadka and A. Bhattarai, Fighting the SARS CoV-2 (COVID-19) Pandemic with Soap, Preprints, 2020, 2020050060 Search PubMed , https://www.preprints.org/manuscript/202005.0060/v2.
- U. Mirastschijski, R. Dembinski and K. Maedler, Front. Med., 2020, 7, 1–4 CrossRef PubMed.
- C. Scheller, F. Krebs, R. Minkner, I. Astner, M. Gil-Moles and H. Wätzig, Electrophoresis, 2020, 41, 1137–1151 CrossRef CAS PubMed.
- Y. Y. Zuo, W. E. Uspal and T. Wei, ACS Nano, 2020, 14, 16502–16524 CrossRef CAS PubMed.
- J. A. Whitsett and T. Alenghat, Nat. Immunol., 2015, 16, 27–35 CrossRef CAS PubMed.
- R. J. Jose and A. Manuel, Lancet Respir. Med., 2020, 8, e46–e47 CrossRef CAS PubMed.
- F. Coperchini, L. Chiovato, L. Croce, F. Magri and M. Rotondi, Cytokine Growth Factor Rev., 2020, 53, 25–32 CrossRef CAS PubMed.
- D. de Moraes, B. V. B. Paiva, S. S. Cury, J. P. A. Junior, M. A. da Silva Mori and R. F. Carvalho, Aging Dis., 2021, 12, 42–49 CrossRef PubMed.
- S. Han and R. K. Mallampalli, Ann. Am. Thorac. Soc., 2015, 12, 765–774 CrossRef PubMed.
- M. Griese, Viral Immunol., 2002, 15, 357–363 CrossRef CAS PubMed.
- K. Pramod, S. Kotta, U. S. Jijith, A. Aravind, M. Abu Tahir, C. S. Manju and H. V. Gangadharappa, Med. Hypotheses, 2020, 143, 110081 CrossRef CAS PubMed.
- S. M. Amin Arefi, C. W. Tony Yang, D. D. Sin and J. J. Feng, Biomicrofluidics, 2020, 14, 044117 CrossRef CAS PubMed.
- Q. Hu, X. Bai, G. Hu and Y. Y. Zuo, ACS Nano, 2017, 11, 6832–6842 CrossRef CAS PubMed.
- S. R. Bonam, N. G. Kotla, R. A. Bohara, Y. Rochev, T. J. Webster and J. Bayry, Nano Today, 2021, 36, 101051 CrossRef CAS PubMed.
- D. Q. Arick, Y. H. Choi, H. C. Kim and Y. Y. Won, Adv. Colloid Interface Sci., 2015, 225, 218–228 CrossRef CAS PubMed.
- S. S. Raesch, S. Tenzer, W. Storck, A. Rurainski, D. Selzer, C. A. Ruge, J. Perez-Gil, U. F. Schaefer and C.-M. Lehr, ACS Nano, 2015, 9, 11872–11885 CrossRef CAS PubMed.
- F. Mousseau and J.-F. Berret, Soft Matter, 2018, 14, 5764–5774 RSC.
- A. J. Thorley, P. Ruenraroengsak, T. E. Potter and T. D. Tetley, ACS Nano, 2014, 8, 11778–11789 CrossRef CAS PubMed.
- L.-P.-A. Thai, F. Mousseau, E. Oikonomou, M. Radiom and J.-F. Berret, ACS Nano, 2020, 14, 466–475 CrossRef CAS PubMed.
- C. Garcia-Mouton, A. Hidalgo, A. Cruz and J. Pérez-Gil, Eur. J. Pharm. Biopharm., 2019, 144, 230–243 CrossRef PubMed.
- C. A. Ruge, J. Kirch, O. Cañadas, M. Schneider, J. Perez-Gil, U. F. Schaefer, C. Casals and C.-M. Lehr, Nanomedicine, 2011, 7, 690–693 CrossRef CAS PubMed.
- G. Hu, B. Jiao, X. Shi, R. P. Valle, Q. Fan and Y. Y. Zuo, ACS Nano, 2013, 7, 10525–10533 CrossRef CAS PubMed.
- M. R. Miller, J. B. Raftis, J. P. Langrish, S. G. McLean, P. Samutrtai, S. P. Connell, S. Wilson, A. T. Vesey, P. H. B. Fokkens, A. J. F. Boere, P. Krystek, C. J. Campbell, P. W. F. Hadoke, K. Donaldson, F. R. Cassee, D. E. Newby, R. Duffin and N. L. Mills, ACS Nano, 2017, 11, 4542–4552 CrossRef CAS PubMed.
- K. P. Y. Hui, M.-C. Cheung, R. A. P. M. Perera, K.-C. Ng, C. H. T. Bui, J. C. W. Ho, M. M. T. Ng, D. I. T. Kuok, K. C. Shih, S.-W. Tsao, L. L. M. Poon, M. Peiris, J. M. Nicholls and M. C. W. Chan, Lancet Respir. Med., 2020, 8, 687–695 CrossRef CAS PubMed.
- T. H. Mogensen, Clin. Microbiol. Rev., 2009, 22, 240–273 CrossRef CAS PubMed.
- J. M. Connors and J. H. Levy, Blood, 2020, 135, 2033–2040 CrossRef CAS PubMed.
- J. J. Meiring, P. J. A. Borm, K. Bagate, M. Semmler, J. Seitz, S. Takenaka and W. G. Kreyling, Part. Fibre Toxicol., 2005, 2, 1–13 CrossRef PubMed.
- A. J. Omlor, D. D. Le, J. Schlicker, M. Hannig, R. Ewen, S. Heck, C. Herr, A. Kraegeloh, C. Hein, R. Kautenburger, G. Kickelbick, R. Bals, J. Nguyen and Q. T. Dinh, Small, 2017, 13, 1603070 CrossRef PubMed.
- J. Lipka, M. Semmler-Behnke, R. A. Sperling, A. Wenk, S. Takenaka, C. Schleh, T. Kissel, W. J. Parak and W. G. Kreyling, Biomaterials, 2010, 31, 6574–6581 CrossRef CAS PubMed.
- C. Schleh, U. Holzwarth, S. Hirn, A. Wenk, F. Simonelli, M. Schäffler, W. Möller, N. Gibson and W. G. Kreyling, J. Aerosol Med. Pulm. Drug Delivery, 2013, 26, 24–30 CrossRef CAS PubMed.
- S. Hussain, J. A. J. Vanoirbeek, S. Haenen, V. Haufroid, S. Boland, F. Marano, B. Nemery and P. H. M. Hoet, Biomed Res. Int., 2013, 2013, 1–6 Search PubMed.
- M. Li, H. Zhou, W. Jiang, C. Yang, H. Miao and Y. Wang, Nano Today, 2020, 35, 101007 CrossRef CAS.
- S. M. Moghimi and D. Simberg, Nano Today, 2017, 15, 8–10 CrossRef CAS PubMed.
- L. Talamini, M. B. Violatto, Q. Cai, M. P. Monopoli, K. Kantner, Ž. Krpetić, A. Perez-Potti, J. Cookman, D. Garry, C. P. Silveira, L. Boselli, B. Pelaz, T. Serchi, S. Cambier, A. C. Gutleb, N. Feliu, Y. Yan, M. Salmona, W. J. Parak, K. A. Dawson and P. Bigini, ACS Nano, 2017, 11, 5519–5529 CrossRef CAS PubMed.
- K. Niikura, T. Matsunaga, T. Suzuki, S. Kobayashi, H. Yamaguchi, Y. Orba, A. Kawaguchi, H. Hasegawa, K. Kajino, T. Ninomiya, K. Ijiro and H. Sawa, ACS Nano, 2013, 7, 3926–3938 CrossRef CAS PubMed.
- A. S. Pitek, A. M. Wen, S. Shukla and N. F. Steinmetz, Small, 2016, 12, 1758–1769 CrossRef CAS PubMed.
- K. Ezzat, M. Pernemalm, S. Pålsson, T. C. Roberts, P. Järver, A. Dondalska, B. Bestas, M. J. Sobkowiak, B. Levänen, M. Sköld, E. A. Thompson, O. Saher, O. K. Kari, T. Lajunen, E. Sverremark Ekström, C. Nilsson, Y. Ishchenko, T. Malm, M. J. A. Wood, U. F. Power, S. Masich, A. Lindén, J. K. Sandberg, J. Lehtiö, A.-L. Spetz and S. E. L. Andaloussi, Nat. Commun., 2019, 10, 2331 CrossRef PubMed.
- A. Saito, K. Kuronuma, K. Moniwa, K. Kodama, S. Takahashi, H. Takahashi and H. Chiba, Res. Sq. Prepr., 2020, 1–17 Search PubMed.
- Y. Yao, J. Cao, Q. Wang, Q. Shi, K. Liu, Z. Luo, X. Chen, S. Chen, K. Yu, Z. Huang and B. Hu, J. Intensive Care, 2020, 8, 49 CrossRef PubMed.
- T. P. Velavan and C. G. Meyer, Int. J. Infect. Dis., 2020, 95, 304–307 CrossRef CAS PubMed.
- M. Franchini, C. Glingani, C. Del Fante, M. Capuzzo, V. Di Stasi, G. Rastrelli, L. Vignozzi, G. De Donno and C. Perotti, Vox Sang., 2020, vox.13003 Search PubMed.
- Y. Wu, Z. Feng, P. Li and Q. Yu, Clin. Chim. Acta, 2020, 509, 220–223 CrossRef CAS PubMed.
- J. Zackova Suchanova, A. Hejtmankova, J. Neburkova, P. Cigler, J. Forstova and H. Spanielova, Bioconjugate Chem., 2020, 31, 1575–1585 CrossRef CAS PubMed.
- A. S. Popel and P. C. Johnson, Annu. Rev. Fluid Mech., 2005, 37, 43–69 CrossRef PubMed.
- J. Tan, A. Thomas and Y. Liu, Soft Matter, 2012, 8, 1934–1946 RSC.
- D. A. Fedosov and G. Gompper, Soft Matter, 2014, 10, 2961–2970 RSC.
- Z. Liu, Y. Zhu, R. R. Rao, J. R. Clausen and C. K. Aidun, Comput. Fluids, 2018, 172, 609–620 CrossRef.
- G. Fullstone, J. Wood, M. Holcombe and G. Battaglia, Sci. Rep., 2015, 5, 1–13 Search PubMed.
- M. Nowak, T. D. Brown, A. Graham, M. E. Helgeson and S. Mitragotri, Bioeng. Transl. Med., 2020, 5, 1–11 Search PubMed.
- R. D. Hartschuh, S. P. Wargacki, H. Xiong, J. Neiswinger, A. Kisliuk, S. Sihn, V. Ward, R. A. Vaia and A. P. Sokolov, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2008, 78, 021907 CrossRef CAS PubMed.
- W. H. Roos, I. L. Ivanovska, A. Evilevitch and G. J. L. Wuite, Cell. Mol. Life Sci., 2007, 64, 1484–1497 CrossRef CAS PubMed.
- W. H. Roos and G. J. L. Wuite, Adv. Mater., 2009, 21, 1187–1192 CrossRef CAS.
- J. W. Myerson, B. Braender, O. Mcpherson, P. M. Glassman, R. Y. Kiseleva, V. V. Shuvaev, O. Marcos-Contreras, M. E. Grady, H.-S. Lee, C. F. Greineder, R. V. Stan, R. J. Composto, D. M. Eckmann and V. R. Muzykantov, Adv. Mater., 2018, 30, 1802373 CrossRef PubMed.
- X. Yi and H. Gao, Nanoscale, 2017, 9, 454–463 RSC.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |