An electrochemical biosensor for simultaneous detection of breast cancer clinically related microRNAs based on a gold nanoparticles/graphene quantum dots/graphene oxide film†
Chammari
Pothipor
abc,
Jaroon
Jakmunee
 acd,
Suwussa
Bamrungsap
acd,
Suwussa
Bamrungsap
 e and
Kontad
Ounnunkad
e and
Kontad
Ounnunkad
 *acdf
*acdf
aDepartment of Chemistry, Faculty of Science, Chiang Mai University, Chiang Mai 50200, Thailand. E-mail: suriyacmu@yahoo.com; kontad.ounnunkad@cmu.ac.th
bThe Graduate School, Chiang Mai University, Chiang Mai, 50200, Thailand
cCenter of Excellence for Innovation in Chemistry, Faculty of Science, Chiang Mai University, Chiang Mai, 50200, Thailand
dResearch Center on Chemistry for Development of Health Promoting Products from Northern Resources, Chiang Mai University, Chiang Mai, 50200, Thailand
eNational Nanotechnology Center (NANOTEC), National Science and Technology Development Agency (NSTDA), Pathum Thani 12120, Thailand
fCenter of Excellence in Materials Science and Technology, Chiang Mai University, Chiang Mai 50200, Thailand
First published on 26th April 2021
Abstract
A label-free multiplexed electrochemical biosensor based on a gold nanoparticles/graphene quantum dots/graphene oxide (AuNPs/GQDs/GO) modified three-screen-printed carbon electrode (3SPCE) array is successfully constructed to detect miRNA-21, miRNA-155, and miRNA-210 biomarkers for the first time. Redox species (anthraquinone (AQ), methylene blue (MB), and polydopamine (PDA)) are used as redox indicators for anchoring capture miRNA probes, which hybridize with the complementary targets, miRNA-21, miRNA-155, and miRNA-210, respectively. After three target miRNAs are present, the square wave voltammetry (SWV) scan displays three well-separated peaks. Each peak indicates the presence of one miRNA, and its intensity quantitatively correlates with the concentration of the corresponding target analyte. This phenomenon results in the substantial decline of the SWV peak current of the redox probes. The developed AuNPs/GQDs/GO-based biosensor reveals excellent performance for simultaneous miRNA sensing. It offers a wide linear dynamic range from 0.001 to 1000 pM with ultrasensitive low detection limits of 0.04, 0.33, and 0.28 fM for the detection of miRNA-21, miRNA-155, and miRNA-210, respectively. It also presents high selectivity and applicability for the detection of miRNAs in human serum samples. This multiplex label-free miRNA biosensor has great potential for applications in breast cancer diagnosis.
Introduction
MicroRNA (miRNA) is a small single-stranded non-coding RNA that plays significant roles in several cellular processes such as differentiation, proliferation, metabolism, and apoptosis.1–5 Abnormal miRNA expression levels in body fluids have been associated with the occurrence and progression of numerous cancers like pancreatic cancer,6,7 colorectal cancer,8,9 lung cancer,10,11 and breast cancer.12–14 One miRNA can control gene expression in many various organisms and is related to various diseases, while one disease is often associated with the expression of multiple miRNAs.15 For example, the analysis of miRNA expression profiling in lung, pancreatic, colon, breast, and thyroid tumors reveals a highly increased miRNA-155 expression in patients compared to normal people.16–18 In addition, many studies have found that miRNA-21 plays an important role in different human diseases, including ovarian cancer, hepatocellular carcinoma (HCC) tumor, breast cancer and cardiovascular disease.19,20 However, the detection of a single miRNA biomarker is not sufficient for an accurate cancer diagnosis due to inadequate data. Moreover, it is quite indistinguishable to use only one miRNA marker to indicate the disease state and type.21,22 Therefore, the simultaneous assay of multiple miRNA tumor markers is required to improve the diagnostic precision.23,24 Numerous research studies have supported an overexpression of miRNA-21, miRNA-155, and miRNA-210 as biomarkers for early-stage breast cancer detection14,25,26 Therefore, the detection of these cancer biomarkers is desired. A number of electrochemical methods for miRNA investigation have been developed based on the construction of the hybridization structure27 followed by signal amplification with enzymes,28,29 hybridization chain reaction (HCR),30–32 polymerase chain reaction (PCR),33 and redox nanoparticles.34–36 Although these methods provide high sensitivity and specificity, they require complex operation and time-consuming processes.34 The label-free electrochemical technique with outstanding advantages such as high selectivity, high sensitivity, simple operation, and rapid analysis features has been recognized as a powerful tool for the multiplexed detection of cancer biomarkers.23,37,38 The direct, quantitative, and multiplexed electrochemical detection of the target miRNAs can be performed via the conductivity change at the electrode surface before and after the target miRNA-probe hybridization using distinguishable redox tags.15,39–41In order to improve the sensitivity, specificity, and stability of electrochemical biosensors, nanomaterials have been utilized for increasing the surface area and improving the speed of electron transfer at the electrodes.42,43 Gold nanoparticles (AuNPs) are widely used as a kind of suitable electrode material, due to their high chemical stability, large surface to volume ratios, good biocompatibility, excellent catalytic ability, and outstanding electrical conductivity enhancement.44–47 For electrochemical biosensing, AuNPs have been applied for electrode coating to further amplify the electric signal. In combination with other conductive materials, AuNPs can support functionalization of DNA capture probes, enzymes, antibodies, or electroactive molecules.48 Interestingly, graphene oxide (GO) is also a broadly employed electrode material. Its surface holds numerous hydroxyl groups and carboxylic groups, and it has high dispersibility in water and is suitable for biomedical analysis.49 The occurrence of these functional groups makes GO sheets strongly hydrophilic. It allows the combination with many types of inorganic nanoparticles, including metal oxides, semiconducting nanoparticles, noble metals, and quantum dots (QDs), to improve the performance of the sensors.50,51 In addition, graphene quantum dots (GQDs) have been utilized to construct miRNA biosensors.52 They are a zero-dimensional nanomaterial obtained from graphene which have features of high electrical conductivity and a large surface area.53 Attractively, the carboxylic groups at the edge of GQDs make them easy to immobilize with various polymeric, organic, inorganic, or biological species.52 Therefore, their composites formed by combinations of the functional nanomaterials could improve electrochemical sensors’ performance.54,55
Inspired by the abovementioned points, we investigate a AuNPs/GQDs/GO based electrochemical biosensor for label-free multiplexed detection of miRNA-21, miRNA-155, and miRNA-210 for the first time. In this strategy, three miRNA probes are assembled on the modified three-working area electrode. The redox dyes (AQ, MB and PDA) are used as signal indicators with distinguishable electrochemical signals. The AuNPs/GQDs/GO composite with outstanding electron transfer ability, high surface area, and ease of biomolecule immobilization is used with the aim to improve the analytical performance of the electrochemical miRNA biosensor. Furthermore, the proposed multiplex label-free miRNA biosensor would make the detection simple and cost-effective. Quantification of miRNA-21, miRNA-155 and miRNA-210 is simultaneously performed by measuring decreases in the AQ, MB and PDA electrochemical signals, respectively. The peak current responses of the redox species are related to the concentration of the target miRNAs, which hybridize with the probes immobilized on the electrode. The performance of the present biosensor for simultaneous miRNA detection is investigated. The sensor shows high sensitivity, selectivity, reproducibility, and stability resulting in effective and accurate breast cancer diagnosis.
Experimental section
Materials and reagents
Potassium ferrocyanide (K4Fe(CN)6, 98.5%) and potassium ferricyanide (K3Fe(CN)6, 98.5%) were purchased from Merck (Germany). Tris(2-carboxyethyl) phosphine hydrochloride (TCEP-HCl) and 10 mM tris(hydroxymethyl)-HCl (Tris-HCl) containing 1.0 mM ethylenediaminetetraacetic acid (EDTA) were obtained from Fisher Scientific (USA). Gold(III) chloride trihydrate (HAuCl4·3H2O, ≥99.9%), phosphate buffered saline tablets (PBS), potassium chloride (KCl, 99.0%), 6-mercapto-1-hexanol (MCH, 97.0%), trisodium citrate dihydrate (Na3C6H5O7, 99.0%), methylene blue hydrate (MB, >97.0), anthraquinone-2-carboxylic acid (AQ, 98.0), dopamine hydrate (DA, 99.5%), human serum from a human male (AB plasma, USA origin, lot: SLBS6544), graphene quantum dots (GQDs, aqua green luminescent, 1.0 mg mL−1 in H2O), and graphite powder were purchased from Sigma Aldrich (USA). All aqueous solutions were prepared with 18 MΩ deionized water (DI). Capture miRNA probes, miRNA targets, single-base mismatch (1 MM) miRNAs, triple-base mismatch (3 MM) miRNAs, and non-complementary miRNAs were purchased from Sigma Aldrich (USA).Instruments and apparatus
A plasma cleaner (PDC-32G, Harrick Plasma, USA) was utilized to treat the three-screen-printed carbon electrode (3SPCE) array before modification. A three-electrode system was used for electrochemical experiments, which consisted of an Ag/AgCl (3 M NaCl) reference electrode, a platinum wire auxiliary electrode, and a 3SPCE working electrode. Cyclic voltammetry (CV) and square wave voltammetry (SWV) measurements were performed on an Emstat 3 (Plamsens, Netherlands) controlled by PSTrace 5.6 software. The electrochemical impedance spectroscopy (EIS) measurement was carried out with an Autolab potentiostat/galvanostat (Metrohm, Netherlands) controlled by NOVA 2.1 software. The CV measurements were performed between −0.4 V and +0.8 V in 0.10 M KCl solution containing 5.0 mM [Fe(CN)6]3−/4− at a scan rate of 50 mV s−1. The EIS technique was conducted from the frequency of 0.1 Hz to 100 kHz with an open circuit potential of 0.2 V. A thermomixer (ThermoMixer C, Eppendorf, Germany) was employed for mixing and incubating bioconjugate samples. A high-speed centrifuge (5430R Eppendorf, Germany) was utilized for the purification of the AuNP solution. The morphologies of the nanomaterials were characterized by scanning electron microscopy (SEM, JEOL JSM-6335F, Japan).Preparation of modified electrodes
GO was synthesized by a triple exfoliation procedure adapted from a modified Hummers’ method.56,57 The citrate stabilized AuNPs were prepared by citrate reduction of HAuCl4.58 The PDA redox probe was prepared using the oxidative self-polymerization of dopamine in Tris buffer (10 mM, pH 8.5).59 In the preparation of modified electrodes, the 3SPCE consisting of W1, W2, and W3 working electrodes was cleaned using O2 plasma at 400 millitorr pressure for 1 min. After that, a solution containing 2.0 mg mL−1 GO and 1.0 mg mL−1 GQDs was dropped onto the 3SPCE. The modified 3SPCE was dried in air at room temperature. The AuNP (OD = 1) solution was dropped onto GQDs/GO/SPCEs for assembling miRNA capture probes and then dried in air at room temperature. The signaling redox probes (10 mM of AQ, MB, and PDA) were dropped on W1, W2, and W3, respectively, of the AuNPs/GO/GQDs/3SPCE for 30 min. Finally, the resultant electrode was washed with DI water several times and kept at 4 °C when not in use.Preparation of the electrochemical biosensor
The fabrication and detection processes of the multiplexed miRNA biosensor are schematically illustrated in Fig. 1. The sequences of all miRNAs are listed in Table S1.† To fabricate a three-miRNA probe-modified 3SPCE array, the thiol-modified miRNA probes (20 μL, 10 μM) were treated with 20 μL of 10 mM TCEP to reduce the disulfide bonds at room temperature for 2 h. Then, 1.0 μL of the 5.0 μM reduced miRNA-21, miRNA-155, and miRNA-210 capture probe solutions were consequently immobilized onto the surface of each working electrode of the dye/AuNPs/GO/GQD modified 3SPCE at room temperature for 1 h. The resulting miRNA capture probe-covered electrode was washed with PBS solution, and then added with 1.0 μL of 20 μM MCH (dissolved in 50% ethanol) to block non-specific binding sites for 30 min. Next, the SWV response was recorded as a baseline response after the electrode was rinsed with DI water several times.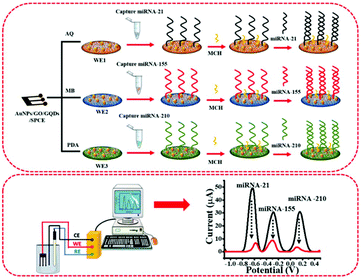 | ||
| Fig. 1 The diagram of AuNPs/GQDs/GO-based biosensors for the detection of miRNA-21, miRNA-155, and miRNA-210 related to breast cancer. | ||
Simultaneous electrochemical detection of miRNAs
For the simultaneous miRNA quantitative detection, 1.0 μL of target miRNA-21, miRNA-155, and miRNA-210 solutions with different concentrations (0.001, 0.01, 0.1, 1, 10, 100 and 1000 pM) were dropped onto each designed electrode surface, which was incubated for 1 h. After further washing the electrode with PBS solution to remove unspecific adsorbed miRNA, the electrochemical responses of the electrode in contact with PBS were recorded using SWV. SWV curves were acquired by scanning the potential from −1.1 V to +0.5 V with a step potential of 4 mV, a frequency of 15 Hz, and an amplitude potential of 25 mV.Results and discussion
Characterization of the modified electrodes
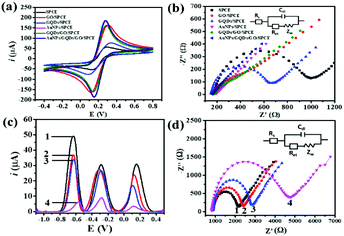
The morphologies of nanomaterials produced in this study are investigated through their SEM images. Fig. 2 shows SEM images of the bare SPCE, GQDs/SPCE, GO/SPCE, GQDs/GO/SPCE, and AuNPs/GQDs/GO/SPCE. The latter two are made of optimized 70% GO in the GQDs/GO composite (Fig. S1†). It is observed that the surfaces of the bare SPCE and GQDs/SPCE are rough and possess some porosity (Fig. 2(a) and (b), respectively). In Fig. 2(c), the characteristics of GO are observed with irregularly crumpled and wrinkled sheet-like structures, which can enlarge the electrode surface.60Fig. 2(d) displays the morphology of the GO/GQD nanocomposite similar to that of the GO structure. To improve the biocompatibility of the GO/GQD nanocomposite, AuNPs are employed for further modification (Fig. 2(e) and (f)). It can be obviously observed that AuNPs are thoroughly distributed on the surface of the GQDs/GO/SPCE.CV and EIS analytical tools are employed to study the electrochemical behavior of the as-prepared 3SPCE and the modified 3SPCEs. The electrochemical experiments are performed in 0.10 M KCl solution containing 5.0 mM [Fe(CN)6]3−/4−. The modification of the 3SPCE surface with nanomaterials is investigated as shown in Fig. 3. The bare 3SPCE and GO-, GQD-, GO/GQD-, AuNP-, and AuNPs/GO/GQD-modified 3SPCEs exhibit a pair of redox peaks. CVs of bare and modified 3SPCE surfaces are presented in Fig. 3(a). The oxidation peak current (ip) and the peak-to-peak separation (ΔEp) of the bare SPCE are 72 μA and 0.34 V, respectively. After the GQDs, GO, AuNPs, and GO/GQDs are coated on 3SPCEs, the ipa increases to 93, 189, 209, and 188 μA, respectively, and ΔEp values are lowered down to 0.25, 0.18, 0.13, and 0.14 V, respectively. The AuNPs/GQDs/GO/nanocomposite modified 3SPCE surface exhibits the highest ip value (218 μA), and lowest ΔEp (0.12 V) due to the good electrocatalytic and electronic properties of AuNPs and GQDs/GO deposited on the 3SPCE surface.61–63 This behavior results from the synergetic effect of AuNPs, GQDs, and GO. Therefore, modification of AuNPs/GO/GQDs on the 3SPCE would offer a large electroactive surface area to improve electrical conductivity and the functionalization of miRNA capture probes. In addition, the EIS curves of these electrode surfaces are shown in Fig. 3(b). The charge transfer resistance (Rct) value of the bare 3SPCE is estimated as 782 Ω. In the case of the GQDs/SPCE, the Rct value decreased to 504 Ω. GO, GQDs/GO, AuNPs, and AuNPs/GQDs/GO modified electrodes exhibit a straight line implying exceptional low resistance because they can effectively accelerate the electron transfer through the electrode surface. These results suggest that the AuNPs/GQDs/GO modified electrode can improve electrochemical performance for miRNA detection.
The CV responses of the developed electrochemical platform are assessed with different scan rates in the range of 10 to 100 mV s−1 in 0.10 M KCl solution having 5.0 mM [Fe(CN)6]3−/4−. Fig. S2(a)† illustrates that with increasing the scan rate, the anodic peak current is shifted to more positive potential while the cathodic peak current is shifted to the negative potential. Fig. S2(b)† shows the linearity of ipa and ipc values with respect to the square root of scan rates, allowing regression coefficient (R2) values of 0.9964 and 0.9964, respectively, suggesting that the electrochemical process is a diffusion-controlled electron-transfer process.64,65
The effect of different modified-SPCEs on responsive signal intensities with three redox indicators (AQ, MB, and PDA) is investigated from the SWV voltammograms in 10 mM PBS (Fig. S3†). Eight types of 3SPCEs modified with AQ-MB-PDA (Fig. S3(a)†), AQ-MB-PDA/GO (Fig. S3(b)†), AQ-MB-PDA/GQDs (Fig. S3(c)†), AQ-MB-PDA/AuNPs (Fig. S3(d)†), AQ-MB-PDA/GQDs/GO (Fig. S3(e)†), AQ-MB-PDA/AuNPs/GO (Fig. S3(f)†), AQ-MB-PDA/AuNPs/GQDs (Fig. S3(g)†), and AQ-MB-PDA/AuNPs/GQDs/GO (Fig. S3(h)†) are prepared under the same test conditions. The positions of the SWV peaks are distinguishable, which offers multiple miRNA detection at the same time. Three SWV peaks of the dyes on bare and one- or two-material modified 3SPCEs are observed in the voltammograms and are well separated from each other (Fig. S3(a) to (g)†). Fig. S3(h)† demonstrates that the composite of AuNPs, GQDs, and GO obviously enhances the responsive signals of the three redox species. This may be attributed to its highly specific surface area and accelerating electron transfer feature of AuNPs, GQDs, and GO. Therefore, the AuNPs/GQDs/GO modified 3SPCE is selected for further experiments.
Fabrication of the biosensor
To fabricate our biosensor, the optimized 3SPCE modified with a AuNPs/GQDs/GO film is used. Fig. 3(c) illustrates the SWV oxidation peaks of the dyes/AuNPs/GQDs/GO modified 3SPCE at potentials of −0.644, −0.296, and 0.111 V for AQ, MB, and PDA, respectively (curve 1). After the dyes/AuNP/GQDs/GO coated 3SPCE is immobilized with three miRNA probes corresponding to miRNA-21, miRNA-155, and miRNA-210, respectively, the SWV curve is recorded. From curve 2 (Fig. 3(c)), immobilization of the capture miRNAs with their nonconductive structure and negative charge causes the obviously reduced peak currents of the redox probes.39 When the MCH is used to block non-specific sites on the electrode surface, the redox peak currents slightly decrease (curve 3, Fig. 3(c)). Then, these 3SPCE surfaces are hybridized with 100 pM miRNA targets for 1 h (optimum hybridization time), and the lowest current values are observed (curve 4, Fig. 3(c)), suggesting that the double-strand structure of RNA-RNA duplexes hinders the electron transfer.27,66 Additionally, EIS is also employed to investigate the target–probe hybridization process to support the SWV results (Fig. 3d). It can be observed that the dyes/GO/GQDs/3SPCE presents a small half circle with a Rct of 1277 Ω (curve 1, Fig. 3d). The capture miRNA probe functionalized surface's Rct value is determined as 1534 Ω (curve 2, Fig. 3d). After blocking non-specific sites of the electrode surface with MCH, the Rct increased up to 1976 Ω (curve 3, Fig. 3d). This Rct value increases about 2 times after the hybridization as 3668 Ω (curve 4, Fig. 3d), which is attributed to many miRNA targets hybridizing on the electrode. The negatively charged miRNAs restrict the negative charge of [Fe(CN)6]3−/4− to the electrode surface due to the electrostatic repulsion as well as the steric hindrance effect.38,66,67 Overall, SWV and EIS test results are consistent and confirm chemical changes at the electrode surface, resulting from both the biosensor fabrication and the hybridization event.Optimization of the experimental conditions
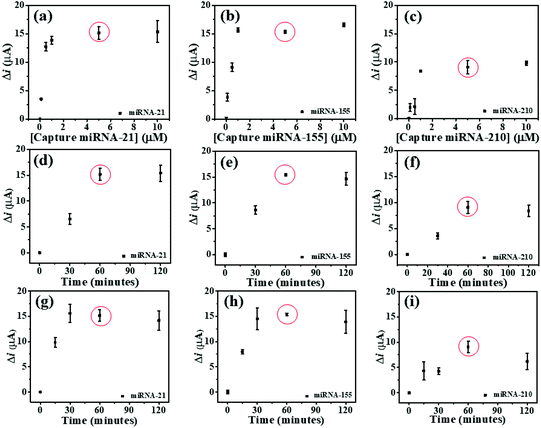
To study the optimal conditions, the miRNA capture probe concentration, the incubation time of miRNA capture probes, and the hybridization time are analyzed. Consequently, the SWV response differences (Δi) before and after the hybridization of the individual miRNA capture probe and its complementary miRNA targets are determined at the potentials of AQ, MB, and PDA. These Δi values are employed to optimize such conditions. To find the saturated amount of the probes on the modified 3SPCE, the miRNA capture probes at concentrations of 0.1, 0.5, 1, 5, and 10 μM are applied for the hybridizations with 10 pM target miRNAs as presented in Fig. 4. The ΔimiRNA-21 and ΔimiRNA-210 values increase with the increase of the miRNA capture probe concentration and reach plateaus at 5.0 μM (Fig. 4(a) and (b)), while the ΔimiRNA-155 is constant when the concentration of the miRNA-155 capture probe reaches 1.0 μM (Fig. 4(c). In practical use, the optimal concentration must be the same. Therefore, 5.0 μM miRNA capture probe concentration is selected for further experiments. The effect of incubation time of miRNA capture probes is also studied via the hybridizations with 10 pM target miRNAs. The incubation time of miRNA capture probes from 30 to 120 min is used. The Δi values increase speedily with increasing the incubation time ranging from 30 to 60 min. They then remain nearly unchanged after the time exceeds 60 min. Thus, the incubation time of 60 min is employed (Fig. 4(d), (e), and (f)) for miRNA probe immobilization. The effect of miRNA hybridization time is also examined. 10 pM miRNA target solutions are incubated with the miRNA capture probe-modified electrode for different periods of time from 15 to 120 min. In Fig. 4(g), the ΔimiRNA-21 value obtained after hybridization of the miRNA-21 target increases with increasing the hybridization time, and the highest Δi value is obtained at 30 min. In the case of miRNA-155 and miRNA-210, both Δi values increase with increasing the hybridization time from 15 to 60 min and begin to level off at 60 min (Fig. 4(h), and (i)), indicating that the hybridization reaction is completed within 60 min. This result indicates that increasing hybridization time provides higher efficiency of the binding between the miRNA capture probes and miRNA target. Therefore, our approach requires 60 min to provide the highest efficiency of the hybridization process and it is selected for the analysis of target miRNAs by this biosensor.Analytical performance of the biosensor
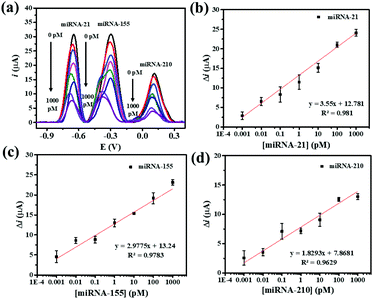
Under the optimum assay conditions, the SWV peak currents decrease with the concentration of miRNA-21, miRNA-155, and miRNA-210 in the range of 0.001 to 1000 pM (Fig. 5(a)). The peak current of AQ is found to be corresponding to the concentration of miRNA-21. The calibration curve fits a regression equation of ΔimiRNA-21 = 12.781 + 3.55 [miRNA-21] (pM) with a correlation coefficient R2 = 0.981 (Fig. 5(b)). The redox peak current from MB is related to the concentration of miRNA-155 (Fig. 5(c)). The calibration curve fits a regression equation of ΔimiRNA-155 = 13.24 + 2.9775 [miRNA-155] (pM) with a correlation coefficient R2 = 0.9783. The peak current of PDA is proportionally correlated with the concentration of miRNA-210 (Fig. 5(d)). The linear curve fits a regression equation of ΔimiRNA-210 = 7.8671 + 1.8293 [miRNA-210] (pM) with a correlation coefficient (R2 = 0.9783). The limits of detection (LODs) for detecting miRNA-21, miRNA-155, and miRNA-210 are calculated to be 0.04, 0.33, and 0.28 fM, respectively. The wide linear range and LODs suggest that our proposed method is useful for simultaneous electrochemical detection of the target miRNAs.The analytical properties of the biosensor toward multiplex miRNA detection are compared with those of other electrochemical biosensors as listed in Table 1. Using the developed AuNPs/GQDs/GO-based biosensor offers wider linear dynamic ranges. Moreover, the sensor exhibits relatively low LODs that is comparable to those of the first two sensors. However, the obtained LODs are higher than those of the third entry. Furthermore, the design of the present label-free biosensor is simple and requires no labeling molecules, thus reducing the time for the fabrication of the biosensors and the determination, cost, and operational complexity of the assay. With these advantages, the developed label-free electrochemical AuNPs/GQDs/GO-based biosensor is potentially useful as a new efficient analytical tool for the simultaneous detection of miRNAs in clinical cancer screening tests and diagnostics.
| Methods | Materials | Signal Amplifier | Targets | Time for the fabrication of the biosensors and the determination | Detection range | Detection limit | Ref. |
|---|---|---|---|---|---|---|---|
| DPV | AuNP-MMBs/MAuE | MB | miRNA-182 | 62 h 30 min | 5–600 fM | 0.20 fM | 68 |
| Fc | miRNA-381 | 1–800 fM | 0.12 fM | ||||
| DPV | rGO/PTH/SPCE | Au/TMC/Fe3O4 | miRNA-106a | 20 h 15 min | 0.1–5000 fM | 0.06 fM | 69 |
| CdSe@CdsQD /TMC/Fe3O4 | let-7a | 0.1–5000 fM | 0.02 fM | ||||
| SWV | TDN/depAu/GCE | Fc | miRNA-21 | 21 h | 1.0 × 10−7–10 nM | 18.9 aM | 70 |
| MB | miRNA-155 | 1.0 × 10−7–10 nM | 39.6 aM | ||||
| DPV | TBAPy-MA-COF-COOH/AuE | AgNCs@AuNPs | miRNA-155 | 4 h 30 min | 0.01–1000 pM | 6.7 fM | 71 |
| Cu2O@AuNPs | miRNA-122 | 0.01–1000 pM | 1.5 fM | ||||
| SWV | AuNPs/GQDs/GO/SPCE | AQ | miRNA-21 | 4 h 30 min | 0.001–1000 pM | 0.04 fM | This work |
| MB | miRNA-155 | 0.001–1000 pM | 0.33 fM | ||||
| PDA | miRNA-210 | 0.001–1000 pM | 0.28 fM |
Multiplex capability of the biosensor
The multiplex capability of the biosensor is studied by comparing the SWV responses of the detection in the sample solutions between with and without all three miRNA analytes including with only one miRNA target. The SWV responses obtained are present in Fig. S4.† Firstly, in the absence of the target miRNA-21, miRNA-155, and miRNA-210, a high SWV response is observed (Fig. S4(a–d),† solid lines). Then, in the solution containing all targets, miRNA-21, miRNA-155, and miRNA-210, decreases in SWV peaks of AQ, MB, and PDA at −0.644 V, −0.296 V, and 0.111 V are observed (Fig. S4(a),† dashed line) with simultaneous monitoring. Moreover, the biosensor is incubated with the sample solution containing a single target type including individual miRNA-21, miRNA-155, and miRNA-210, respectively. For the solution containing miRNA-21, only the SWV peak of AQ decreases (Fig. S4(b),† dashed line). The SWV peak of MB reduces when the electrode is incubated with the solution containing only miRNA-155 while the SWV peaks of AQ and PDA remain unchanged (Fig. S4(c),† dashed line). Besides, when the solution comprising only miRNA-210 is applied on the electrode, a SWV peak of PDA declines (Fig. S4(d),† dashed line) whereas the other SWV peaks show a negligible change. The result indicates the excellent feasibility of our biosensor for multiple miRNA detection without cross reaction.Selectivity, stability, and reproducibility of the proposed miRNA biosensor
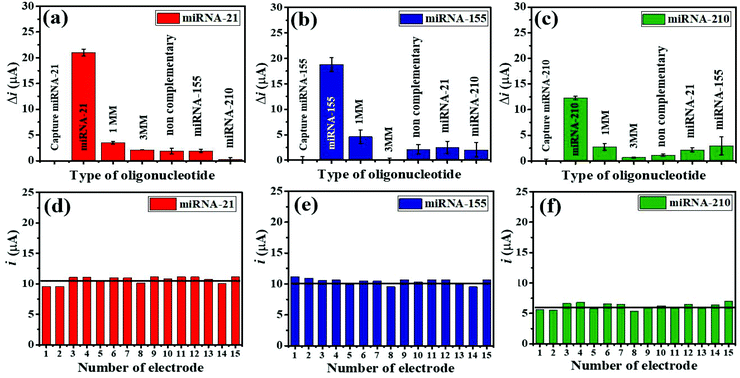
The selective binding of the three miRNA assays has been confirmed using closely related miRNA sequences. The detection responses of 100 pM miRNAs are determined and compared with the responses of 1.0 μM interfering sequences (1 MM sequence, 3 MM sequence, and non-complementary miRNAs) (Fig. 6(a–c)). As displayed in Fig. 6(a), (b), and (c), the highest Δi values are obtained with only three target miRNAs. The Δi values of both their mismatched sequences and non-complementary sequences, and their non-targets are significantly lower as compared to those of the targets. Fig. 6(a) presents that the Δi value of a complementary miRNA-21 target is 5.95 times higher than that of the 1 MM sequence, while the responses of the 3 MM, non-complementary, miRNA-155, and miRNA-210 sequences are negligible. Fig. 6(b) shows that the Δi value of a complementary miRNA-155 target is 4.07 times higher than that of the 1 MM sequence, and the responses of others can be ignored. Fig. 6(c) depicts that the Δi value of a perfectly complementary miRNA-210 target is 4.54 times higher than that of the 1 MM sequence whilst the responses of the 3 MM sequence and non-complementary sequences are as low as background. It is noted that the developed electrochemical miRNA sensor possesses an excellent specificity and outstanding discrimination of similar miRNA sequences.To study the reproducibility of the miRNA biosensor, each miRNA target is tested on the 15 fabricated electrodes individually using 100 pM for all three targets under similar conditions. Fig. 6(d–f) show the reproducibility of the SWV response for simultaneous miRNA-21 (Fig. 6(d)), miRNA-155 (Fig. 6(e)), and miRNA-210 (Fig. 6(f)) detection. The average relative standard deviations (RSDs) are 8.40% for miRNA-21, 4.51% for miRNA-155, and 9.43% for miRNA-210, indicating that the proposed multiplex miRNA biosensor has acceptable precision and fabrication protocol.
Furthermore, for the stability test, the miRNA-21, miRNA-155, and miRNA-210 biosensors are kept at 4 °C when they are not in use. After 3 weeks of storage, 84.3%, 85.9%, and 89.5% of the initial response values are retained for the detection of miRNA-21, miRNA-155, and miRNA-210, respectively (Fig. 7(a), (b), and (c)). Therefore, the lifetime of the miRNA sensor is estimated to be at least 21 days (the value decrease is no more than 20%), indicating good stability of the proposed miRNA assay in practical applications.
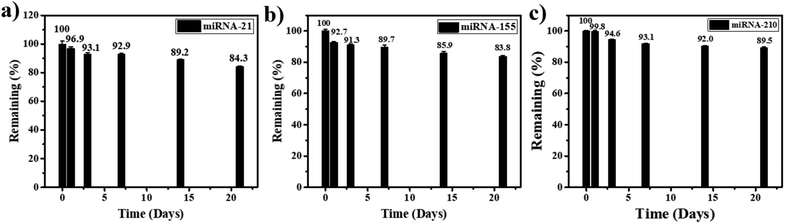 | ||
| Fig. 7 Stability test of the proposed biosensor in the detection of (a) miRNA-21, (b) miRNA-155, and (c) miRNA-210 when the sensors are stored for 3 weeks. | ||
Detection of miRNAs in human serum samples
To demonstrate the ability of the biosensor for multiplexed miRNA detection, miRNA-21, miRNA-155, and miRNA-210 in 50% diluted human serum samples with PBS (pH 7.4) are determined. The four concentrations of miRNA-21, miRNA-155, and miRNA-210 (0.01, 1.0, 10, and 1000 pM) are added into the diluted human serum sample for the recovery tests. The results of the recoveries are summarized in Table 2. The recoveries are achieved in the range of 98.51–112.2% for miRNA-21, 108.5–109.2% for miRNA-155, and 103.8–110.0% for miRNA-210. This result indicates that the measurement and recoveries of miRNAs in the serum samples using the fabricated biosensor are satisfactory. Therefore, the developed label-free biosensor can possibly be used to accurately detect multiple miRNAs in clinical samples.| Sample no. | Standard value (pM) | Found value (pM) | Recovery % | ||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA-21 | miRNA-155 | miRNA-210 | miRNA-21 | miRNA-155 | miRNA-210 | miRNA-21 | miRNA-155 | miRNA-210 | |
| 1 | 0.01 | 0.01 | 0.01 | 0.0109 | 0.01085 | 0.0110 | 109.0 | 108.5 | 110.0 |
| 2 | 1 | 1 | 1 | 1.045 | 1.074 | 1.038 | 104.5 | 107.4 | 103.8 |
| 3 | 10 | 10 | 10 | 9.851 | 10.92 | 1.055 | 98.51 | 109.2 | 105.5 |
| 4 | 1000 | 1000 | 1000 | 1122.11 | 1086.41 | 1095.55 | 112.2 | 108.6 | 109.6 |
Conclusions
A label-free electrochemical biosensor using the newly designed AuNPs/GQDs/GO modified 3SPCE platform is successfully developed for the simultaneous detection of multiple miRNA tumor markers. Three redox species (AQ, MB, and PDA) attached on the AuNPs/GQDs/GO-modified 3SPCE are used for distinguishable signals in the multiplex biosensor and the AuNPs/GQDs/GO composite is a signal enhancer. The biosensor shows a linear range of detection from 0.001 to 1000 pM with the LODs of 0.04, 0.33, and 0.28 fM for miRNA-21, miRNA-155, and miRNA-210, respectively. With high sensitivity and multiplexing capability, the fabricated biosensor could improve diagnostic accuracy for early breast cancer detection. Furthermore, this biosensor also demonstrates a high selectivity which can distinguish the miRNA target from other interfering oligonucleotides, acceptable stability, and reproducibility. The sensor is not only promising for breast cancer and other cancer diagnoses clinically but also benefits and expands knowledge in biomedical research.Abbreviation
| SWV | Square wave voltammetry |
| DPV | Differential pulse voltammetry |
| AuNP-MMBs | Gold nanoparticle-coated magnetic microbeads |
| rGO | Reduced graphene oxide |
| PTH | Polythiophene |
| TMC | Trimethylene carbonate |
| Fe3O4 | Magnetite |
| CdSe@CdsQD | CdSe@Cds quantum dot |
| CHIT | Chitosan |
| GCE | Glassy carbon electrode |
| TB | Toluidine blue |
| PB | Prussian blue |
| TBAPy-MA-COF-COOH | 1,3,6,8-Tetra(4-carboxylphenyl)pyrene and melamine |
| AuE | Au electrode |
| MAuE | Magnetic gold electrode |
| Cu2O@AuNPs | Shell-encoded AuNPs with Cu2O |
| AgNCs | Ag nanoclusters |
| TDN | Tetrahedron DNA nanostructure |
| Fc | Ferrocene |
| MB | Methylene blue |
Author contributions
Chammari Pothipor: Investigation, result analysis, and writing – original draft. Jaroon Jakmunee: Writing – review and editing. Suwussa Bamrungsap: Writing – review and editing. Kontad Ounnunkad: Conceptualization, methodology, validation, resources, writing – original draft, writing – review and editing, visualization, supervision, project administration, and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
C. P. would like to thank for the scholarship from the Thailand Graduate Institute of Science and Technology (TGIST) 2017, and National Science and Technology Development Agency (NSTDA), Thailand (Grant No. TG-55-10-60-025D). This research work was partially supported by Chiang Mai University (CMU). K. O. would like to thank the Research Center on Chemistry for Development of Health Promoting Products from Northern Resources, the Graduate School (CMU), and the Center of Excellence for Innovation in Chemistry (PERCH-CIC) and the Department of Chemistry, Faculty of Science (CMU), and Center of Excellence in Materials Science and Technology (CMU). This research was supported by the Program Management Unit for Human Resources & Institutional Development, Research and Invitation, NXPO [Grant Number: B16F640001].References
- H. Zhao, M. Wang, X. Xiong, Y. Liu and X. Chen, Talanta, 2020, 210, 120677 CrossRef CAS PubMed.
- J. Chen, T. Fan, Y. Chen, L. Ye, C. Zhang, F. Liu, Y. Qin, Y. Tan and Y. Jiang, Biosens. Bioelectron., 2020, 169, 112631 CrossRef CAS PubMed.
- J. Bao, C. Hou, Y. Zhao, X. Geng, M. Samalo, H. Yang, M. Bian and D. Huo, Talanta, 2019, 196, 329–336 CrossRef CAS PubMed.
- Q. Wang, B.-C. Yin and B.-C. Ye, Biosens. Bioelectron., 2016, 80, 366–372 CrossRef CAS PubMed.
- C. E. Condrat, D. C. Thompson, M. G. Barbu, O. L. Bugnar, A. Boboc, D. Cretoiu, N. Suciu, S. M. Cretoiu and S. C. Voinea, Cells, 2020, 9, 276 CrossRef CAS PubMed.
- A. Z. Daoud, E. J. Mulholland, G. Cole and H. O. McCarthy, BMC Cancer, 2019, 19, 1130 CrossRef CAS PubMed.
- R. Słotwiński, G. Lech and S. M. Słotwińska, Cent. Eur. J. Immunol., 2018, 43, 314–324 CrossRef PubMed.
- A. J. Schetter, H. Okayama and C. C. Harris, Cancer J., 2012, 18, 244–252 CrossRef CAS PubMed.
- B. Chen, Z. Xia, Y.-N. Deng, Y. Yang, P. Zhang, H. Zhu, N. Xu and S. Liang, Open Biol., 2019, 9, 180212–180212 CrossRef CAS PubMed.
- K.-L. Wu, Y.-M. Tsai, C.-T. Lien, P.-L. Kuo and J.-Y. Hung, Int. J. Mol. Sci., 2019, 20(7), 1611 CrossRef CAS PubMed.
- X. Wu, M. G. Piper-Hunter, M. Crawford, G. J. Nuovo, C. B. Marsh, G. A. Otterson and S. P. Nana-Sinkam, J. Thorac. Oncol., 2009, 4, 1028–1034 CrossRef PubMed.
- H.-Y. Loh, B. P. Norman, K.-S. Lai, N. M. A. N. A. Rahman, N. B. M. Alitheen and M. A. Osman, Int. J. Mol. Sci., 2019, 20(19), 4940 CrossRef CAS PubMed.
- R. Singh and Y.-Y. Mo, Cancer Biol. Ther., 2013, 14, 201–212 CrossRef CAS PubMed.
- E. Zografos, F. Zagouri, D. Kalapanida, R. Zakopoulou, A. Kyriazoglou, K. Apostolidou, M. Gazouli and M.-A. Dimopoulos, Oncotarget, 2019, 10, 7156–7178 CrossRef PubMed.
- R. D'Agata and G. Spoto, Anal. Bioanal. Chem., 2019, 411, 4425–4444 CrossRef PubMed.
- S. Mattiske, R. J. Suetani, P. M. Neilsen and D. F. Callen, Cancer Epidemiol., Biomarkers Prev., 2012, 21, 1236–1243 CrossRef CAS PubMed.
- R. Bayraktar and K. Van Roosbroeck, Cancer Metastasis Rev., 2018, 37, 33–44 CrossRef PubMed.
- X. Zhang, M. Li, K. Zuo, D. Li, M. Ye, L. Ding, H. Cai, D. Fu, Y. Fan and Z. Lv, J. Clin. Endocrinol. Metab., 2013, 98, E1305–E1313 CrossRef CAS PubMed.
- Z. Shabaninejad, F. Yousefi, A. Movahedpour, Y. Ghasemi, S. Dokanehiifard, S. Rezaei, R. Aryan, A. Savardashtaki and H. Mirzaei, Anal. Biochem., 2019, 581, 113349 CrossRef CAS PubMed.
- D. Bautista-Sánchez, C. Arriaga-Canon, A. Pedroza-Torres, I. A. De La Rosa-Velázquez, R. González-Barrios, L. Contreras-Espinosa, R. Montiel-Manríquez, C. Castro-Hernández, V. Fragoso-Ontiveros, R. M. Álvarez-Gómez and L. A. Herrera, Mol. Ther. – Nucleic Acids, 2020, 20, 409–420 CrossRef PubMed.
- D. Wang, N. Gan, J. Zhou, P. Xiong, Y. Cao, T. Li, D. Pan and S. Jiang, Sens. Actuators, B, 2014, 197, 244–253 CrossRef CAS.
- A. Brecht and R. Abuknesha, TrAC, Trends Anal. Chem., 1995, 14, 361–371 CAS.
- X. Chen, X. Jia, J. Han, J. Ma and Z. Ma, Biosens. Bioelectron., 2013, 50, 356–361 CrossRef CAS.
- D. Zeng, Z. Wang, Z. Meng, P. Wang, L. San, W. Wang, A. Aldalbahi, L. Li, J. Shen and X. Mi, ACS Appl. Mater. Interfaces, 2017, 9, 24118–24125 CrossRef CAS PubMed.
- L.-X. Yan, X.-F. Huang, Q. Shao, M.-Y. Huang, L. Deng, Q.-L. Wu, Y.-X. Zeng and J.-Y. Shao, RNA, 2008, 14, 2348–2360 CrossRef CAS PubMed.
- A. Bahrami, A. Aledavood, K. Anvari, S. M. Hassanian, M. Maftouh, A. Yaghobzade, O. Salarzaee, S. ShahidSales and A. Avan, J. Cell. Physiol., 2018, 233, 774–786 CrossRef CAS PubMed.
- Y.-X. Chen, W.-J. Zhang, K.-J. Huang, M. Zheng and Y.-C. Mao, Analyst, 2017, 142, 4843–4851 RSC.
- L. Liu, Y. Gao, H. Liu and N. Xia, Sens. Actuators, B, 2015, 208, 137–142 CrossRef CAS.
- N. Xia, Y. Zhang, X. Wei, Y. Huang and L. Liu, Anal. Chim. Acta, 2015, 878, 95–101 CrossRef CAS PubMed.
- Q. Guo, Y. Yu, H. Zhang, C. Cai and Q. Shen, Anal. Chem., 2020, 92, 5302–5310 CrossRef CAS PubMed.
- L. Jirakova, R. Hrstka, S. Campuzano, J. M. Pingarrón and M. Bartosik, Electroanalysis, 2019, 31, 293–302 CrossRef CAS.
- L. Liu, C. Song, Z. Zhang, J. Yang, L. Zhou, X. Zhang and G. Xie, Biosens. Bioelectron., 2015, 70, 351–357 CrossRef CAS PubMed.
- W. L. Ang, X. Y. Seah, P. C. Koh, C. Caroline and A. Bonanni, ACS Appl. Nano Mater., 2020, 3, 5489–5498 CrossRef CAS.
- W. Zhu, X. Su, X. Gao, Z. Dai and X. Zou, Biosens. Bioelectron., 2014, 53, 414–419 CrossRef CAS PubMed.
- D. Feng, L. Li, J. Zhao and Y. Zhang, Anal. Biochem., 2015, 482, 48–54 CrossRef CAS PubMed.
- Z. Wang, N. Liu and Z. Ma, Biosens. Bioelectron., 2014, 53, 324–329 CrossRef CAS PubMed.
- S. Kasturi, Y. Eom, S. R. Torati and C. Kim, Ind. Eng. Chem. Res., 2020, 93, 186–195 CrossRef.
- A. Salimi, B. Kavosi and A. Navaee, Measurement, 2019, 143, 191–198 CrossRef.
- D. Zhu, W. Liu, D. Zhao, Q. Hao, J. Li, J. Huang, J. Shi, J. Chao, S. Su and L. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 35597–35603 CrossRef CAS PubMed.
- E. Kesici, E. Eksin and A. Erdem, Sensors, 2018, 18, 2868 CrossRef PubMed.
- J.-Y. Park and S.-M. Park, Sensors, 2009, 9, 9513–9532 CrossRef CAS PubMed.
- M. Azimzadeh, M. Rahaie, N. Nasirizadeh, M. Daneshpour and H. Naderi-Manesh, Nanomed. J., 2017, 2, 36–48 CAS.
- N. Xia and L. Zhang, Materials, 2014, 7, 5366–5384 CrossRef PubMed.
- P. Jiang, Y. Wang, L. Zhao, C. Ji, D. Chen and L. Nie, Nanomaterials, 2018, 8, 977 CrossRef PubMed.
- C. Coutinho and Á. Somoza, Anal. Bioanal. Chem., 2019, 411, 1807–1824 CrossRef CAS.
- P. A. Rasheed and N. Sandhyarani, Microchim. Acta, 2017, 184, 981–1000 CrossRef.
- P. M. Tiwari, K. Vig, V. A. Dennis and S. R. Singh, Nanomaterials, 2011, 1(1), 31–63 CrossRef CAS PubMed.
- C. Coutinho and Á. Somoza, Anal. Bioanal. Chem., 2019, 411, 1807–1824 CrossRef CAS.
- H. Beitollai, M. Safaei and S. Tajik, Int. J. Nano Dimens., 2019, 10, 125–140 Search PubMed.
- S. K. Krishnan, E. Singh, P. Singh, M. Meyyappan and H. S. Nalwa, RSC Adv., 2019, 9, 8778–8881 RSC.
- M. Hasanzadeh, A. Karimzadeh, S. Sadeghi, A. Mokhtarzadeh, N. Shadjou and A. Jouyban, J. Mater. Sci.: Mater. Electron., 2016, 27, 6488–6495 CrossRef CAS.
- T. Hu, L. Zhang, W. Wen, X. Zhang and S. Wang, Biosens. Bioelectron., 2016, 77, 451–456 CrossRef CAS.
- B. D. Mansuriya and Z. Altintas, Materials, 2019, 13, 96 CrossRef PubMed.
- A. I. Ruiz-Carmuega, C. Garcia-Hernandez, J. Ortiz, C. Garcia-Cabezon, F. Martin-Pedrosa, Á. Sastre-Santos, M. A. Rodríguez-Perez and M. L. Rodriguez-Mendez, Nanomaterials, 2019, 9(11), 1506 CrossRef CAS PubMed.
- J. Wang, J. Wen and H. Yan, Chem. – Asian J., 2021, 16, 114–128 CrossRef CAS.
- C. Pothipor, N. Kungwan, J. Jakmunee and K. Ounnunkad, Chem. Lett., 2015, 44, 800–802 CrossRef CAS.
- W. Jumpathong, J. Jakmunee and K. Ounnunkad, Anal. Sci., 2016, 32, 323–328 CrossRef CAS PubMed.
- A. A. Volkert, V. Subramaniam, M. R. Ivanov, A. M. Goodman and A. J. Haes, ACS Nano, 2011, 5, 4570–4580 CrossRef CAS PubMed.
- Y. Liu, K. Ai and L. Lu, Chem. Rev., 2014, 114, 5057–5051 CrossRef CAS PubMed.
- M. Azimzadeh, M. Rahaie, N. Nasirizadeh, K. Ashtari and H. Naderi-Manesh, Biosens. Bioelectron., 2016, 77, 99–106 CrossRef CAS PubMed.
- B. Liu and L. Lu, Microchim. Acta, 2019, 186, 262 CrossRef PubMed.
- J. Tang, R. Huang, S. Zheng, S. Jiang, H. Yu, Z. Li and J. Wang, Microchem. J., 2019, 145, 899–907 CrossRef CAS.
- Z. Shekari, H. R. Zare and A. Falahati, Microchim. Acta, 2019, 186, 530 CrossRef CAS PubMed.
- S. Jain, S. Verma, S. P. Singh and S. N. Sharma, Biosens. Bioelectron., 2019, 127, 135–141 CrossRef CAS PubMed.
- S. Jain, S. Verma, S. P. Singh and S. N. Sharma, Biosens. Bioelectron., 2019, 127, 135–141 CrossRef CAS PubMed.
- L. Cui, J. Hu, C.-c. Li, C.-m. Wang and C.-y. Zhang, Biosens. Bioelectron., 2018, 122, 168–174 CrossRef CAS PubMed.
- A. R. Cardoso, F. T. C. Moreira, R. Fernandes and M. G. F. Sales, Biosens. Bioelectron., 2016, 80, 621–630 CrossRef CAS PubMed.
- J. Wang, Z. Lu, H. Tang, L. Wu, Z. Wang, M. Wu, X. Yi and J. Wang, Anal. Chem., 2017, 89, 10834–10840 CrossRef CAS PubMed.
- M. Daneshpour, B. Karimi and K. Omidfar, Biosens. Bioelectron., 2018, 109, 197–205 CrossRef CAS PubMed.
- S. Xu, Y. Chang, Z. Wu, Y. Li, R. Yuan and Y. Chai, Biosens. Bioelectron., 2020, 149, 111848 CrossRef CAS.
- Z. Cao, F. Duan, X. Huang, Y. Liu, N. Zhou, L. Xia, Z. Zhang and M. Du, Anal. Chim. Acta, 2019, 1082, 176–185 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an00436k |
| This journal is © The Royal Society of Chemistry 2021 |

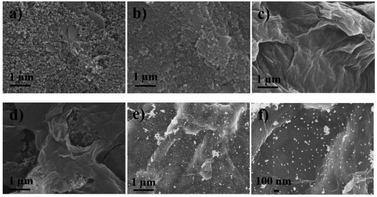
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and (f) high magnification (×50
000) and (f) high magnification (×50