A separable nanodevice enables multilayer imaging of diverse biomarkers for precise diagnosis†
Sha
Yu
 a,
Yao
Sun
b,
Jingyi
Cai
a,
Yao
Sun
b,
Jingyi
Cai
 a,
Yuanzhen
Zhou
a,
Yuanzhen
Zhou
 *a and
Jun-Jie
Zhu
*a and
Jun-Jie
Zhu
 *ab
*ab
aSchool of Chemistry and Chemical Engineering, Xi’an University of Architecture and Technology, Xi’an 710055, China. E-mail: zhouyuanzhen@xauat.edu.cn
bState Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, China. E-mail: jjzhu@nju.edu.cn
First published on 21st February 2023
Abstract
An acid-driven separable nanodevice was designed for multilayer imaging of diverse biomarkers with different spatial distributions in living cells. The proposed nanodevice can simultaneously perform in situ imaging of the intracellular microRNAs and extracellular pH, affording a new approach to develop a precise imaging system for disease diagnosis.
Accurate diagnosis of cancer is of great significance for its early prevention and ultimate intervention. The abnormal expression of some biomarkers, including extracellular factors,1 surface membrane proteins,2 enzymes and microRNAs (miRNAs),3,4 is often directly associated with the occurrence and progression of cancer. Therefore, the precise quantification and monitoring of these cancer-associated biomarkers in living cells will hold great promise for the early diagnosis and progression prediction of cancer. So far, fluorescence imaging as a noninvasive in situ analysis technology with high sensitivity, high temporal and spatial resolution, can reflect the real dynamic change of these biomarkers in living cells without complex sample pretreatment,5,6 which offers an appealing approach for visual evaluation of cancer progression. In particular, with the development of DNA nanotechnology, numerous fluorescent nanoprobes based on enzyme-free signal amplification strategies, including hybridization chain reaction (HCR) and catalytic hairpin assembly (CHA), have been devised for the highly sensitive imaging and detection of intracellular biomarkers with low abundance.7,8
It has been found that the progression of one kind of cancer is typically accompanied by the simultaneous changes of multiple biomarkers. For instance, microRNA-21 (miR-21) and microRNA-141 (miR-141) are all abnormally expressed in breast cancer.9 Cardiac troponin I (cTnI), N-terminal prohormone of brain natriuretic peptide (NTProBNP) and neutrophil gelatinase-associated lipocalin (NGAL) have been demonstrated to be used as biomarkers for cardiorenal syndrome (CRS).10 Thus, the accuracy and reliability of pathological information obtained only from single target detection are always limited. By contrast, simultaneous analysis of multiple biomarkers in living cells is vital to understand their roles in cancer progression and improve the accuracy of early diagnosis.11 In this regard, numerous strategies for simultaneous in situ imaging of biomarkers in cancer cells have been developed. Tang and co-workers first proposed a multicolor fluorescence nanoprobe for simultaneous imaging of three types of cancer-related mRNAs in living cells, which can offer more comprehensive and reliable information for cancer diagnosis and effectively prevent false positive results.12 Moreover, various in situ amplification strategies based on DNA nanotechnology and fluorescent signal enhancement systems have been demonstrated to be an effective means for improving the sensitivity of intracellular multiple target simultaneous imaging.13,14
As compared to the simultaneous imaging of multiple targets with the same spatial distribution in living cells, simultaneous imaging of diverse biomarkers located at different parts of cells from the cell membrane to the cytoplasm or nucleus can not only accurately localize the lesion site of the origin of cancer, but also acquire multilayer information about the physiological status of cancer cells in real time and provide more accurate information for cancer early diagnosis and drug evaluation.15 For instance, a deformable satellite nanocapsule has achieved simultaneously quantitative detection of both telomerase activity in the cytoplasm and the degree of DNA methylation in the nucleus, and also provides a new approach for drug evaluation based on the relationship of these two biomarkers.16 The multilayer imaging results of cytoplasmic survivin mRNA and transmembrane glycoprotein mucin 1 (MUC1) in the cell membrane have also been visualized by the multicolor DNA nanoprobe, which can realize more precise cancer diagnosis and metastasis monitoring.17 In addition, the extracellular abnormal factors, especially the extracellular pH, are proposed to be one of the significant indications of malignant tumors, which plays crucial roles in physiological and pathological processes, including tumor metastasis and chemotherapy resistance.18,19 Therefore, the precise and real-time monitoring of the dynamic change of extracellular pH can provide critical information for an in-depth understanding of the pathological processes and early diagnosis of cancer.
Herein, we have designed a separable i-motif tethered deoxyribonucleic acid-gold particle (I-DNA-Au) nanodevice based on the like-catalytic hairpin assembly (LCHA) reaction for simultaneous imaging of extracellular pH and intracellular microRNA-125a (miR-125a). As shown in Scheme 1, the I-DNA-Au nanodevice mainly involves two modules: the A/C hybrid that is formed by hybridizing hairpin A (A) and FAM-labelled strand C (FAM-C), and the B/I hybrid by the composition of the TAMRA, cholesterol-labelled i-motif sequence (I) and the hairpin B (B), all of them are immobilized on the same gold nanoparticles (AuNPs) via Au–S covalent bonds and the initial fluorescence is all quenched. Under the acidic tumor microenvironment (pH 6.2–6.8), the conformation of the i-motif sequence can be reorganized by partial hybridization of cytosine and protonated cytosine (C–H·C+),20 resulting in the separation of the I-DNA-Au nanodevice into a quadruple-helix structure and the DNA-Au nanodevice. The quadruple-helix structure can be inserted into the cell membrane by cholesterol via hydrophobic interaction, consequently imaging the extracellular pH via the recovered TAMRA fluorescence. Whereas, the DNA-Au nanodevice will be internalized into the MDA-MB-231 cells in virtue of the excellent cellular uptake ability of AuNPs and activated by the target miR-125a for in situ imaging in living cells with high sensitivity via the LCHA reaction. Briefly, the miR-125a first interacts with the toehold on A (domain I) and forms a complex A/C/miR-125a, resulting in the exposure of the active site on A (domain II). Then the B promptly binds to domain II (A/C/miR-125a/B) and triggers the strand displacement reaction to form the A/B hybrid and the aggregated AuNPs, leading to the release of miR-125a and C as well as the recovery of FAM fluorescence. The recovered fluorescence is used for in situ imaging of miR-125a, while the regenerated miR-125a continues to initiate the next new cycle for signal amplification. Therefore, the proposed nanodevice can be employed for the simultaneous imaging of extracellular pH and intracellular miRNAs, which holds remarkable potential in engineering multidimensional bioimaging systems for visual discrimination of different cell lines and precise clinical diagnosis.
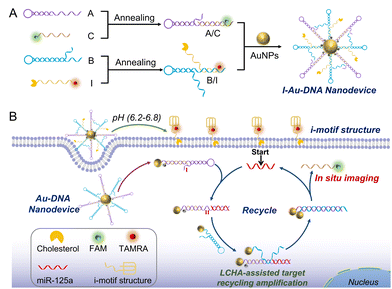 | ||
| Scheme 1 (A) The construction of the acidic-driven separable I-DNA-Au nanodevice. (B) The I-DNA-Au nanodevice for multilayer imaging of extracellular pH and intracellular miR-125a. | ||
The feasibility of the LCHA reaction is first confirmed by the native polyacrylamide gel electrophoresis (PAGE). As shown in Fig. 1(A), the DNA sequences of A, B, C and the miR-125a are used as controls (lanes 1, 2, 3, and 4, respectively). The lagged band can be clearly observed after the A hybridizes with C (that is A/C hybrid, lane 5) and B (that is A/B hybrid, lane 8), respectively. After incubation of the A/C hybrid with B, there are no new bands, indicating that these two DNA components are inactive in the absence of the target and the designed DNA probes have little signal leakage. In addition, the A/C hybrid and B have similar base pairs, thus the mixture still exhibits one band in lane 6. In contrast, upon further addition of miR-125a, a new band of A/B hybrid can be observed (lane 7). These results demonstrate that the designed DNA probes based on the LCHA reaction are successful, which will provide a feasible way to fabricate the DNA-Au nanodevice with high sensitivity. Moreover, the pH responsiveness of the I-DNA-Au nanodevice is assessed in PBS buffer via fluorescence spectroscopy (Fig. S1, ESI†). As shown in Fig. 1(B), the fluorescence intensity of TAMRA gradually increases with decreasing pH from 7.4 to 6.0, indicating that TAMRA-I can be released by acid stimulation and the I-DNA-Au nanodevice possesses excellent pH responsiveness. This result is also verified by circular dichroism (CD) spectra (Fig. S2, ESI†).
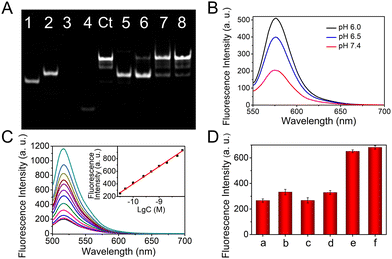 | ||
Fig. 1 (A) 12% native PAGE characterization of DNA-Au nanodevice probes (detailed rearrangement process and description see ESI†): lane 1, A; lane 2, B; lane 3, C; lane 4, miR-125a; lane control (Ct), A/B (annealing); lane 5, A/C; lane 6, A/C + B; lane 7, A/C + B + miR-125a; lane 8, A/B. (B) The fluorescence spectra of the I-DNA-Au nanodevice in PBS buffer with diverse pHs. (C) Fluorescence spectra of the nanodevice in the presence of miR-125a at different concentrations. (inset) Corresponding calibration curve of fluorescence intensity versus the logarithm of miR-125a concentration, where the regression equation is FL = 276.67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C + 3165.68 (R2 = 0.99). (D) The selective evaluation of this nanodevice. The a is blank, the concentration of miR-125a is 1 nM, while the concentration of the other miRNAs is 10 nM. Error bars represent standard deviations from three repeated measurements. C + 3165.68 (R2 = 0.99). (D) The selective evaluation of this nanodevice. The a is blank, the concentration of miR-125a is 1 nM, while the concentration of the other miRNAs is 10 nM. Error bars represent standard deviations from three repeated measurements. | ||
The fabrication of the I-DNA-Au nanodevice is characterized by UV-Vis spectroscopy, as shown in Fig. S3A (ESI†), the as-prepared AuNPs exhibit a typical UV-Vis absorption spectrum with a maximum wavelength at 521 nm. Remarkably, a distinct absorption peak at 260 nm is observed and the characteristic peak of AuNPs is red-shifted to 525 nm after the A/C and B/I immobilization on the AuNPs, indicating the successful fabrication of the I-DNA-Au nanodevice. Next, the reaction process is investigated by using transmission electron microscopy (TEM) monitoring the morphology and dispersity of AuNPs (Fig. S3B, ESI†). Obviously, the I-Au-DNA nanodevice shows better dispersity compared with the bare AuNPs due to the DNA modification. After incubation with miR-125a, it is clear to observe the aggregation of AuNPs, whereas the miR-21 has almost no impact on the dispersion of AuNPs, which is in accordance with the results recorded in dynamic light scattering (DLS) analysis (Fig. S3C, ESI†). All of these results demonstrate the feasibility of the constructed nanodevice, which can only be activated by the target miRNA, thus effectively avoiding the nonspecific signal and improving the accuracy of the analysis.
Then, the analysis performance of the proposed nanodevice for miR-125a is investigated by fluorescence spectroscopy. The reaction time of LCHA is optimized as 2 h (Fig. S4, ESI†). As displayed in Fig. 1(C), a gradually increased fluorescence intensity at 516 nm can be clearly observed as the miR-125a concentration increased from 10 pM to 10 nM, and the detection limit is calculated to be 8.5 pM (S/N = 3, see ESI†). In addition, the excellent selectivity is demonstrated by the treatment of the nanodevice with miRNA-125b (miR-125b, b), miR-21 (c), one-base mismatched miR-125a (d), a mixture of miRNAs (e) and miR-125a (f) (Fig. 1(D) and Fig. S5, ESI†). In addition, the stability is also verified by monitoring the TAMRA and FAM fluorescence intensity after the nanodevice is incubated with 10% fetal bovine serum (FBS) for different times (Fig. S6, ESI†). All of these results demonstrate that the designed I-DNA-Au nanodevice has excellent responsiveness to pH and target miRNA, which will provide a robust nanoplatform for in situ imaging of the extracellular pH and intracellular miRNAs and precision biological analysis.
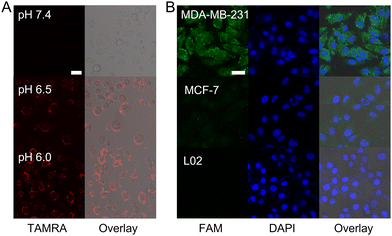
Next, the imaging performance of the I-DNA-Au nanodevice in living cells is evaluated via confocal laser scanning microscopy (CLSM). The cytotoxicity of this nanodevice against the target MDA-MB-231 cells, human breast cancer cells (MCF-7 cells) and human normal liver cells (L02 cells) is first assessed using the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay, which exhibits more than 90% cell viability even when the concentration of the nanodevice reached 8 nM, thus demonstrating the excellent biocompatibility of this nanodevice (Fig. S7, ESI†). The CLSM imaging results demonstrate that the cholesterol-modified nanodevice has excellent anchoring performance on the cell membrane and the modification of cholesterol has no effect on the fluorescence of TAMRA, thus establishing the feasibility of the nanodevice for extracellular pH sensing (Fig. S8, ESI†). Thereafter, the pH responsiveness in living cells of this nanodevice was explored by monitoring the red fluorescence intensity in MDA-MB-231 cells that treated with different pH buffers. As shown in Fig. 2(A), a significant increase of fluorescence intensity on the cell membrane is observed with the extracellular pH ranging from 7.4 to 6.0, which indicates that the designed i-motif sequence can sense the minimal changes in pH, thus achieving in situ imaging of the extracellular pH with high sensitivity.
In addition, the specific response of the nanodevice to intracellular target miRNA is investigated by using CLSM as well (Fig. 2(B)). After the nanodevice is internalized into the MDA-MB-231 cells, the strong green fluorescence of FAM can be recovered under the trigger of miR-125a. A very weak fluorescence is generated in MCF-7 cells due to the low expression level of miR-125a. Whereas, there is no fluorescence in the negative control group of L02 cells. These results demonstrate that the nanodevice has excellent specificity for the target, which can discriminate the different cancer cell lines by imaging the expression level of target miRNAs. Moreover, the MDA-MB-231 cells are treated with miR-125a and the antisense miR-125a (that is complementary to miR-125a) to up-regulate and down-regulate the miR-125a expression level, respectively, followed by incubation with the nanodevice for in situ imaging (Fig. S9, ESI†). Compared to the untreated group, the fluorescence intensity is increased in the miR-125a-treated group and decreased in the antisense miR-125a-treated one. Therefore, this nanodevice can be used to monitor the change of gene expression level in cancer cells due to its high sensitivity via LCHA-assisted target recycling.
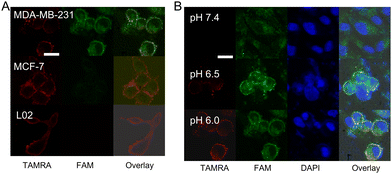
The feasibility of this nanodevice for simultaneous imaging of these two biomarkers with different spatial distributions is further explored (Fig. 3(A)). After treatment of the target MDA-MB-231 cells with this nanodevice in mediums at pH 6.5, an intense TAMRA fluorescence and FAM fluorescence appeared in the same single cell, indicating that the nanodevice can be activated by the targets located at different parts of cells and generate non-interfering fluorescent signals. In addition, this nanodevice is also used to treat MCF-7 cells and L02 cells under the same conditions, respectively. An intense red fluorescence is observed in both MCF-7 cells and L02 cells, indicating the imaging of the extracellular pH by using this nanodevice only depended on the pH, and not on the cell line. However, only a weak green fluorescence is observed in MCF-7 cells and the FAM fluorescence signal is hardly observed in L02 cells. All of these results indicate that the designed nanodevice has the capacity to simultaneously image the extracellular pH and intracellular miR-125a, which can provide multilayer information for distinguishing different cancer cell lines and more precise cancer diagnosis.
Furthermore, the specific response location of the nanodevice in MDA-MB-231 cells is explored by staining the nucleus with 4′,6-diamidino-2-phenylindole (DAPI) for the colocalization experiment (Fig. 3(B)). The red fluorescence of TAMRA is observed at the cell membrane and its intensity increased gradually with the decrease of the pH from 7.4 to 6.0, which is consistent with the response curve in PBS buffer. Whereas, the strong green fluorescence of FAM is observed at the nuclear periphery, indicating that miR-125a is predominantly distributed in the cytoplasm of MDA-MB-231 cells.21 The specificity of the nanodevice for simultaneous imaging in living cells is confirmed in MCF-7 cells and L02 cells (Fig. S10, ESI†). All of these CLSM imaging results demonstrate that the designed nanodevice can not only in situ image targets with different spatial distributions in living cells, but also sense the subtle changes of extracellular pH at the cellular level with high sensitivity.
In summary, we developed an acid-driven separable I-DNA-Au nanodevice based on the pH-sensitive i-motif structure and LCHA-assisted target amplification strategy for in situ multilayer imaging of diverse biomarkers with different locations in a single living cell. The proposed nanodevice could achieve the simultaneous imaging of extracellular pH and cytoplasmic miR-125a in MDA-MB-231 cells, which solved the challenge of simultaneous analysis of different targets in the tumor microenvironment and cytoplasm, and provided multilayer information of spatial distribution and expression levels for different biomarkers. Thus, we believe that the designed nanodevice will serve as a powerful tool for the simultaneous analysis of various biomarkers in living cells and precise disease diagnosis.
This research was funded by the Young Scientists Fund of the National Natural Science Foundation of China (22104118), the National Natural Science Foundation of China (22274125) and the State Key Laboratory of Analytical Chemistry for Life Science (SKLACLS2204).
Conflicts of interest
There are no conflicts to declare.Notes and references
- S. Ling, X. Yang, C. Li, Y. Zhang, H. Yang, G. Chen and Q. Wang, Angew. Chem., Int. Ed., 2020, 59, 7219–7223 CrossRef CAS PubMed.
- C. Liu, X. Xu, B. Li, B. Situ, W. Pan, Y. Hu, T. An, S. Yao and L. Zheng, Nano Lett., 2018, 18, 4226–4232 CrossRef CAS PubMed.
- S. Cai, T. Pataillot-Meakin, A. Shibakawa, R. Ren, C. L. Bevan, S. Ladame, A. P. Ivanov and J. B. Edel, Nat. Commun., 2021, 12, 3515–3527 CrossRef CAS PubMed.
- A. Git, H. Dvinge, M. Salmon-Divon, M. Osborne, C. Kutter, J. Hadfield, P. Bertone and C. Caldas, RNA, 2010, 16, 991–1006 CrossRef CAS PubMed.
- H. Xiao, W. Zhang, P. Li, W. Zhang, X. Wang and B. Tang, Angew. Chem., Int. Ed., 2020, 59, 4216–4230 CrossRef CAS PubMed.
- W. Zhu, M. Kang, Q. Wu, Z. Zhang, Y. Wu, C. Li, K. Li, L. Wang, D. Wang and B. Z. Tang, Adv. Funct. Mater., 2020, 31, 2007026 CrossRef.
- Y. Zhang, Z. Chen, Y. Tao, Z. Wang, J. Ren and X. Qu, Chem. Commun., 2015, 51, 11496–11499 RSC.
- Z. Jiang, H. Wang, X. Zhang, C. Liu and Z. Li, Anal. Methods, 2014, 6, 9477–9482 RSC.
- H. Xu, Z. Zhang, Y. Wang, X. Zhang, J.-J. Zhu and Q. Min, Anal. Chem., 2022, 94, 6329–6337 CrossRef CAS PubMed.
- Y. Su, S. Xu, J. Zhang, X. Chen, L.-P. Jiang, T. Zheng and J.-J. Zhu, Anal. Chem., 2019, 91, 864–872 CrossRef CAS PubMed.
- S. Li, Q. Li, W. Chen, Z. Song, Y. An, P. Chen, Y. Wu, G. Wang, Y. He and Q. Miao, Small, 2022, 18, e2201334 CrossRef PubMed.
- N. Li, C. Chang, W. Pan and B. Tang, Angew. Chem., Int. Ed., 2012, 51, 7426–7430 CrossRef CAS PubMed.
- P. Yuan, X. Mao, S. S. Liew, S. Wu, Y. Huang, L. Li and S. Q. Yao, ACS Appl. Mater. Interfaces, 2020, 12, 57695–57709 CrossRef CAS PubMed.
- W. Zhou, D. Li, R. Yuan and Y. Xiang, Anal. Chem., 2019, 91, 3628–3635 CrossRef CAS PubMed.
- R.-C. Qian, Y. Cao, L.-J. Zhao, Z. Gu and Y.-T. Long, Angew. Chem., Int. Ed., 2017, 56, 4802–4805 CrossRef CAS PubMed.
- S. Huang, C. Ma, J. Lin, W. Wang, Y. Xu, X. Wu, J.-R. Zhang and J.-J. Zhu, CCS Chem., 2020, 2, 1231–1244 Search PubMed.
- P. Gao, R. Wei, Y. Chen, X. Liu, J. Zhang, W. Pan, N. Li and B. Tang, Anal. Chem., 2021, 93, 13734–13741 CrossRef CAS PubMed.
- A. I. Hashim, X. Zhang, J. W. Wojtkowiak, G. V. Martinez and R. J. Gillies, NMR Biomed., 2011, 24, 582–591 CrossRef PubMed.
- A. Morandi and S. Indraccolo, Biochim. Biophys. Acta, 2017, 1868, 1–6 CAS.
- S. Modi, S. M. G., D. Goswami, G. D. Gupta, S. Mayor and Y. Krishnan, Nat. Nanotechnol., 2009, 4, 325–330 CrossRef CAS PubMed.
- B. Fu, W. Liu, C. Zhu, P. Li, L. Wang, L. Pan, K. Li, P. Cai, M. Meng, Y. Wang, A. Zhang, W. Tang and M. An, Int. J. Biol. Sci., 2021, 17, 3104–3117 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cc05825a |
| This journal is © The Royal Society of Chemistry 2023 |