Open Access Article
Open Access ArticleNanoscale antiadhesion properties of sophorolipid-coated surfaces against pathogenic bacteria†
Claire
Valotteau
a,
Niki
Baccile
 b,
Vincent
Humblot
b,
Vincent
Humblot
 c,
Sophie
Roelants
de,
Wim
Soetaert
d,
Christian V.
Stevens
c,
Sophie
Roelants
de,
Wim
Soetaert
d,
Christian V.
Stevens
 f and
Yves F.
Dufrêne
f and
Yves F.
Dufrêne
 *ag
*ag
aLouvain Institute of Biomolecular Science and Technology, Université catholique de Louvain, Croix du Sud, 4-5, bte L7.07.06., B-1348 Louvain-la-Neuve, Belgium. E-mail: Yves.Dufrene@uclouvain.be
bSorbonne Université, Centre National de la Recherche Scientifique, Laboratoire de Chimie de la Matière Condensée de Paris, LCMCP, F-75005 Paris, France
cSorbonne Université, Centre National de la Recherche Scientifique, Laboratoire de Réactivité de Surface, LRS, F-75005 Paris, France
dGhent University, Centre for Industrial Biotechnology and Biocatalysis (InBio.be), Coupure Links 653, B-9000 Gent, Belgium
eBio Base Europe Pilot Plant, Rodenhuizekaai 1, B-9000 Gent, Belgium
fSynBioC Research Group, Department of Green Chemistry and Technology, Faculty of Bioscience Engineering, Ghent University, Coupure links 653, B-9000 Gent, Belgium
gWalloon Excellence in Life sciences and Biotechnology (WELBIO), B-1300 Wavre, Belgium
First published on 25th March 2019
Abstract
A current challenge in nanomedicine is to develop innovative strategies to fight infections caused by multiresistant bacterial pathogens. A striking example is antiadhesion therapy, which represents an attractive alternative to antibiotics to prevent and treat biofilm-associated infections on medical devices. By means of single-cell force nanoscopy, we demonstrate that sophorolipid (SL) biosurfactants feature unusually strong antiadhesion properties against Staphylococcus aureus and Escherichia coli, two nosocomial pathogens involved in catheter-related infections, which represent a major public health problem worldwide. We find that the nanoscale adhesion forces of single bacteria are much weaker on SL monolayers than on abiotic alkanethiol monolayers. The remarkable antifouling efficacy of SL-surfaces is likely to involve repulsive hydration forces associated with sophorose headgroups. We also show that, owing to their surfactant properties, soluble SLs block bacterial adhesion forces towards abiotic surfaces. Collectively, our single-cell experiments demonstrate that sophorolipids exhibit strong and versatile antiadhesion properties, making them promising candidates to design anti-infective biomaterials.
Conceptual insightsMany bacterial pathogens attach to biomaterials such as catheters, and grow to form biofilms. These surface-associated multicellular communities cause infections that are difficult to eradicate. Antiadhesion therapy represents an attractive alternative to antibiotics to prevent and treat biofilm infections. Using single-cell experiments, we show that glycolipids feature unusual high-efficiency and broad-range antiadhesive properties against Gram-positive and Gram-negative pathogens, making them ideally suited to fight biomaterial-associated infections. Our study show that glycosylated substrates, classically used to favor protein and cell interactions, exhibit strong antifouling properties that could be used to design anti-infective biomaterials. We anticipate that force nanoscopy will become an important tool in nanomedicine to test the efficiency of bioinspired antifouling compounds. |
Introduction
Many bacterial pathogens attach to biomaterials such as catheters, and grow to form biofilms.1,2 These surface-associated multicellular communities cause infections that are difficult to treat because bacteria within biofilms are inherently more resistant to antibiotics. Bacterial adhesion to medical materials involves a wealth of molecular interactions, that are either specific (adhesin–ligand interactions) or non-specific (hydrophobic and electrostatic interactions).3An attractive strategy to fight biomaterial-related infections is the use of surface coatings with anti-infective properties, relying either on biocidal or antiadhesive activities.4–7 The advantage of antiadhesion surfaces is that they do not involve the spread of antibacterial compounds in the environment, therefore limiting the development of resistant strains. Effective antiadhesion coatings should be able to inhibit both specific (e.g. lectin-sugar) and non-specific (e.g. hydrophobic) adhesive interactions. End-grafted poly(ethylene glycol) (PEG) chains represent a popular strategy for the design of antifouling biomaterials.7,8 An interesting alternative is the use of carbohydrate compounds, which feature antiadhesion properties, low toxicity and can be produced in large amounts.9,10 Among these, microbial glycolipids are biosurfactants11–13 well-known for their antimicrobial properties,14 which involve both biocidal15,16 and antiadhesive effects.15,17–19
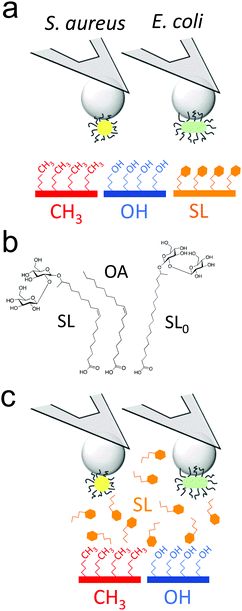
Sophorolipids (SLs) are glycolipids featuring strong antimicrobial action,20 yet the underlying molecular mechanism remain controversial.15,16,21–23 Using single-cell force nanoscopy,24,25 we measure the nanoscale adhesive forces between single bacteria and monolayers of alkanethiols or deacetylated C18:1 acidic, where the carboxyl group has been modified to allow binding onto gold surfaces (Fig. 1a).16,22 We focus on the Gram-positive bacterium Staphylococcus aureus, a nosocomial pathogen frequently implicated in the colonization of biomaterials such as central venous catheters and prosthetic joints,26 as well as on the uropathogenic Gram-negative Escherichia coli, known to be commonly involved in catheter-associated urinary tract infections.5 We compare the passivating activity of SL-surfaces with that of surfaces modified with sophorose-free oleic acid (OA) which mimics only the aliphatic chain of sophorolipids or with a C18:0 SL-derivative having a fully saturated aliphatic chain (SL0) (Fig. 1b). We also show the ability of soluble SLs to inhibit bacterial adhesion forces towards alkanethiol monolayers (Fig. 1c). Our results are of clinical significance as they open up new avenues for the design of infection-resistant biomaterials.
Results and discussion
Sophorolipid-surfaces show strong antiadhesion activity against S. aureus
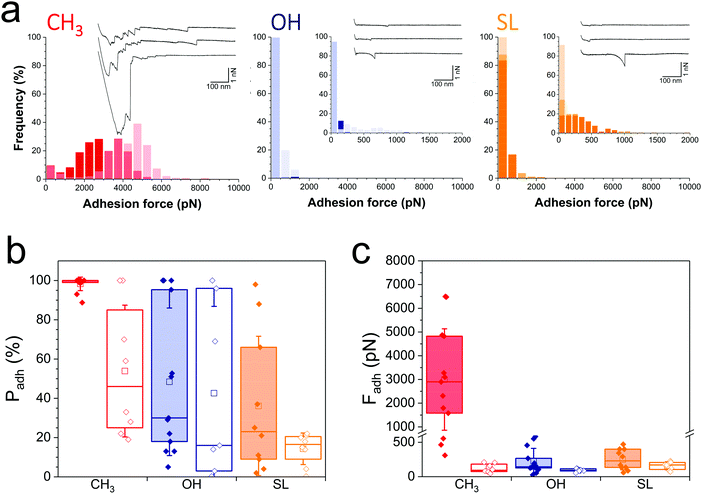
We first sought to compare the nanoscale adhesive interactions between S. aureus and alkanethiol or SL monolayers (Fig. 2). By means of single-cell force spectroscopy,27,28 we investigated the forces between single Newman wild-type (WT) bacteria and hydrophobic methyl-terminated monolayers (water contact angle ∼110°).29 Hydrophobic surfaces were used because hydrophobic interactions represent one of the driving forces for the adhesion of bacterial pathogens to biomaterials.30Fig. 2a (left panel) shows the adhesion forces measured between single S. aureus bacteria and hydrophobic monolayers (three replicates; for data on more cells see Fig. 2b and c, red filled bars). Essentially all force curves (adhesion probability of 98 ± 2%; mean ± s.d. from 13 cells) showed large adhesion events of 3220 ± 2067 pN magnitude (mean ± s.d., n = 2825 adhesive curves from 13 cells). Adhesion profiles generally featured multiple sequential peaks (inset in Fig. 2a, left panel) with long extensions (258 ± 187 nm, n = 2825), suggesting that cell detachment lead to the stretching of large cell surface molecules. We postulated that these adhesion signatures are associated with the binding and unfolding of hydrophobic domains from cell surface proteins. To test this hypothesis, we studied the adhesion of a mutant deficient in sortase A,31 the enzyme responsible for anchoring surface proteins to peptidoglycan (hereafter referred to as Newman ΔsrtA). As can be seen in Fig. 2b and c (red open bars) and Fig. S1a (ESI†) (red histogram), the interaction between Newman ΔsrtA cells and hydrophobic surfaces lead to much lower adhesion probability (54 ± 34%, 8 cells) and adhesion forces (119 ± 64 pN, n = 489 adhesive curves from 8 cells), suggesting that the strong forces of Newman WT bacteria are due to protein-dependent hydrophobic adhesion. Supporting this notion, adhesion between Newman WT cells and hydrophilic hydroxyl-terminated monolayers (water contact angle ∼30°)29 (Fig. 2a, middle panel in blue, and Fig. 2c, blue filled bars) is much weaker (adhesion forces of 226 ± 185 pN, n = 2021 adhesive curves from 13 cells) than on hydrophobic surfaces, and only featured single adhesive events. Even weaker adhesive forces (92 ± 22 pN, n = 320 curves from 7 cells) were observed with Newman ΔsrtA cells (Fig. 2c and Fig. S1, ESI,† blue), suggesting the involvement of surface proteins in hydrophilic adhesion. These results show that bacterial cell surface proteins are engaged in strong hydrophobic interactions with surfaces, while they also modestly contribute to hydrophilic adhesion. We speculate that, under force, hydrophobic protein domains are unfolded and exposed, thereby promoting hydrophobic adhesion. The ∼3000 pN forces are much larger than that those generally associated with single proteins at similar loading rates (∼50–200 pN),32 suggesting they reflect the binding and unfolding of a large number of adhesins. The ∼250 nm extensions correspond to elongated polypeptide chains of ∼700 amino acids, which is in the range expected for staphylococcal adhesins.26 Besides, we note that for both WT and ΔsrtA strains, the adhesion probability on hydrophilic surfaces was substantial (48 ± 38% and 43 ± 43% from 13 WT and 7 ΔsrtA cells, respectively), pointing to the involvement of glycopolymers such as teichoic acids and peptidoglycan in hydrophilic interactions.Analysis of WT bacterial adhesion forces on SL monolayers (water contact angle ∼50°)22 (Fig. 2a, right panel; Fig. 2b and c, orange filled bars) revealed adhesion forces of 249 ± 149 pN (n = 1186 adhesive curves from 10 cells) much lower than on hydrophobic surfaces (Fig. 2b and c, red filled bars), and in the range of those on hydrophilic surfaces (Fig. 2b and c, blue filled bars). In addition, the binding probability of the Newman WT strain was lower on SL surfaces (36 ± 35%) than on hydrophilic surfaces (Fig. 2b). The Newman ΔsrtA strain showed even lower binding probability (Fig. 2b and Fig. S1a, ESI,† orange), which suggests that SL targets not only surface proteins but also, to some extent, other components like teichoic acids and peptidoglycan. Overall, SL-surfaces display weaker adhesion than hydrophobic and hydrophilic surfaces, indicating that the structure of the glycolipid is critical for its antiadhesion activity.
Sophorolipid-surfaces show a broad spectrum of antiadhesion properties
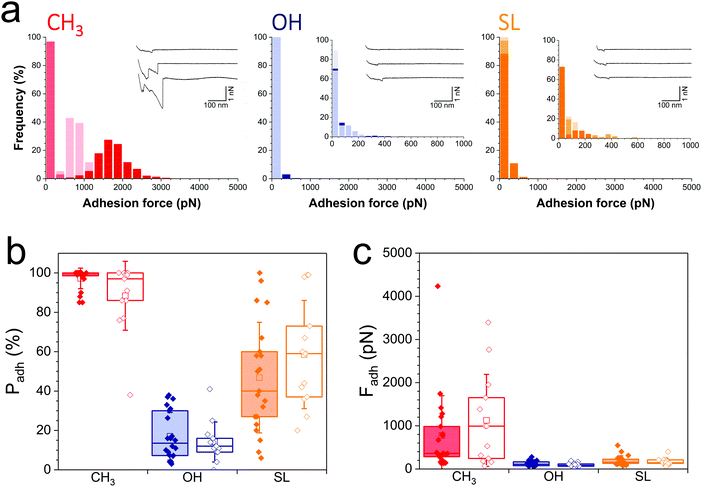
During infection, the Gram-negative uropathogenic E. coli bacterium specifically binds to urinary epithelial cells using FimH, a widely-investigated mannose-binding protein expressed on cell surface pili. Whether FimH also contributes to non-specific adhesion to abiotic surfaces is unclear. Interaction forces between cells overexpressing FimH (E. coli FimH+) and hydrophobic monolayers (Fig. 3a, left panel) featured adhesion events of 774 ± 724 pN magnitude (n = 2956 adhesive curves from 20 cells; Fig. 3c, red filled bar), which, although substantial, are clearly weaker than for S. aureus (Fig. 2). Also, mostly single adhesion peaks were observed (inset in Fig. 3a, left panel), leading us to believe that the molecular origin of hydrophobic binding differed in the two species. The nanoscale adhesive properties of E. coli cells which poorly express FimH (E. coli FimH−, Fig. S1b, ESI†) were similar to those of E. coli FimH+ cells (Fig. 3a), meaning FimH was not engaged in hydrophobic adhesion (Fig. 3b and c, filled vs. open red bars). Adhesion of both FimH+ and FimH− strains was almost abrogated on hydrophilic monolayers (17 ± 12% and 14 ± 10%, from 20 FimH+ and 13 FimH− cells, Fig. 3b filled vs. open blue bars), confirming that strong adhesion is mostly hydrophobic in nature. Of note, rupture distances of E. coli FimH+ cells were rather short on all substrates (379 ± 430 nm and 247 ± 317 nm, n = 3863 and 1154 adhesive curves from 20 cells, for hydrophobic and hydrophilic surfaces, respectively). This is in sharp contrast with the interaction between E. coli FimH+ cells and mannose-coated surface, which showed elongated force plateaus (up to ∼5000 nm in length) reflecting pili unfolding.33 All together, our observations show that FimH is not engaged in non-specific adhesion to surfaces, pointing to the role of other surface components such as outer membrane proteins and lipopolysaccharides.Notably, SL-surfaces featured very weak adhesion forces (Fig. 3a, right panel), without any substantial differences between the FimH+ (192 ± 114 pN, n = 2059 adhesive curves from 20 cells) and FimH− (169 ± 84 pN, n = 1868 adhesives curves from 13 cells) strains (Fig. 3c, orange bars). The binding probability appeared similar for both strains (47 ± 28% and 59 ± 27%, Fig. 3b, orange bars). Hence, while FimH interacts with mannosyl derivatives, it does not specifically bind to sophorose units. Similarly, others sugar-binding proteins such as Concanavalin A were also unable to bind sophorose.34,35
Collectively, these single-cell experiments demonstrate that SL-coated surfaces exhibit a broad spectrum of antiadhesion activity, being able to prevent the adhesion of both Gram-positive and Gram-negative pathogens. SLs interfere with S. aureus adhesion primarily by blocking protein-dependent hydrophobic forces, and to some extent by limiting glycopolymer interactions via e.g. teichoic acids and peptidoglycan. For E. coli, SL surfaces are insensitive to carbohydrate-binding protein FimH, and largely inhibit non-specific hydrophobic forces.
Molecular origin of the passivating effect of sophorolipids
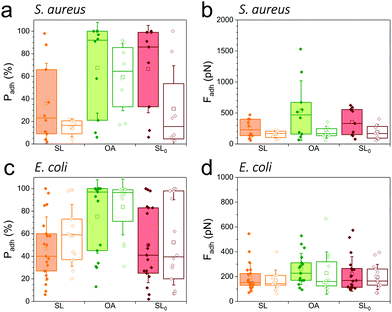
As SLs are amphiphilic molecules, we wondered whether the structure of the carbohydrate group and of the aliphatic chain play important roles in their antiadhesive behavior. To test this, we probed bacterial adhesion forces towards substrates modified with sophorose-free oleic acid (OA) and with a fully saturated aliphatic chain C18:0 sophorolipid-derivative (SL0) (Fig. 4; water contact angles of SL, SL0 and OA monolayers: 48 ± 2°, 71 ± 2° and 55 ± 2°).22 For all strains, the adhesion frequency was much higher on OA-surfaces (68 ± 40%, 59 ± 30%, 75 ± 33%, and 84 ± 25% from 10 S. aureus WT, 8 ΔsrtA cells, 17 E. coli FimH+ and 20 FimH− cells, respectively; Fig. 4a and c) than on SL-surfaces (36 ± 35%, 14 ± 8%, 47 ± 28% and 58 ± 27% from 10, 8, 20 and 13 cells, respectively; Fig. 4a and c), implying that the sophorose group plays an important role in preventing adhesion. Increased, yet less pronounced, adhesion frequency was also observed on SL0-surfaces (Fig. 4a and c),22 which suggests that the conformation of the aliphatic chain also plays some role. All strains showed rather weak adhesion forces on OA-, SL0- and SL-surfaces (Fig. 4b and d), which may be linked to their similar intermediate hydrophilicity.22Our finding that the antiadhesion activity of SL-surfaces is mainly controlled by the sophorose unit emphasizes the role of the structural and conformational properties of the sugar moiety. We speculate that hydration forces associated with the sophorose headgroup largely contribute to repel bacteria. Hydration repulsion results from the structuring of a thin layer of water on polar surfaces, preventing them to come into tight contact.36 Forces between membranes made of zwitterionic phospholipids feature strong hydration repulsion and water structuring effects which result from the large electric dipoles of the headgroups.37 Uncharged glycolipids show hydration forces as well, though they are weaker as the headgroups only possess small electric dipoles (hydroxyl groups).37 The repelling properties of SLs may also involve repulsive forces associated with oligo(ethylene oxide) chains. Surfaces terminated with short chains (n = 2 to 6 units) of oligo(ethylene oxide) strongly prevent protein adsorption.38,39 The ability to resist fouling has been linked to the high degree of solvation and disorder of the (ethylene glycol) chains. We thus suggest that the antifouling behavior of SL-surfaces involve water-mediated repulsions between SL and cell surfaces.
Soluble sophorolipids inhibit bacterial adhesion to abiotic surfaces
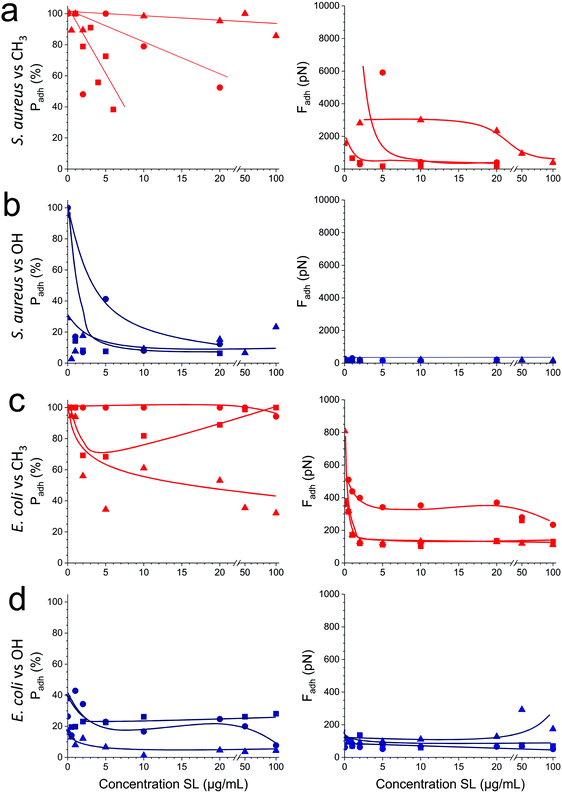
As soluble SLs have been shown to disrupt bacterial biofilms,21 we wondered if SLs in solution can efficiently inhibit the interactions of S. aureus and E. coli with hydrophobic and hydrophilic surfaces. Although there were substantial variations from one cell to another, we found that addition of the glycolipid at a concentration ranging from 1 to 100 μg/ml generally reduced the adhesion probability (Fig. 5, left) and adhesion force (Fig. 5, right) of both strains. SLs showed stronger blocking capacity on S. aureus (Fig. 5a and b) than on E. coli (Fig. 5c and d). Initially, the former strain exhibited stronger adhesiveness, which decreased as the concentration of soluble SLs increased. The antiadhesion activity was more pronounced on hydrophobic surfaces (in red) than on hydrophilic ones (in blue). Interestingly, forces observed on all surfaces at high SL concentration were in the range of forces measured on SL-modified surfaces (∼200 pN). These results show that soluble SLs can be efficiently used to reduce bacterial adhesion on a broad range of materials, regardless of their hydrophobicity. In line with the amphiphilic nature of glycolipids, we propose that the mechanism of action involves a surfactant effect whereby SL molecules accumulate at the interfaces to lower the cell–water and substrate–water interfacial tensions.Conclusions
Antiadhesion therapies against bacterial pathogens are traditionally based on the use of soluble inhibitors targeting the bacteria-host interface.17,40,41 In the prototypical uropathogenic E. coli, a high-affinity inhibitory mannoside targeting FimH was shown to inhibit intestinal colonization by the pathogens, suggesting they could markedly reduce the rate of urinary tract infections.42 Although valuable, these strategies are usually highly specific against a given pathogen, and thus do not exhibit a large spectrum of action. We have shown here that SL glycolipids feature unusual high-efficiency and broad-range antiadhesive properties against Gram-positive and Gram-negative pathogens, making them ideally suited to fight biomaterial-associated infections. SL-surfaces represent an innovative potential application of so-called glycoarrays, i.e. glycosylated substrates that are classically used to favor and/or study the interaction of carbohydrates with proteins and cells.43,44 What we discovered here is that SL-based glycoarrays have strong antifouling properties. We believe that the unexpected high efficacy of SLs as antifouling surfaces involves repulsive forces between the sophorose headgroups and cell surface components, originating from hydration forces such as in phospholipid membranes, and from solvation/disorder forces as in oligo(ethylene oxide) chains. In addition, soluble SLs strongly block bacterial adhesion to solid surfaces, owing to their remarkable surfactant and detergent properties. We anticipate that force nanoscopy will become an important tool in nanomedicine to test the efficiency of bioinspired antifouling compounds.Earlier work has shown that SL-surfaces have biocidal properties.16,22 However, in these experiments bacteria were incubated with SL-surfaces for several hours, whereas here antiadhesive effects were measured within the first seconds of cell–substrate contact. These observations lead us to propose a two step mechanism for the antibacterial activity of SLs, both of which relying on their surfactant properties: at short timescales, antiadhesive effects dominate, both when glycolipids are immobilized and in solution, while the biocidal action comes into play at longer timescales.
Our study is of clinical significance as it offers new prospects for the design of bio-derived antifouling materials. As catheter-associated infections caused by S. aureus ad E. coli represent a common medical problem worldwide, SL-based coatings hold promise for developing innovative anti-infective catheters and other implanted biomaterials. Unlike antibiotics, SLs show a broad range of activities over a wide range of timescales, they are produced at low cost in large amounts, have low toxicity and high biocompatibility, and are not expected to contribute to bacterial resistance.
Methods
Chemicals and surface preparation
Cysteamine (cys), N-hydroxysuccinimide (NHS), 1-(3-(dimethylamino)propyl)-N-ethylcarbodiimide hydrochloride (EDC) and oleic acid (OA) were obtained from Sigma. All solvents were reagent-grade and were used without any further purification. Water was purified with a Milli-Q system (ELGA LabWater). Deacetylated acidic cis C18 sophorolipids (SL) were provided by Bio Base Europe Pilot Plant (Batch 5 in ref. 45). Deacetylated acidic fully saturated C18 sophorolipids (SL0) was derived from the previous one using a Pd-catalyzed hydrogenation reaction described in ref. 46.Sophorolipids (SL, SL0) and oleic acid (OA) were grafted onto flat polycrystalline gold substrates via a self-assembled monolayer of short aminothiols, as described previously.22 Briefly, glass coverslips coated with a thin layer of gold by vacuum evaporation were immersed in an ethanol solution containing 10 mM of cysteamine (cys) for 3 h, rinsed with ethanol, and dried with N2. Substrates and cantilevers were then immersed for 3 h in a solution containing 50 mg mL−1 sophorolipids whose carboxylic acid termination having been previously activated into a succinimide ester using a mixture of EDC and NHS (molar ratio NHS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EDC = 1
EDC = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). Finally, after incubation, sonication and successive rinsing in ultrapure water and ethanol were carried out to remove non-covalently grafted reactants before drying under a flow of N2. These coatings were previously characterized by infrared spectroscopy, X-ray photoelectron spectroscopy and quartz-crystal microbalance.16,22 All these techniques confirm that the glycolipids were efficiently attached to the surface via amide bonds, forming a layer of around 30 Å.
2). Finally, after incubation, sonication and successive rinsing in ultrapure water and ethanol were carried out to remove non-covalently grafted reactants before drying under a flow of N2. These coatings were previously characterized by infrared spectroscopy, X-ray photoelectron spectroscopy and quartz-crystal microbalance.16,22 All these techniques confirm that the glycolipids were efficiently attached to the surface via amide bonds, forming a layer of around 30 Å.
Hydrophobic and hydrophilic substrates were prepared by immersing gold-coated substrates in ethanol solutions containing 1 mM 1-dodecanethiol (Sigma-Aldrich; 98%) or 1 mM 11-mercapto-1-undecanol (Sigma-Aldrich; 97%) overnight, rinsing them with ethanol, and drying them under N2.
Bacterial strains and growth conditions
Wild-type (WT) S. aureus strain Newman and its mutant (ΔsrtA) deficient in sortase A, the enzyme responsible for anchoring CWA proteins to peptidoglycan, were cultured in Trypticase soy broth (TSB) overnight at 37 °C under agitation, then washed twice in prewarmed TSB and grown again at 37 °C to exponential phase (until an optical density at 600 nm of 0.3 was reached). E. coli strains UTI89 WT (FimH−) and the FimH UTI89 strain engineered for continuous fimbriation (FimH+) were cultivated overnight in lysogeny broth (LB) at 37 °C under agitation. Before AFM experiments, cells were harvested by centrifugation, washed twice in phosphate-buffered saline (PBS), and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in PBS.
100 in PBS.
Single-cell force spectroscopy
Colloidal probes were obtained by attaching a single silica microsphere (6.1 μm in diameter; Bangs Laboratories) with a thin layer of UV-curable glue (NOA 63; Norland Edmund Optics) onto triangular tipless cantilevers (NP-O10; Bruker) and using a NanoWizard III AFM System (JPK Instruments). These colloidal probes were immersed for 1 h in Tris-buffered saline (pH 8.5) containing 4 mg mL−1 dopamine hydrochloride (Sigma), rinsed in Tris-buffered saline (pH 8.5), and used directly for cell-probe preparation. The nominal spring constant of the colloidal probe cantilever was ∼0.06 N m−1, as determined by the thermal noise method.For cell probe preparation, 50 μL of a diluted cell suspension was deposited into a Petri dish where the functionalized surfaces were glued on double tape at a distinct location, and the system was filled with 3 mL of PBS. The colloidal probe was brought into contact with an isolated bacterium and retracted to attach the bacterial cell; proper attachment of the cell on the colloidal probe was checked using optical microscopy. The cell probe was then moved towards the surfaces to measure cell-surface interactions. Force curves were recorded at room temperature using an applied force of 0.25 nN, a constant approach–retraction speed of 1.0 μm s−1, and a contact time of 100 ms. For inhibition with soluble SLs experiments, the sophorolipids were added at increasing concentrations from 1 μg mL−1 up to 100 μg mL−1. Data were analyzed using the Data Processing software from JPK Instruments. Adhesion force and distance rupture histograms were obtained by calculating the maximum adhesion force and rupture distance of the last peak for each curve. The adhesion probability is calculated as the percentage of curves displaying adhesive events over the total number of recorded curves.
Author contributions
C. V., N. B., V. H. and Y. F. D. designed the experiments, analyzed the data and wrote the article. C. V. collected the data. S. R., C. S. and W. S. produced and purified the glycolipids. All authors discussed the results and commented the article.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
Work at the Université catholique de Louvain was supported by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement No [693630]), the National Fund for Scientific Research (FNRS), the FNRS-WELBIO (Grant no. WELBIO-CR-2015A-05), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Y. F. D. is a Research Director at the FNRS. We thank Joan Geoghegan and Stéphane Vincent for providing us with bacterial strains.References
- J. W. Costerton, P. S. Stewart and E. P. Greenberg, Science, 1999, 284, 1318–1322 CrossRef CAS PubMed.
- R. Kolter and E. P. Greenberg, Nature, 2006, 441, 300–302 CrossRef CAS PubMed.
- H. J. Busscher, W. Norde and H. C. Van Der Mei, Appl. Environ. Microbiol., 2008, 74, 2559–2564 CrossRef CAS PubMed.
- D. Campoccia, L. Montanaro and C. R. Arciola, Biomaterials, 2013, 34, 8533–8554 CrossRef CAS PubMed.
- D. M. Siddiq and R. O. Darouiche, Nat. Rev. Urol., 2012, 9, 305–314 CrossRef CAS PubMed.
- H. J. Busscher, H. C. van der Mei, G. Subbiahdoss, P. C. Jutte, J. J. van den Dungen, S. A. Zaat, M. J. Schultz and D. W. Grainger, Sci. Transl. Med., 2012, 4, 153rv110 Search PubMed.
- I. Francolini, C. Vuotto, A. Piozzi and G. Donelli, APMIS, 2017, 125, 392–417 CrossRef PubMed.
- D. L. Elbert and J. A. Hubbell, Annu. Rev. Mater. Sci., 1996, 26, 365–394 CrossRef CAS.
- M. Morra, Biomacromolecules, 2005, 6, 1205–1223 CrossRef CAS PubMed.
- V. Gadenne, L. Lebrun, T. Jouenne and P. Thebault, Colloids Surf., B, 2013, 112, 229–236 CrossRef CAS PubMed.
- S. S. Cameotra and R. S. Makkar, Curr. Opin. Microbiol., 2004, 7, 262–266 CrossRef CAS PubMed.
- P. Singh and S. S. Cameotra, Trends Biotechnol., 2004, 22, 142–146 CrossRef CAS PubMed.
- L. R. Rodrigues and J. A. Teixeira, Adv. Exp. Med. Biol., 2010, 672, 75–87 CrossRef CAS.
- M. Ines and G. Dhouha, Carbohydr. Res., 2015, 416, 59–69 CrossRef CAS PubMed.
- A. d. J. Cortés-Sánchez, H. Hernández-Sánchez and M. E. Jaramillo-Flores, Microbiol. Res., 2013, 168, 22–32 CrossRef PubMed.
- C. Valotteau, I. M. Banat, C. A. Mitchell, H. Lydon, R. Marchant, F. Babonneau, C.-M. Pradier, N. Baccile and V. Humblot, Colloids Surf., B, 2017, 157, 325–334 CrossRef CAS PubMed.
- I. Ofek, D. L. Hasty and N. Sharon, FEMS Immunol. Med. Microbiol., 2003, 38, 181–191 CrossRef CAS PubMed.
- S. Kesel, A. Mader, P. Seeberger, O. Lieleg and M. Opitz, Appl. Environ. Microbiol., 2014, 80, 5911–5917 CrossRef CAS PubMed.
- A. L. Flores-Mireles, J. N. Walker, M. Caparon and S. J. Hultgren, Nat. Rev. Microbiol., 2015, 13, 269 CrossRef CAS PubMed.
- E. I. P. Delbeke, M. Movsisyan, K. M. Van Geem and C. V. Stevens, Green Chem., 2016, 18, 76–104 RSC.
- M. A. Díaz De Rienzo, P. Stevenson, R. Marchant and I. M. Banat, FEMS Microbiol. Lett., 2016, 363, fnv224 CrossRef PubMed.
- C. Valotteau, C. Calers, S. Casale, J. Berton, C. V. Stevens, F. Babonneau, C.-M. Pradier, V. Humblot and N. Baccile, ACS Appl. Mater. Interfaces, 2015, 7, 18086–18095 CrossRef CAS PubMed.
- C. Pontes, M. Alves, C. Santos, M. H. Ribeiro, L. Gonçalves, A. F. Bettencourt and I. A. Ribeiro, Int. J. Pharm., 2016, 513, 697–708 CrossRef CAS PubMed.
- J. Xiao and Y. F. Dufrene, Nat. Microbiol., 2016, 1, 16186 CrossRef CAS PubMed.
- Y. F. Dufrene, ACS Nano, 2017, 11, 19–22 CrossRef CAS PubMed.
- T. J. Foster, J. A. Geoghegan, V. K. Ganesh and M. Hook, Nat. Rev. Microbiol., 2014, 12, 49–62 CrossRef CAS PubMed.
- A. Beaussart, S. El-Kirat-Chatel, R. M. A. Sullan, D. Alsteens, P. Herman, S. Derclaye and Y. F. Dufrêne, Nat. Protoc., 2014, 9, 1049–1055 CrossRef CAS PubMed.
- A. Beaussart, S. El-Kirat-Chatel, P. Herman, D. Alsteens, J. Mahillon, P. Hols and Y. F. Dufrêne, Biophys. J., 2013, 104, 1886–1892 CrossRef CAS PubMed.
- D. Alsteens, E. Dague, P. G. Rouxhet, A. R. Baulard and Y. F. Dufrêne, Langmuir, 2007, 23, 11977–11979 CrossRef CAS PubMed.
- R. J. Doyle, Microbes Infect., 2000, 2, 391–400 CrossRef CAS PubMed.
- S. K. Mazmanian, G. Liu, E. R. Jensen, E. Lenoy and O. Schneewind, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 5510–5515 CrossRef CAS PubMed.
- D. J. Muller, J. Helenius, D. Alsteens and Y. F. Dufrene, Nat. Chem. Biol., 2009, 5, 383–390 CrossRef PubMed.
- A. Beaussart, M. Abellán-Flos, S. El-Kirat-Chatel, S. P. Vincent and Y. F. Dufrêne, Nano Lett., 2016, 16, 1299–1307 CrossRef CAS PubMed.
- I. J. Goldstein, R. N. Iyer, E. E. Smith and L. L. So, Biochemistry, 1967, 6, 2373–2377 CrossRef CAS PubMed.
- R. N. Iyer and I. J. Goldstein, Immunochemistry, 1973, 10, 313–322 CrossRef CAS PubMed.
- V. A. Parsegian and T. Zemb, Curr. Opin. Colloid Interface Sci., 2011, 16, 618–624 CrossRef CAS.
- M. Kanduc, A. Schlaich, A. H. de Vries, J. Jouhet, E. Marechal, B. Deme, R. R. Netz and E. Schneck, Nat. Commun., 2017, 8, 14899 CrossRef CAS PubMed.
- K. L. Prime and G. M. Whitesides, J. Am. Chem. Soc., 1993, 115, 10714–10721 CrossRef CAS.
- K. Prime and G. Whitesides, Science, 1991, 252, 1164–1167 CrossRef CAS PubMed.
- A. M. Krachler and K. Orth, Virulence, 2013, 4, 284–294 CrossRef PubMed.
- J. A. Geoghegan, T. J. Foster, P. Speziale and Y. F. Dufrene, Trends Microbiol., 2017, 25, 512–514 CrossRef CAS PubMed.
- C. N. Spaulding, R. D. Klein, S. Ruer, A. L. Kau, H. L. Schreiber, Z. T. Cusumano, K. W. Dodson, J. S. Pinkner, D. H. Fremont, J. W. Janetka, H. Remaut, J. I. Gordon and S. J. Hultgren, Nature, 2017, 546, 528–532 CrossRef CAS PubMed.
- L. K. Mahal, Chem. Biol., 2004, 11, 1602–1604 CrossRef CAS PubMed.
- N. Laurent, J. Voglmeir and S. L. Flitsch, Chem. Commun., 2008, 4400–4412, 10.1039/B806983M.
- P. Dhasaiyan, P. Le Griel, S. Roelants, E. Redant, I. N. Van Bogaert, S. Prevost, B. L. Prasad and N. Baccile, ChemPhysChem, 2017, 18, 643–652 CrossRef CAS PubMed.
- A. S. Cuvier, J. Berton, C. V. Stevens, G. C. Fadda, F. Babonneau, I. N. Van Bogaert, W. Soetaert, G. Pehau-Arnaudet and N. Baccile, Soft Matter, 2014, 10, 3950–3959 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9nh00006b |
| This journal is © The Royal Society of Chemistry 2019 |