Chemical tools for discriminating single nucleotide variants: from design principles to clinical applications
Dan
Huang†
ab,
Yao
Liu†
a,
Guan Alex
Wang†
a,
Sitong
Lv
a,
Yun
Tan
a and
Feng
Li
 *a
*a
aCollege of Chemistry, Sichuan University, Chengdu, Sichuan 610064, China. E-mail: windtalker_1205@scu.edu.cn
bDepartment of Laboratory Medicine, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, China
First published on 12th November 2025
Abstract
Small variations in nucleic acids, such as single-nucleotide variants (SNVs), can have a profound phenotypic impact and are essential and often confirmatory biomarkers for disease diagnosis. Because of the subtle structural and energetic difference between an SNV and its wild-type (WT) counterpart, accurate discrimination of minute SNVs in complex biological and clinical samples, especially in the presence of high concentrations of WT sequences, presents a formidable analytical challenge. In this review, we provide a comprehensive overview of three mainstream chemical tools for recognizing and discriminating SNVs, with an emphasis on their underlying thermodynamic, kinetic, and enzymatic principles. We also discuss two emerging clinical applications of SNV discrimination tools in the point-of-care diagnosis of infectious diseases and precision management of cancer, which have enabled numerous recent innovations in assay development and device fabrication. By illustrating the design principles and clinical applications, we hope this review will help guide the best use of chemical tools for detecting, quantifying, and enriching SNVs and inspire new ideas, technological advances, and engineering strategies for addressing ongoing clinical challenges.
1. Introduction
Subtle genetic alterations play a pivotal role in shaping biological evolution and driving pathogenesis.1 Even a single nucleotide difference can exert a profound impact on gene expression,2–7 RNA splicing,8–10 and protein folding,11–13 leading to phenotypic changes,14–17 resistance to drugs,18–22 and cancer.23–25 For instance, a single base substitution in the haemoglobin subunit beta gene (GAG to GTG) results in the substitution of glutamic acid with valine. This seemingly minor change induces haemoglobin polymerization, ultimately leading to the development of sickle cell anemia.26 In oncology, many somatic single nucleotide variants (SNVs) act as key driver mutations in the growth and metastasis of tumors. For example, SNVs in codon 12 of the KRAS gene, such as G12D, G12V, and G12C, are commonly observed and are associated with worse overall survival in many KRAS-mutated cancers.27 SNVs in the BRCA1 gene are known to significantly increase the lifetime risk of breast cancer by nearly 6-fold.28 In addition to inherent diseases and cancer, the evolution of pathogenic microbials also relies on mutations. Single nucleotide mutations in the Mycobacterium tuberculosis (MTB) rpoB gene, such as S450L, are the most prevalent mutations leading to rifampicin (RIF) resistance.29 The single D614G substitution in the SARS-CoV-2 genome dominated the global strains in 2021 by enhancing viral fitness.30 Because of their essential roles in pathogenesis, SNVs are ideal disease biomarkers offering confirmatory information for diagnosing genetic disorders, guiding cancer treatment, evaluating gene therapy, tracking antimicrobial resistance, and advancing precision medicine.31–37The profound biological and clinical implications of SNVs drive the development of diverse chemical tools for their precise detection and quantification within complex biological and clinical specimens.34,38–45 However, this pursuit presents formidable analytical challenges. Firstly, the minimal perturbation of the overall physicochemical properties of nucleic acids by a single base substitution renders the discrimination between SNVs and wild-type (WT) sequences exceptionally difficult. Thermodynamically, the free energy differences (ΔΔG°) between a SNV and its WT counterpart only range between 1.5 and 7 kcal mol−1.46 Because of the subtle ΔΔG°, the design of hybridization probes that selectively recognize SNVs requires exquisite fine-tuning of the probe affinity, the process of which involves a trade-off between binding affinity and sequence selectivity. Secondly, clinical scenarios, such as the analysis of cell-free DNA (cfDNA) in blood samples, frequently demand the detection and quantification of rare SNVs (often down to 0.01% allele frequency) within a vast excess of WT background.34,47 This overwhelming concentration disparity between a SNV and its WT can readily mask the intrinsic ΔΔG°, where erroneous hybridization with the abundant WT sequences obscures the detection signals from minute SNVs. Low-abundance SNVs also pose a significant challenge to high-throughput sequencing technologies, requiring high sequencing depths that escalate data volume, computational burden, upfront cost, and turnaround time. Lastly, the inherent biological diversity of SNVs complicates assay design and clinical applications. The prevalence of phenotypically silent synonymous mutations necessitates the development of technologies capable of selectively detecting pathogenic SNVs while tolerating adjacent silent variants.48–51 Furthermore, personalized diagnostics, particularly in oncology, require multiplexed detection of diverse, patient-specific SNV profiles.35,52–57 Similarly, tracking pathogen evolution (e.g., SARS-CoV-2 variants or antimicrobial resistance) often mandates the simultaneous identification of multiple co-occurring mutations.58–63 Consequently, these intertwined challenges in achieving sufficient specificity, sensitivity, and multiplexing capability for diverse SNVs drive persistently the innovation and iterative advancement of chemical tools and bioanalytical methods for discriminating SNVs.
In this review, we aim to elucidate the fundamental basis of chemical tools for recognizing and discriminating single nucleotide mutations by examining their underlying thermodynamic, kinetic, or enzymatic principles, thereby providing a comprehensive analysis of recognition mechanisms and assay design strategies. Specifically, we focus on three main streams of chemical tools (Fig. 1), including (1) hybridization probes for affinity-based selective recognition, (2) enzyme-assisted tools for selective catalysis, and (3) emerging CRISPR technology offering multiple recognition modes for SNV discrimination. Building upon the in-depth understanding of molecular mechanisms for diverse assay designs, we further emphasize on innovations in SNV discrimination tools for two emerging clinical applications, including (1) the field-deployable tests and devices for detecting and screening pathogenic microbial variants and drug-resistant strains in point-of-care (POC) settings and (2) enrichment tools enabling the high-throughput capturing and sequencing of large panels of SNVs for cancer diagnosis and prognosis. For the sake of consistency, this review uses SNVs as the terminology to refer to all targeted single nucleotide mutations and WTs as the spurious counterparts. It is worth noting that single nucleotide polymorphism (SNP) is typically used in the literature for SNVs present at frequencies of at least 1% in a population. While the same discrimination principle also underpins many advanced imaging techniques, such as single molecular fluorescence in situ hybridization (smFISH)64,65 and spatial transcriptomics with single-base resolutions,66 these applications have been extensively reviewed elsewhere and thus are not the primary scope of this review.39,41
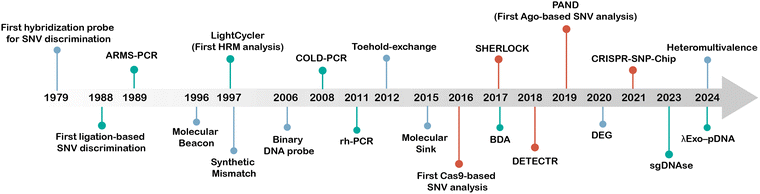 | ||
| Fig. 1 A timeline outlining the critical development and milestones for hybridization (blue), enzyme (green), and CRISPR (red) based tools for discriminating SNVs. | ||
2. Hybridization-based tools for SNV discrimination
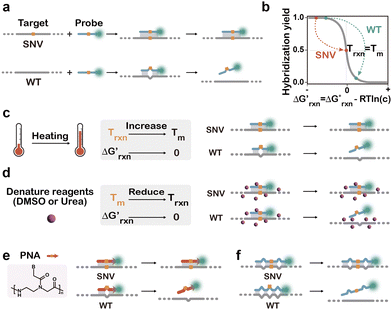
In 1979, Wallace et al. reported the observation that single-base mismatch could alter the thermal stability of short DNA duplexes, opening the possibility to discriminate SNVs using synthetic DNA hybridization probes.67 The detection of clinically relevant SNVs was first achieved by Conner et al. in 1984, where a sickle cell βS-globin allele was detected by a 32P-labeled synthetic DNA hybridization probe at elevated temperatures.68 While early successes were achieved mainly using denaturing gradient gels, numerous innovations have been made to advance the denaturing strategies and probe designs, as well as assay format and signal readout systems.69–72 Fundamentally, the discrimination of a SNV from its WT counterpart relies on their thermodynamic difference where a hybridization probe binds selectively to SNVs rather than WTs. The need for fine tuning hybridization affinities drives the development of various denaturation-based strategies and innovative hybridization probes for SNV discrimination. Besides thermodynamics-based strategies, kinetics-based SNV discrimination, where a hybridization probe binds more rapidly to SNVs over WTs, has also been extensively explored, offering an alternative design route for many nonequilibrium hybridization techniques with single nucleotide resolutions.2.1. Denaturation-assisted hybridization probes
When discriminating SNVs, the hybridization reaction between a correct target (C) and the hybridization probe (P) can be simplified as C + P → CP. For a spurious WT target (S) containing a single nucleotide mutation, the reaction can be expressed as S + P→ SP (Fig. 2a). By establishing a mathematical model, Zhang and colleges demonstrated that the reaction yields for forming the hybridization products (CP or SP) are quantitatively determined by the concentration adjusted reaction free energy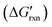 (Fig. 2b).46 As hybridization probes are typically designed to be 15 nt or longer to avoid cross-hybridization with scramble sequences,
(Fig. 2b).46 As hybridization probes are typically designed to be 15 nt or longer to avoid cross-hybridization with scramble sequences, 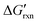 for both SNVs and WTs are often highly negative, leading to nearly 100% reaction yields for both CP and SP at room temperature or 37 °C. Under these conditions, the hybridization probe is unable to discriminate SNVs and the discrimination factor (DF), a quantitative selectivity metric defined as the yields between CP and SP at the same target concentration, is equal to 1. To gain the sequence selectivity at single nucleotide resolution, one must increase
for both SNVs and WTs are often highly negative, leading to nearly 100% reaction yields for both CP and SP at room temperature or 37 °C. Under these conditions, the hybridization probe is unable to discriminate SNVs and the discrimination factor (DF), a quantitative selectivity metric defined as the yields between CP and SP at the same target concentration, is equal to 1. To gain the sequence selectivity at single nucleotide resolution, one must increase 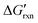 via the increase of standard hybridization free energy
via the increase of standard hybridization free energy 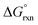 , which serves as the thermodynamic basis for many denature-based methods for SNV discrimination.73,74
, which serves as the thermodynamic basis for many denature-based methods for SNV discrimination.73,74
Practically, the optimal hybridization reaction temperature (Trxn) can be estimated based on the melting temperature (Tm), referring to the temperature at which half of the hybridization products denature to single-stranded forms. When Trxn approaches Tm, 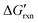 approaches 0 and the difference between the reaction yields for the correct and spurious hybridization products becomes significant. Therefore, operating the hybridization reaction near Tm has become a rule of thumb for most denaturation-based strategies to obtain optimal sequence selectivity for SNV analysis meanwhile maintaining sufficient binding affinity (Fig. 2c).75–77 Besides elevating Trxn to match Tm, chemical denaturation offers an alternative strategy to reduce the Tm of hybridization products, enabling SNV discrimination at lower temperatures. Denaturing chemicals, such as DMSO, urea, and formamide, disrupt hydrogen bonding and hydrophobic interactions, thereby destabilizing base pairing and reducing duplex stability (Fig. 2d).78,79 This approach enables the finetuning of probe affinities at constant temperature and is ideal for detection systems that are sensitive to temperatures.77
approaches 0 and the difference between the reaction yields for the correct and spurious hybridization products becomes significant. Therefore, operating the hybridization reaction near Tm has become a rule of thumb for most denaturation-based strategies to obtain optimal sequence selectivity for SNV analysis meanwhile maintaining sufficient binding affinity (Fig. 2c).75–77 Besides elevating Trxn to match Tm, chemical denaturation offers an alternative strategy to reduce the Tm of hybridization products, enabling SNV discrimination at lower temperatures. Denaturing chemicals, such as DMSO, urea, and formamide, disrupt hydrogen bonding and hydrophobic interactions, thereby destabilizing base pairing and reducing duplex stability (Fig. 2d).78,79 This approach enables the finetuning of probe affinities at constant temperature and is ideal for detection systems that are sensitive to temperatures.77
While the use of denaturing chemicals is not always feasible and reproducible, molecular engineering of hybridization probes offers much enhanced predictability and reproducibility for denaturation-based SNV discrimination. Shortening the probe length is a straightforward method to reduce Tm and enhance SNV selectivity, but this approach often compromises binding affinity. As a compensation, chemical modifications, such as locked nucleic acids (LNAs) and peptide nucleic acids (PNAs), were introduced into probe design. With a rigid backbone via a methylene bridge between the 2′-oxygen and 4′-carbon of ribose, LNAs effectively elevate Tm by 2–8 °C per single incorporation, allowing shorter hybridization probes with uncompromised affinities.80–84 PNAs with a sugar-phosphate backbone replaced with N-(2-aminoethyl)-glycine units eliminate electrostatic repulsion with target nucleic acids, and thus achieve higher affinity (Fig. 2e). Moreover, PNA probes generated greater ΔΔG° for SNVs because of larger local distortion of mismatched duplex structures.85–90
Besides shortened hybridization probes, introducing deliberate mismatches into probes can also destabilize hybridization products without altering the probe length (Fig. 2f). For example, Guo et al. demonstrated SNV analysis with enhanced specificity by introducing artificial mismatches into hybridization probes.91 This strategy was then adopted in DNA microarrays, where imperfect matching greatly increased selectivity.92 Lee et al. further proposed a computational algorithm ProDeG to help design mismatch-containing probes in microarrays.93 With rational designs, mismatches could also be incorporated to enhance sequence selectivity and reduce off-target effects. For example, Cisse et al. established a rule of seven, suggesting that seven contiguous base pairs produced rapid annealing of oligonucleotides.94 Therefore, evenly allocating mismatch positions can avoid off-target effects for small interfering RNAs, antisense oligonucleotides, and CRISPR/Cas systems.
2.2. Structured hybridization probes
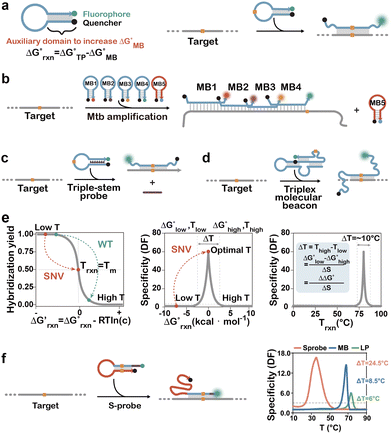
In 1987, Moser and Dervan achieved site-specific DNA recognition with single-nucleotide precision via triple-helix formation.95 Building on this seminal work, Roberts and Crothers introduced the stringency clamp strategy in 1991, establishing one of the earliest classes of structured hybridization probes capable of SNV detection.96 In 1997, Kramer and colleagues conducted rigorous thermodynamic analysis of molecular beacons (MBs), another seminal class of structured probes, and demonstrated that enhanced sequence selectivity was as a general feature of all structurally constrained probes.97 Collectively, these pioneering efforts promoted the rational design and widespread adoption of diverse structured hybridization probes for high-fidelity SNV discrimination.First introduced by Tyagi and Kramer in 1996, MBs represent one of the most successful classes of structured hybridization probes for nucleic acid testing and SNV discrimination.98–101 They adopt a hairpin structure, wherein the loop domain serves as the target-recognition sequence, while the stem duplex provides structural stabilization. Because of the intrinsic folding energy  , hybridization of the target sequence to MBs must disrupt the hairpin structure and is less thermodynamically favourable than standard hybridization, thus allowing for higher specificity to SNVs (Fig. 3a). Moreover, the termini of the stem are chemically conjugated with a fluorophore–quencher pair for signal reporting. Upon hybridization with a complementary target strand, the thermodynamically favored target–probe duplex formation induces structural transition from the hairpin conformation, spatially separating the fluorophore from the quencher and restoring fluorescence. Owing to their inherent specificity and real-time signal transduction capabilities, MBs have been widely employed for SNV analysis in diverse biological and clinical settings. For example, Cepheid's Xpert MTB/RIF took advantage of real-time PCR and five MBs targeting RIF-resistance mutations in a highly variable core of the Mycobacterium tuberculosis (MTB) rpoB gene.100,102–104 This commercial assay effectively covers over 90% RIF-resistant MTB strains and has thus been recommended by the World Health Organization as a preferred test for the rapid screening of MTB infection and RIF resistance in both centralized laboratories and resource-limited settings (Fig. 3b).
, hybridization of the target sequence to MBs must disrupt the hairpin structure and is less thermodynamically favourable than standard hybridization, thus allowing for higher specificity to SNVs (Fig. 3a). Moreover, the termini of the stem are chemically conjugated with a fluorophore–quencher pair for signal reporting. Upon hybridization with a complementary target strand, the thermodynamically favored target–probe duplex formation induces structural transition from the hairpin conformation, spatially separating the fluorophore from the quencher and restoring fluorescence. Owing to their inherent specificity and real-time signal transduction capabilities, MBs have been widely employed for SNV analysis in diverse biological and clinical settings. For example, Cepheid's Xpert MTB/RIF took advantage of real-time PCR and five MBs targeting RIF-resistance mutations in a highly variable core of the Mycobacterium tuberculosis (MTB) rpoB gene.100,102–104 This commercial assay effectively covers over 90% RIF-resistant MTB strains and has thus been recommended by the World Health Organization as a preferred test for the rapid screening of MTB infection and RIF resistance in both centralized laboratories and resource-limited settings (Fig. 3b).
 | ||
| Fig. 3 Structured hybridization probes for SNV discrimination. (a) A representative stem-looped molecular beacon (MB) design for discriminating SNVs. (b) MB-based Expert MTB/RIF assay for detecting multiple drug-resistant SNVs.113 (c) The design of a triplex MB with Hoogsteen hydrogen bonding for enhancing assay specificity.106 (d) The design of triple-stem probes with discontinuous duplex domains for enhancing assay specificity.112 (e) Thermodynamic basis of structured hybridization probes with narrow workable temperature windows.114 (f) The design of entropy-compensate probes for discriminating SNVs over wide temperature ranges.114 Modified with permission, Copyright (20223) Wiley. | ||
Increasing the structural complexity of hybridization probes represents a strategic approach to improve SNV discrimination specificity. A pioneering design involves DNA triplex formation, wherein a third strand stabilizes the duplex stem through Hoogsteen hydrogen bonding.96,105 This configuration elevates the energy barrier for mismatched hybridization, significantly enhancing clamping stringency and SNV specificity. The sensitivity and specificity of triplex MBs can be finely modulated by adjusting the length of the triplex-forming domain (Fig. 3c).106 To maintain robust binding affinity, PNAs were employed to facilitate Hoogsteen interactions by mitigating electrostatic repulsion between strands.107–109 Beyond conventional hybridization, triplex-based molecular beacons have been integrated with advanced technologies such as nanopore sequencing to achieve ultrasensitive SNV detection.110 An alternative structural innovation is the triple-stem probe, where a hairpin is stabilized by three discontinuous duplex domains (Fig. 3d). Soh and colleagues demonstrated that this design markedly improves SNV discrimination efficiency.111,112 Crucially, the inherent thermodynamic stability of both probes and probe–target duplexes enables high specificity across a broad temperature range (up to 60 °C), addressing limitations of conventional molecular beacons.112
To draw a general rule to guide the rational design of structured hybridization probes with wide temperature ranges, Li and colleagues established a theoretical framework to determine the mathematical relationship between the optimal reaction temperature range (ΔTrxn) and thermodynamic parameters, revealing that the high entropy penalty was the lead factor resulting in a narrow temperature range with high assay specificity (Fig. 3e).114 With this insight, the authors further introduced an entropy penalty compensation probe (S-probe) with coded intrinsic disorder (Fig. 3f). The target sequence turned on the S-probe through a toehold-mediated strand displacement and released a long single-stranded domain that compensates the entropy penalty arising from the rigid double helical structure of the hybridization product and the reduction in the number of molecules. Compared to a classic MB with a ΔTrxn of only ∼10 °C, the S-probe effectively expanded ΔTrxn to ∼40 °C, enabling robust discrimination of epidermal growth factor receptor (EGFR) L858R mutation in clinical tissue samples collected from lung cancer patients.
2.3. Multicomponent hybridization probes
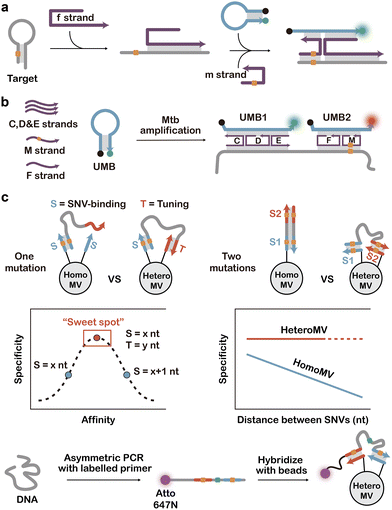
Discrimination of biologically and clinically relevant SNVs often faces challenges caused by the structural and biological complexity of the target sequence. For example, many SNV-containing sequences involve a complex secondary structure, making it difficult for hybridization probes to access the SNV site. Biologically, pathogenic bacteria and viruses are hypervariable; the detection of infectious or drug-resistant strains often required the simultaneous analysis of multiple SNVs within a single target sequence. Toward these challenges, multicomponent hybridization probes demonstrate superior performance, enabling smart discrimination of SNVs in complex nucleic acid targets.Multicomponent hybridization probes were first described by Kolpashchikov and colleagues, where two or more probes were combined to analyse a single target sequence.113,115–122 For a target containing a severe secondary structure, multiple probes acted cooperatively with one MB probe to discriminate the SNV and other probes to disrupt the secondary structure of the target (Fig. 4a). Notably, the selectivity of such multicomponent probes depended not solely on target–probe duplex stability but also on the overall rigidity and structural integrity of the DNA nanostructure. To reinforce complex stability, probes are often designed with 3′- and 5′-termini positioned adjacently, leveraging base-stacking interactions to enhance binding cooperativity.120
 | ||
| Fig. 4 Multicomponent hybridization probes for SNV discrimination. (a) Multicomponent hybridization probes for SNVs located within secondary structures.115 (b) Multicomponent hybridization probes for logic gate-based discrimination of SNVs in complex nucleic acid targets.113 (c) Tuning of binding affinity and sequence selectivity of hybridization probes via a heteromultivalent recognition mechanism, where AND-gate-based detection of multiple SNVs was achieved.125 Modified with permission, Copyright (2024) Springer. | ||
Aside from disrupting the secondary structure, multicomponent probes offer several additional advantages. For example, it enabled the discrimination of SNVs at low and wide temperature ranges via the controlled combination of nonequilibrium hybridization conditions and mismatch-induced increase of equilibration time with respect to that of a fully matched complex.119 It is also possible to integrate signal amplification motifs, such as DNAzymes, into the multicomponent probes to compensate for the compromised assay sensitivity that was initially traded for enhanced specificity for SNV discrimination.123 By harnessing multiple DNA strands, multicomponent probes are also ideal systems for incorporating the molecular logic gate function to achieve smart discrimination of multiple SNVs within a single target sequence. For example, Kolpashchikov and colleagues achieved the detection of MTB RIF-resistant mutations using two MBs with the assistance of five unlabelled probes to achieve an OR-gate-based discrimination of hotspot SNVs (Fig. 4b), whereas conventional Xpert MTB/RIF test relied on five independently labelled MBs.113
Once co-conjugated on the surfaces of micro- or nanoparticles, multicomponent probes demonstrated unique heteromultivalent binding properties to target nucleic acid sequences. This property was first reported by Salaita and colleagues who patterned six DNA probes on a single gold nanoparticle surface for a 90 nt DNA target and found an increase in the affinity of up to 50 orders of magnitude compared to homomultivalent nanoparticles.124 They further demonstrated recently that heteromultivalency also enabled enhanced detection of SNVs by co-conjugating a SNV-binding probe (S) and a tuning probe (T) on a microparticle (Fig. 4c).125 Thermodynamically, the binding through heteromultivalence could be modelled as a cooperative two-step process where the initial hybridization event concentrated the target and thus enhanced the secondary hybridization. With cooperativity, the authors demonstrated that heteromultivalency could control the binding strength more precisely than monovalent binding, enabling near-maximum discrimination of SNVs. Moreover, customizing the spacer length and binding orientation allowed the highly cooperative binding to two SNVs located on the same target sequence. The AND-gate-based detection of Q498R and Y505H mutations of SARS-CoV-2 enabled ∼200-fold higher binding to the genome sequence of omicron strain over the original strain.
2.4. Toehold-exchange probes
Although structured and multicomponent hybridization probes can enhance sequence selectivity via the reduction of the thermodynamic favourability of hybridization, they only work under optimal experimental conditions with well-controlled temperature and salinity. A robust hybridization probe that discriminates SNVs across diverse conditions without retuning is vastly more useful. To achieve this goal, Zhang et al. established a theoretical framework for the analysis of nucleic acid hybridization specificity, providing a benchmark for evaluating the performance of hybridization probes.46 Through the theoretical analysis, they established a set of rules for robust hybridization reactions for discriminating SNVs, including (1) a little change in concentration-adjusted standard free energy , (2) no net change in the number of nucleic acid molecules in solution, and (3) no net change in the number of paired bases. Guided by the thermodynamic criteria, the authors further introduced toehold-exchange probes, where the hybridization of a duplex probe to the correct target was initiated at the short single-stranded domain termed forward toehold (Fig. 5a).126 This reaction further proceeded through a branch migration process and was completed via the spontaneous dissociation of the reverse toehold to release a single-stranded protector. The toeholds allowed the forward and reverse reactions to proceed with fast kinetics and
, (2) no net change in the number of nucleic acid molecules in solution, and (3) no net change in the number of paired bases. Guided by the thermodynamic criteria, the authors further introduced toehold-exchange probes, where the hybridization of a duplex probe to the correct target was initiated at the short single-stranded domain termed forward toehold (Fig. 5a).126 This reaction further proceeded through a branch migration process and was completed via the spontaneous dissociation of the reverse toehold to release a single-stranded protector. The toeholds allowed the forward and reverse reactions to proceed with fast kinetics and 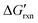 of the reaction was close to 0 when the forward and reverse toeholds were of comparable length (Fig. 5b). Therefore, toehold-exchange offered exceptional specificity to SNV discrimination and meanwhile highly robust against varying environmental factors.
of the reaction was close to 0 when the forward and reverse toeholds were of comparable length (Fig. 5b). Therefore, toehold-exchange offered exceptional specificity to SNV discrimination and meanwhile highly robust against varying environmental factors.
 | ||
| Fig. 5 Toehold-exchange probes for SNV discrimination. (a) Representative toehold-exchange reactions for discriminating SNVs. (b) Thermodynamic basis of toehold-exchange for tuning the selectivity of nucleic acid recognition. (c) Stoichiometric tuning approach to achieve the fine-grained control of probe binding affinity, so that optimal trade-off between sensitivity and specificity can be achieved.127 Reprinted with permission, Copyright (2015) Springer. (d) An anti-toehold approach to achieve the fine-tuning of probe affinity and selectivity via two orthogonal stoichiometrically tunable species.128 Modified with permission, Copyright (2024) American Chemical Society. (e) M-probe design for analyzing long complex nucleic acid targets with multiple and repetitive SNVs.129 Reprinted with permission, Copyright (2017) Springer. | ||
The selectivity of toehold-exchange probes to SNVs is highly tunable by controlling the number of base pairs in both forward and reverse toeholds. Nevertheless, this approach is a coarse-grained tuning, as the rate constant of the strand exchange could change by an order of magnitude with a single base adjustment. To achieve a fine-grained tuning for obtaining optimal trade-off between assay sensitivity and specificity, Zhang and colleagues proposed a stoichiometric tuning strategy by manipulating the concentration of the protector sequence of the duplex probe (Fig. 5c).127 In this strategy, the protector molecule acted as a molecular competitor species that competed with the target for binding to the complementary probe, which allowed the continuous tuning of the hybridization yields from 0% to 100%. This approach was practically highly useful, as it allowed on-the-fly tuning of sensitivity and specificity without the need for re-designing the probe. Recently, Li and colleagues introduced an anti-toehold design into toehold-exchange, so that each hybridization reaction released both a protector and an anti-toehold sequence.128 The two auxiliary species further increased the flexibility for fine-grained tuning of assay specificity and sensitivity, where highly specific SNV discrimination could be achieved with minimal loss of assay sensitivity (Fig. 5d).
The length of most hybridization probes ranges typically from 20 nt to 50 nt, making them not suitable for analyzing SNVs in complex nucleic acid targets that are long, repetitive and/or hypervariable. To address this challenge, Zhang and colleagues invented an M-probe that integrated multiple probe segments, each targeting one section of a potentially long target sequence (Fig. 5e).129 The modular nature of M-probe circumvented the synthesis limitations of oligonucleotides, allowing toehold-exchange with targets of 500 nt long. Moreover, the junctions between the probe segments tolerated up to 7 nt of sequence variation without significantly altering the binding affinity, while single nucleotide variation at other locations resulted in observable signal reduction. Remarkably, the M-probe design successfully enabled the determination of the exact triplet repeat expansion number in the Huntington's gene of genomic DNA using quantitative PCR (qPCR).
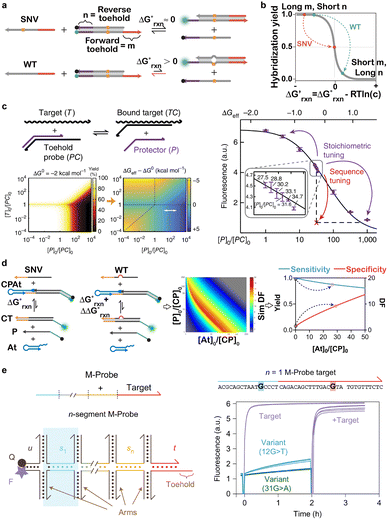
Because toehold-exchange is also a fundamental building block in dynamic DNA nanotechnology, it also offers exceptional programmability to design and operate natural or synthetic machineries for ultrasensitive and ultraspecific SNV discrimination. Riboregulators are RNA-based machineries that can be genetically encoded to translate nucleic acid sensing events into reporter proteins synthesized by the cell or in cell-free transcription–translation systems. To enable such a powerful machinery for analyzing SNVs, Green and colleagues developed a de novo-designed riboregulator termed a single-nucleotide-specific programmable riboregulatory (SNIPR) that was capable of discriminating transcript SNVs in vitro in cell-free systems (Fig. 6a).130 SNIPR was designed to contain a core hairpin secondary structure, positioning the ON- and OFF-state configurations of the riboregulatory near chemical equilibrium. The binding of a correct RNA target yielded an OFF- to ON-state transition with a slightly negative free energy via toehold-exchange. In contrast, binding of a spurious target with a single nucleotide mutation led to a positive free energy, thus biasing the riboregulatory toward the OFF-state. Once positioned at the ON-state, SNIPR exposed both the ribosomal binding site (RBS) and the start codon to activate translation of the downstream output genes, such as fluorescent proteins and beta-galactosidase. By further coupling SNIPR with isothermal amplification and paper-based cell-free systems, the authors achieved detection of cancer-causing SNVs from clinical blood samples and identification of virus strains using convenient colorimetric readouts.
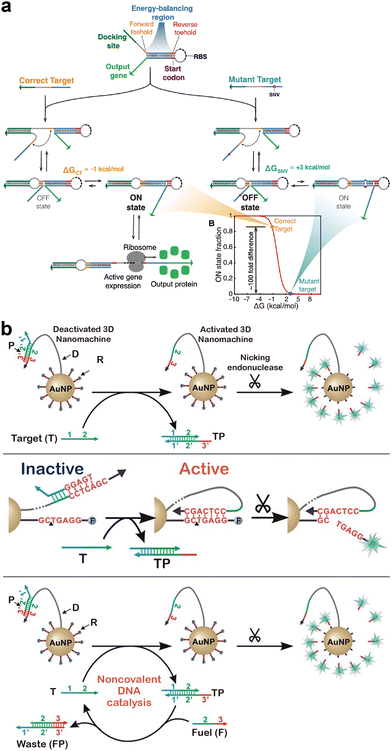 | ||
| Fig. 6 (a) The SNIPR system that translated SNV recognition into the protein expression event via a de novo-designed riboregulator activated by toehold-exchange.130 Reprinted with permission, Copyright (2020) Elsevier. (b) The design of a three-dimensional DNA nanomachine that amplified SNV recognition via the cleavage of hundreds of fluorogenic substrates using a toehold-exchange-activated stochastic DNA walker.131 Reprinted with permission, Copyright (2018) Royal Society of Chemistry. | ||
In addition to naturally occurring machineries, toehold-exchange has also been integrated into synthetic DNA nanomachines for amplified detection of SNVs. For example, Li and colleague engineered a nicking endonuclease-powered three-dimensional DNA nanomachine (3DDN) with toehold-exchange-based target activation to achieve highly sensitive and specific discrimination of SNVs (Fig. 6b).131 3DDN was constructed by co-conjugating a DNA walker molecule containing a nicking recognition site and hundreds of DNA reporters containing nicking cleavage sites on the same AuNP. Therefore, once activated via toehold-exchange, each SNV could trigger the release of hundreds of reporters and thus amplify the detection signal.
Toehold-exchange can also be used to trigger the SNV-specific formation of DNA nanostructures, generating structure-selective detection signals for SNV discrimination. Recently, Keyser and colleagues demonstrated an example of this, where RNA/DNA origami nanotechnology was combined with glass nanopore sensing for SNV detection in complex RNA structures (Fig. 7).132 In this approach, the target RNA molecule served as a scaffold and hybridized with designed DNA probes to form a stable RNA/DNA structure, protecting RNA from degradation. To detect SNVs, a toehold domain was designed adjacent to the target site, inducing the binding and folding of DNA probes through toehold-exchange. A single mismatch would lead to failure of assembly and incomplete RNA/DNA origami. By sensing the shape differences between SNVs and WTs using the ∼10 nm nanopore, distinct ionic current patterns were generated, allowing SNV discrimination in 16S rRNA between E. coli and S. typhi.
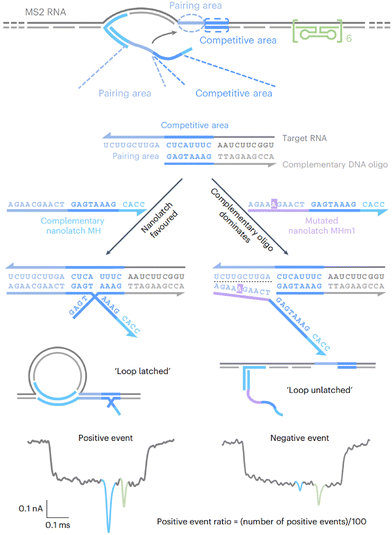 | ||
| Fig. 7 Detection of SNVs in complex RNA targets using mutation-specific RNA/DNA assembly and nanopore sensing.132 Reprinted with permission, Copyright (2025) Springer. | ||
2.5. SNV discrimination in double-stranded DNA
As hybridization probes rely on Watson–Crick base paring to bind and discriminate SNVs, they can only act on ssDNA. Nevertheless, most genomic markers and nucleic acid amplicons are double-stranded. Therefore, double-strand to single-strand conversion is often involved for hybridization-based SNV discrimination, which is typically achieved through asymmetric amplification, thermal or chemical denaturation, and enzymatic degradation of the antisense oligonucleotide (Fig. 8a). In pursuing more efficient methods for SNV discrimination in dsDNA, Kamenetskii et al. introduced the design of homopyrimidine PNA openers, which induce localized strand separation by forming intramolecular PNA/DNA triplex structures at designated SNV sites (Fig. 8b).108,133,134 Compared to conventional denaturation methods, this partial denaturation approach was readily compatible with subsequent target recognition without the need for complicated clean-up steps and was particularly advantageous for one step SNV analysis or imaging. Kelley and colleagues also combined thermal denaturation with PNA clamps to selectively deplete WT sequences for the detection of low-abundant SNVs in clinical samples (Fig. 8c).135 This clutch probe strategy was further combined with an ultrasensitive nanostructured electrochemical platform, enabling the detection of clinically relevant SNVs at frequencies as low as 0.01%.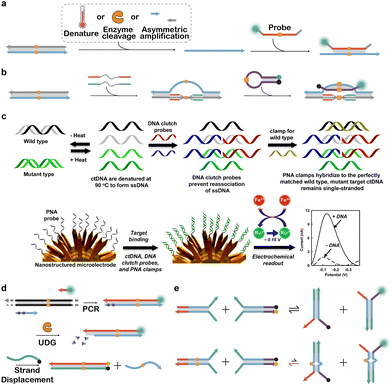 | ||
| Fig. 8 Strategies for discriminating SNVs in double-stranded DNA (dsDNA). (a) Classic workflows for analyzing SNVs in dsDNA, which involve the generation of single-stranded DNA (ssDNA) via denaturation, enzymatic digestion, or asymmetric nucleic acid amplification. (b) In situ generation of an ssDNA window by partially denaturing dsDNA using PNAs as molecular clamps.108 (c) A DNA clutch assay for the discrimination of low-abundant SNVs, which involved the generation of ssDNA using short synthetic clutch probes and depletion of WT sequences using a PNA clamp. Nanostructured electrodes were further used to amplify the detection signal for SNVs.135 Reprinted with permission, Copyright (2016) American Chemical Society. (d) Direct analysis of double-stranded PCR amplicons using toehold-exchange, where a toehold domain was created by cleaving the deoxyuracils via uracil-DNA glycosylase (UDG).136 (e) SNV discrimination via four-stranded toehold-exchange, where both target dsDNA and hybridization probes were engineered with two toehold domains for highly specific discrimination of SNVs.137 | ||
Beyond dsDNA to ssDNA conversion, efforts have also been made to directly analyze SNVs in dsDNA via toehold-exchange probes. For example, Ellis et al. achieved the direct analysis of double-stranded PCR amplicons using toehold-exchange.136 To do so, they replaced deoxythymidine with deoxyuracil in one PCR primer and then treated PCR amplicons using uracil-DNA glycosylase (UDG). As UDG excised uracil bases, a toehold domain was created on the dsDNA amplicon upon UDG treatment, allowing subsequent toehold-exchange and specific discrimination of SNVs (Fig. 8d). Zhang et al. introduced a double-stranded toehold-exchange probe featuring two toeholds on each strand of the duplex probe (Fig. 8e).137 Two corresponding toeholds were also engineered to dsDNA via unbalanced PCR, enabling double-stranded toehold exchange. Based on this mechanism, the detection of each SNV generated two thermodynamically destabilizing mismatch bubbles rather than single mismatch formed during typical hybridization-based assays. Using this probe, the authors successfully discriminated RIF-resistant SNVs in a 198 bp subsequence of the Escherichia coli rpoB gene.
For clinical scenarios such as cancer liquid biopsy, the detection of a minute amount of SNVs is severely interfered by high concentrations of the WT counterpart. Because the detection signals increased monotonically as a function of target concentration for nearly all hybridization probes, the same signal may arise from a minute amount of SNVs or from high concentrations of the WT interference, making the discrimination of low-abundant SNVs highly challenging. In the face of this challenge, a transformative breakthrough came from Li and colleagues who introduced the concept and design strategies of a DNA equalizer gate (DEG).138 By re-engineering pathways of dsDNA renaturation using rationally designed hybridization probes, the DEG redefined the quantitative relationship between the target concentration and the detection signal from a monotonic sigmoidal curve to a unimodal curve (Fig. 9a). This mathematical transformation drastically suppressed the detection signals of high concentrations of WT interferences but had little impact on the low-abundant SNV and thus greatly expanded the concentration window for SNV discrimination. In a subsequent study, they further established a theoretical model for the DEG and predicted that complete elimination of interfering signals could be achieved by increasing probe components. Guided by this prediction, Huang et al. developed a post-amplification SNV-specific DNA assembly (PANDA) method based on the principle of DEG (Fig. 9b), achieving the ultraspecific detection of EGFR SNVs in tissue and plasma samples collected from 108 lung cancer patients.139 The success in DEG and PANDA directed a new design principle for enhancing sequence selectivity of hybridization reactions, which inspired many recent research efforts aiming to improving the specificity for SNV discrimination by reshaping the reaction pathways of SNV discrimination. For example, Zhang and colleagues reprogrammed the reaction pathways of hybridization reactions by introducing two competitive hybridization probes with overlapping domains containing the SNV site.140 By independently monitoring the binding of the two competitive probes, the authors achieved ratiometric readouts that were highly sensitive to low-abundant SNVs meanwhile suppressed by high-abundant WT interferences.
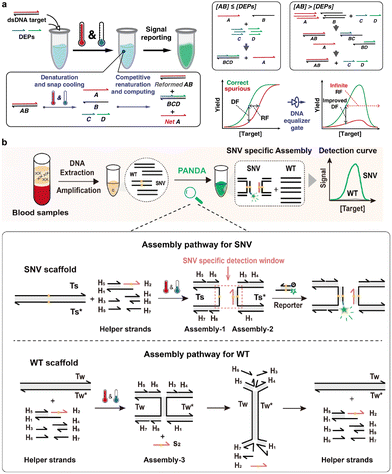 | ||
| Fig. 9 (a) The design of a DNA equalizer gate (DEG) that converted a dsDNA target into a ssDNA for subsequent SNV discrimination using short synthetic DNA equalizer probes (DEPs). DEPs discriminated low-abundant SNVs by re-engineering the reaction pathways of DNA renaturation, where detection signals of high abundant WT were effectively suppressed.138 Reprinted with permission, Copyright (2020) Springer. (b) Engineering post-amplification SNV-specific DNA assembly (PANDA) via the principle of DEG, enabling highly specific detection of cancer-related low-abundant SNVs in clinical blood samples of lung cancer patients.139 Reprinted with permission, Copyright (2024) American Chemical Society. | ||
2.6. Kinetics-based hybridization probes
In addition to the difference in binding free energy at equilibrium, a single nucleotide mismatch between a target sequence and a hybridization probe also alters the kinetics of hybridization. Similar to the small ΔΔG°, the difference in the rate constant between a SNV and its WT counterpart is also small and thus needs to be enhanced to ensure sufficient sequence selectivity. Competitive hybridization between two distinctively labelled hybridization probes, with one for the SNV and the other for the WT, is a commonly used strategy in qPCR and DNA microarrays to enhance the kinetic difference between fully matched and mismatched hybridization events.141–143 Competitive hybridization also serves as the fundamental basis of rational designed molecular sinks that drastically enhance the specificity for SNV discrimination. For example, Xiao and colleagues constructed a theoretical model for competitive DNA hybridization and revealed that assay sensitivity and specificity were inversely correlated over the length and concentration of the sink probes.144 The model further suggested that the trade-off between sensitivity and specificity was a result of the inverse correlation between thermodynamics and kinetic trap. Building on this understanding, they further invented a 4-way strand exchange led competitive DNA testing (SELECT) system that broke the inverse correlation and thus allowed enhanced specificity with long sink probes with little influence on assay sensitivity (Fig. 10a).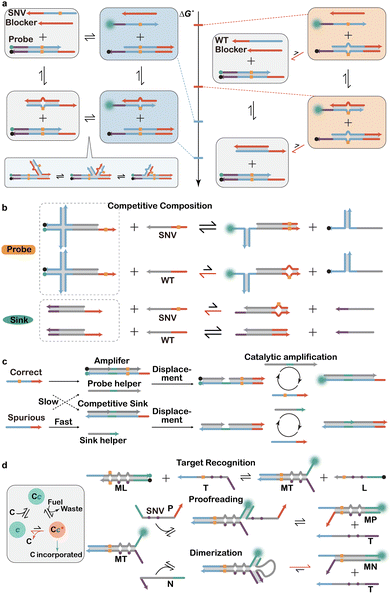 | ||
| Fig. 10 Discriminating SNVs through kinetics-based hybridization probes. (a) A rationally designed blocker serving as a molecular sink to competitively bind to the WT interference.144 (b) Toehold-exchange-based sink design for the competitive blockage of WT binding to the toehold-exchange probe.140 (c) Expanding the molecular sink with a non-covalent DNA circuit to achieve amplified discrimination of SNVs.146 (d) A synthetic kinetic proof-reading system for enhancing the specificity of SNV discrimination.147 | ||
Rational designed molecular sinks have also been integrated with toehold-exchange probes to further enhance the specificity for SNV discrimination. For example, Zhang and colleagues described the design of X-probes where a toehold-exchange probe for a SNV was supplied with a toehold-exchange sink for the corresponding WT.140 They demonstrated both in silico and experimentally that an optimal combination of thermodynamic parameters of the probe and the sink existed attaining an extremely sequence selectivity for the SNV (Fig. 10b). To accurately and quantitatively emulate the hybridization process, they further constructed a kinetic model by assuming a universal forward toehold binding rate of 3 × 105 M−1 s−1 and calculating the reverse rate constants based on the equilibrium constants. These kinetic simulations thus suggested optimal reaction free energies for both the probe and the sink, which in turn guided the sequence-level design. Remarkably, this simulation-informed competition compositions achieved a median 890-fold selectivity for 44 cancer-related SNVs, representing nearly 30-fold improvement over existing hybridization-based assays.
A toehold-exchange-based molecular sink has been integrated into several advanced DNA reaction networks for the analysis of challenging SNVs with small ΔΔG°. For example, Li and colleagues recently integrated a DEG and a molecular sink to achieve the utlraspecific detection of challenging pharmacogenetic SNVs, such as CYP2C19 mutations.145 Advanced sink approaches have also been achieved via catalytic DNA circuits with complex kinetic behaviors and signal amplification capacity. For example, Chen and Seelig described an engineered kinetic amplification mechanism for SNV discrimination via an entropy-driven catalytic DNA circuit (Fig. 10c).146 Operated by toehold-exchange, this catalytic DNA circuit harnessed the target to initiate the first strand displacement reaction to expose a hidden toehold domain that mediated the secondary strand displacement reaction with a rational designed fuel strand to produce the final catalytic product and released the target strand. As the target strand acted essentially as a catalyst to enable multiple turnovers, signal amplification could be achieved. While the DNA circuit was designed to preferentially amplify a signal in the presence of a correct target, a sink was also introduced to preferentially bind and inactivate the spurious target containing a single nucleotide mismatch. The authors demonstrated that this system was quadratically better than discrimination due to competitive hybridization alone and meanwhile provided at least 10-fold better sensitivity than standard hybridization probes.
Beyond sink approaches, dynamic DNA reactions with even more complex kinetic behaviors have been recently employed to design hybridization systems with single nucleotide specificity. For example, Ouldridge and colleagues introduced a nature-inspired kinetic proofreading mechanism that was highly specific to SNVs (Fig. 10d).147 Kinetic proofreading is a naturally existing mechanism that employed chemical fuels, such as ATP hydrolysis, to drive an out-of-equilibrium recognition event.148,149 It allowed the examination of the free energy difference between a correct and a spurious target multiple times and thus allowed sensitive discrimination of SNVs. In this synthetic kinetic proofreading system, both toehold-mediated and handhold-mediated strand displacement reactions were designed to drive a dimerization between M and N. This process was initially inhibited by the prehybridized L strand. Upon target recognition, the ML duplex interchanged to MT and was reversibly proofread by the P strand via toehold-exchange. A second monomer N was bound to the MT duplex via handhold and then completed strand displacement to form a dimeric product MN. Like the natural kinetic proofreading, this discrimination mechanism involved the examination of multiple high energy states (ML, MT, and MP) containing single nucleotide mismatches. In addition to this all DNA-based discrimination system, nucleases have also been employed to facilitate the design of dissipative DNA reactions that were sensitive to single nucleotide mutations. Such kinetic systems offered possible solutions to bypass the thermodynamic barriers and trade-offs for discriminating SNVs with little ΔΔG°.
3. Enzyme-assisted tools for SNV discrimination
While hybridization probes achieve high selectivity for SNV recognition by modulating binding affinities, the presence of a single-nucleotide mismatch within the hybridized product also significantly impacts the activity of various nucleic-acid-processing enzymes. For instance, a mismatch at the 3′-end of a primer impedes efficient recognition by DNA polymerase, preventing primer extension and terminating PCR amplification.150 Similarly, mismatches in hybridized complexes hinder the recognition and function of various ligases and nucleases. Selective activation of such enzymes provides an alternative recognition principle beyond simple modulation of probe affinities and is thus widely exploited for SNV discrimination. Furthermore, diverse nucleases also enable selective cleavage of probes in response to SNVs, facilitating the highly specific identification and amplified detection of low-abundant SNVs in clinical or biological samples.151 This section will focus on enzyme-assisted tools for SNV recognition and discrimination, systematically describing their underlying design principles and method development.3.1. PCR-based discrimination of SNVs
With the ability of exponential amplification, high accurate quantification, and ease of use, PCR is used as a gold standard for nucleic acid quantification in both research and clinical laboratories. Because PCR is typically cycled among three temperatures: a high temperature (e.g., 95 °C) for denaturing dsDNA into ssDNA, a low temperature (e.g., 60 °C) for the primer hybridization, and a moderate temperature (e.g., 70 °C) for the primer extension, direct use of hybridization probes, such as molecular beacons and Taqman probes, to discriminate SNVs can be difficult during such thermal cycles. Therefore, efforts have been made to analyze PCR products after amplification using melting analysis, where hybridization probes can be engineered to provide distinct melting profiles between a SNV and a WT. To further push the LOD for low-abundant SNVs, modifications of PCR reagents or thermal cycles have been made to achieve allele-specific amplification, where SNVs of interest are amplified more efficiently than the corresponding WT sequences. Besides qPCR, digital PCR has also emerged and expanded in the past two decades, representing a paradigm shift for SNV analysis from bulk solutions to single molecules.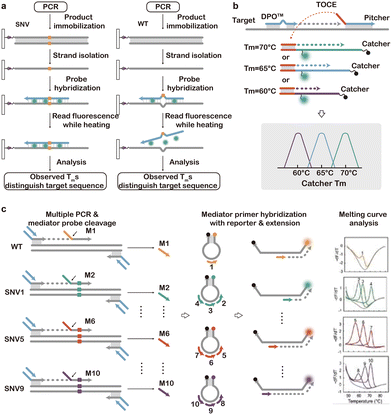 | ||
| Fig. 11 Post-PCR high-resolution melting (HRM) analysis of SNVs. (a) LightCycler for HRM analysis of PCR amplicons produced by the SNV or the WT.152 (b) Multiplexed HRM analysis of SNVs using tagging oligonucleotide cleavage and extension.157 (c) Multiplexed analysis of SNVs using MeltArray with fluorescence and melting profiles as orthogonal signal readouts.158 Modified with permission, Copyright (2022) National Academy of Sciences. | ||
As HRM provides Tm as a quantitative index on top of conventional qPCR readouts, it has also been used to improve the throughput of qPCR, where multiplexed detection of SNVs could be achieved without the need for adding additional fluorescence channels. In 2012, Fu et al. reported a multiplex probe amplification (MPA) technique, where engineered duplex probes with the same fluorescence label but distinct Tm values were introduced.159 In the presence of the target SNV, the corresponding probe was consumed during PCR amplification, which would eventually be reflected in HRM analysis. Using MPA, three SNV types of CYP2C9*2 were successfully analyzed in a single fluorescence detection channel. To further enhance the multiplexity, a commercial tagging oligonucleotide cleavage and extension (TOCE) technology was introduced by Seegene Inc. (Fig. 11b).157 In TOCE, a short DNA fragment was generated during PCR via the 5′ nuclease activity of Taq polymerase. This short fragment then initiated a primer extension converting a coiled DNA probe into a duplex probe with well-designed length and GC content. As such, both fluorescence increase and Tm value could be used orthogonally to expand the multiplexity for high-throughput SNV analysis. Huang et al. recently developed the MeltArray technology, enabling the simultaneous detection of 10 SNVs in codons 12 and 13 of the huma KRAS gene in a single PCR reaction by combining HRM with four distinct fluorescence channels (Fig. 11c).158 Similar to TOCE, MeltArray also employed the 5′-flap endonuclease activity of Taq DNA polymerase to cleave the target-specific mediator probes into mediator primers during the PCR process. These mediator primers bound and extended rationally designed molecular beacons into a series of fluorescent dsDNA duplexes of different length, where each target SNV in a sample is identified by a unique combination of fluorescence color and Tm. By translating the SNV-specific cleavage event into the production of dsDNA barcodes with distinct melting profiles, advanced HRM strategies offer a powerful solution for high-throughput SNV analysis. Nevertheless, as melting profiles of SNV and WT mixtures could be difficult to resolve, HRM is not applicable to clinical scenarios where detection of low-abundant SNVs is required.
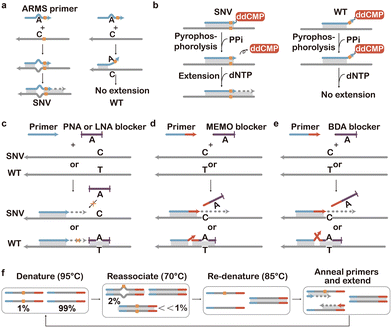 | ||
| Fig. 12 SNV discrimination based on allele-specific PCR. (a) ARMS-PCR that discriminates SNVs by preventing WT extension using the mutation and a synthetic mismatch. (b) Pyrophosporolysis-activated polymerization (PAP for SNV selective activation and extension of the primer.165 (c) Allele-specific polymerization using PNA and LNA blockers.167,169 (d) Allele-specific polymerization using 3′-modified DNA blockers.170 (e) Allele-specific polymerization using blocker displacement amplification (BDA).171 (f) COLD-PCR for allele-specific SNV amplification via programming annealing temperatures.173 | ||
Standard ARMS-PCR can effectively discriminate SNVs down to 1% abundance but does not consistently offer sufficient sequence selectivity below this level. This is because some mismatches are more easily recognized by polymerases, leading to false positive amplification of the WT sequences. To address this issue, allele-specific blockers have been introduced to selectively occupy the binding site of the ARMS primer on the interfering WT sequences. The blockers were typically functionalized with a MGB at the 3′ end to improve binding affinity and prevent polymerase extension.162 Competitive allele-specific Taqman PCR (castPCR) is a highly successful example of this, which has been commercialized by Thermo Fisher for the detection of diverse low-abundant SNVs.163 Besides primers, molecular engineering has also been made to improve the specificity of polymerases. For example, Stadler et al. combined ARMS primers with a modified polymerase (SNPase) with higher extension specificity for the 3′ mismatches.164 Liu and Sommer invented the pyrophosporolysis-activated polymerization (PAP) assay with further improved SNV specificity (Fig. 12b).165 In this assay, the 3′ end of the PAP primer was modified with dideoxynucleotide monophosphate (ddNMP) to prevent direct extension. Only when the PAP primer was fully complementary to the correct template, the terminal ddNMP was removed via pyrophosphorolysis, enabling subsequent primer extension and PCR amplification of the correct template.92,166 The selective removal for matched over unmatched dideoxynucleotides was particularly high, allowing the detection of SNVs with abundances down to 1 in 104–105.
Although gaining high sequence selectivity, the modification of primers inevitably reduced the PCR efficiency and ultimately test sensitivity. Moreover, ARMS primers are specific only until the first spurious extension event generates an amplicon bearing the WT nucleotide and thus are prone to false positive results. To achieve high SNV specificity without modifying primers, numerous blocker PCR approaches have been introduced. As primers for blocker PCR were not allele specific, the primers would amplify both SNV and WT sequences with comparable efficiencies. The sequence selectivity was achieved through rationally designed blockers aimed to suppress WT amplification. Clamping PCR represents one of the earliest reported Blocker PCR techniques, in which Ørum et al. employed a PNA as the blocker.167 A PNA, a synthetic nucleic acid analogue composed of repeating N-(2-aminoethyl)-glycine units linked by peptide bonds, combines high stability, resistance to enzymatic degradation, and strong binding affinity to complementary DNA and RNA sequences.168 The PNA blocker is designed to be fully complementary to the spurious sequence, thereby suppressing spurious amplification during PCR while allowing the amplification of correct targets, enabling the detection of mutations present at frequencies as low as 0.1% (Fig. 12c). In addition to PNAs, other nucleic acid analogues can also function as blockers. LNA-containing oligonucleotides exhibit higher thermostability when hybridized to perfectly matched DNA, whereas any mismatch significantly destabilizes the LNA/DNA duplex. This pronounced sensitivity to mismatches enables LNAs to effectively suppress spurious amplification while selectively enriching correct SNVs at frequencies as low as 0.1%.169
As the synthesis of these non-natural nucleic acids is costly, Lee et al. introduced a low-cost blocker PCR technique known as Mutant Enrichment with 3′-Modified Oligonucleotides (MEMO) (Fig. 12d).170 MEMO operated like PNA/LNA-mediated PCR clamping but replaced PNAs or LNAs with 3′-modified oligonucleotides. In this method, the 3′ end of the blocker was modified with an extension-inhibiting compound such as a C3 spacer, a C6 amine, or a phosphate group. The blocker sequence was designed to be complementary to the spurious sequence and overlaps several bases with the primer neighboring the target mutation site. The blocker competed with the primer for hybridization to the template, and any mismatch significantly reduced its binding affinity to the template, thereby enabling selective amplification of SNVs over their WT counterparts.
Combining a similar principle with toehold-exchange, Zhang and colleagues invented blocker displacement amplification (BDA), where the blocker overlapped with the primer by 6–14 nt (Fig. 12e).171 The exact overlapping domain for each target was determined by comprehensive thermodynamic considerations, providing this approach with temperature robustness at 56 to 64 °C. The 3′ end of the blocker contained a 12–30 nt reverse toehold domain for enhancing the blockage efficiency. Any sequence variation in the template in this region would manifest a mismatch bubble or bulge in the blocker–template duplex, causing the blocker to be displaced by the primer through toehold-exchange. By doing so, multiplex enrichment of multiple mutations within a single target could be achieved. The MEMO and BDA methods have demonstrated the capability to enrich minority alleles with a limit of detection of 0.1% VAF. Nevertheless, BDA was typically constrained to an enrichment region of 12–30 nt, which posed challenges when detecting closely spaced SNVs, often requiring the design of numerous blocker and primer pairs. Recently, Song and collegues addressed this challenge by developing a Long BDA (LBDA) method. They achieved efficient discrimination of many KRAS and NRAS SNVs by extending the overlapping domains and thus the overall length of the blocker.172
In addition to ARMS primers or varying blockers, control over thermal cycles has also been employed to achieve SNV discrimination. Co-amplification at Lower Denaturation temperature PCR (COLD-PCR) is a representative technique that utilizes the slight Tm differences between DNA heteroduplexes and homoduplexes to selectively amplify minority alleles from mixtures of spurious and correct-containing sequences, regardless of the SNV type or position (Fig. 12f).173 COLD-PCR harnesses the principle that each DNA sequence had a critical denaturation temperature (Tc) lower than the Tm, below which PCR efficiency dropped abruptly. Based on this principle, an intermediate annealing temperature was firstly used during PCR cycling to allow cross-hybridization of spurious and correct alleles, resulting in most correct alleles ending up in a mismatch-containing structure (heteroduplex) that had a lower melting temperature than the fully matched structure (homoduplex). Heteroduplexes were then selectively denatured and amplified at Tc, whereas homoduplexes remained double-stranded and did not amplify efficiently. Advantages of COLD-PCR include its simplicity, the preferential amplification of mutant-containing DNA without the need for cumbersome protocols or additional reagents, and the ability to combine with most downstream assays. Several advanced variants of COLD-PCR have been developed to further improve its performance and specificity. For example, ICE-COLD-PCR (Improved and Complete Enrichment COLD-PCR) incorporated a synthetic reference sequence to selectively inhibit WT amplification, so that low-abundant SNVs at 0.1% to 0.5% allele frequencies could be preferentially enriched.174 TT-COLD-PCR (Temperature-Tolerant COLD-PCR) addressed the challenge of precise thermal control by introducing a gradual temperature increase (spanning 2.5–3 °C) during denaturation, thereby enabling the simultaneous enrichment of multiple mutations across a range of Tm values in a single reaction.175
3.2. Ligation-based SNV discrimination
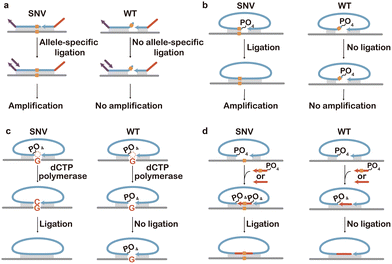
By enzymatically sealing nicks between adjacent residues of a single-stranded break on a double-stranded substrate, ligation is a critical step in living organisms, as well as in many modern molecular biology workflows.38,41 Catalyzed using ligases, a typical ligation process involving the formation of a phosphodiester bond between the 3′ hydroxyl and 5′ phosphate of adjacent DNA residues proceeds in three steps, including (1) the self-adenylation of ligase by reaction with free ATP, (2) the transfer of the adenyl group to the 5′-phosphorylated end of the donor strand, and (3) the formation of the phosphodiester bond via the reaction of the adenylated donor with the adjacent 3′ hydroxyl acceptor.189 Because the formation of the phosphodiester bond relies on the precise base pairing to ensure correct DNA-ligase conformations, the ligation process is highly sensitive to single nucleotide mismatches and gaps near the nick site. Also, the ligation requires the existence of 5′-phosporylation of the donor strand to allow the transfer of the adenyl group. These features make ligation an ideal principle to design chemical tools for discriminating SNVs.39,190,191A straightforward method that harnesses ligation to discriminate SNVs is to position the target SNV adjacent to the nick site, so that spurious hybridization induces DNA helix deformation and the disruption of ligation (Fig. 14a).192–195 By further engineering ligation probes with PCR primers, Schouten et al. invented multiplex ligation-dependent probe amplification (MLPA) for the simultaneous quantification of a large number of nucleic acid sequences.196 By separating and quantifying fluorescently labelled PCR amplicons, the commercialized SALSA MLPA kit (MRC Holland) allowed for the high-throughput determination of up to 60 genomic copy number variations and SNVs in a single reaction. While high concentrations of ligases or long incubation might force mismatch ligation and false positive test results, competitive ligation probes with distinct PCR primers could be used to further enhance the SNV selectivity. For example, Park and colleagues achieved noise-free detection of 15 SNV markers by using competitive ligation probes combined with allele-specific PCR and microarray analysis.197 Allele-specific ligation with competitive probes has also been combined with subsequent strand displacement amplification to enable colorimetric or mass spectrometric readouts.198–200
 | ||
| Fig. 14 Ligation-based SNV discrimination. (a) A representative design of multiplex ligation-dependent probe amplification (MLPA) for SNV discrimination.192–195 (b) A representative design of padlock probes for SNV-specific ligation and circulation.201,202 (c) SNV-specific ligation via filling of a single nucleotide gap.210–213 (d) SNV-specific ligation via sequence-selective filling of a short 5 nt gap.214 | ||
In addition to competitive ligation probes, padlock probes have also been introduced, where the two ends of the single probe were joined by ligation upon hybridization to immediately adjacent target sequences (Fig. 14b).201,202 Compared to two separate probes, padlock probes provide not only the enhanced specificity for SNVs but also the ability to trigger subsequent rolling circle amplification (RCA) via the formation of circular templates.203 Therefore, ligation-based SNV discrimination coupled with padlock probes has been extensively explored over the past few decades, enabling both spectroscopic and imaging readouts for SNV analysis.204–209
To further push the specificity for SNV discrimination, two types of gap-fill ligation strategies were also described. The first type involved the filling of a single nucleotide gap using polymerization, where only target-specific dNTP was supplied (Fig. 14c).210–213 Therefore, the single nucleotide gap could only be filled for the SNV sequence and remained for the WT interference. Consequently, the subsequent ligation and amplification could only occur for the SNV rather than the WT counterpart. This type of gap-fill ligation has been integrated with qPCR, microarray, and electrochemical readouts for genotyping and cancer biopsy.215,216 Nilsson and colleagues described the second type of gap-fill ligation strategy, where a short 5 nt DNA probe was applied to fill the SNV containing gap (Fig. 14d).214 A competitive probe corresponding to the WT sequence was also supplied to enhance the assay specificity. As the competing probe was not phosphorylated, no ligation could be triggered. Upon subsequent RCA, highly sensitive and specific detection of SNVs was achieved in the EGFR gene in fresh frozen and FFPE tumor samples.
3.3. Nuclease-assisted SNV discrimination
While polymerization and ligation-based strategies enable the production of DNA sequences in response to SNV recognition, it is also possible to induce mutation-specific cleavage events via nucleases for SNV discrimination.As the recognition sites of restriction endonucleases are fixed with a typical length ranging from 4 nt to 8 nt, it is critical to match the target sequence with a specific recognition site of a certain enzyme. To enable the detection of targets with no recognition sites, artificial introduction of a restriction site (AIRS) was employed to generate enzyme recognition sites during PCR.222 This method employed a modified primer to change one or more nucleotides for the purpose of creating recognition sites for WT sequences, but not for SNV sequences, thus leading to the cleavage of the WT and the retention of the SNV for subsequent PCR amplification. In addition to PCR, ligation-based strategies, such as amplification via primer ligation at the mutation (APRIL-ATM), have also been employed for the generation of artificial restriction endonuclease sites in target sequences.223 Unlike RFLP, APRIL-ATM cleaved and ligated SNV alleles to an oligonucleotide tail acting as a primer for subsequent PCR amplification. This method applied an initial pre-amplification to the targeted sequence.
Endonuclease IV (Endo IV) is another endonuclease that was engineered for SNV discrimination. Endo IV is a critical DNA repair enzyme that recognizes the abasic sites in DNA strands and cleaves the phosphodiester bond 5′ to the lesion.224 By systematically examining the effect of mismatches on Endo IV activity, Zhao and colleagues demonstrated that Endo IV could effectively discriminate mismatches neighboring the natural abasic site (Fig. 15a).225 Specifically, they found that the introduction of two mismatches at both sides of the abasic nucleotide effectively inhibited the cleavage activity of Endo IV and only mismatch at the 3′ side had no effect on the enzymatic activity. As such, by positioning the targeted SNV at the 5′ side of the abasic nucleotide allows highly sensitive discrimination with DF values ranging from 99 to 860 and the abundance down to 0.1%.
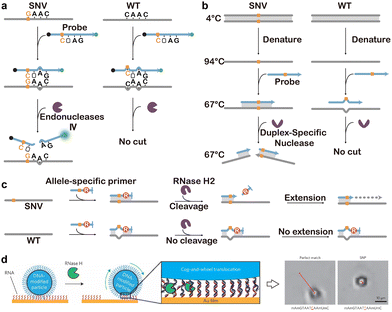 | ||
| Fig. 15 (a) Endonuclease IV-mediated selective cleavage of SNV sequences via the rational introduction of synthetic mismatches.225 (b) Selective cleavage of SNV sequences via duplex-specific nuclease.226 (c) Selective activation of PCR primers containing a SNV-specific ribonucleotide using RNase H2.228 (d) A high-speed DNA-based rolling motor powered by RNase H, converting selective SNV cleavage into the movement of the motor observable using optical microscopy.230 Reprinted with permission, Copyright (2016) Springer. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 common human SNPs with a LOD > 1%.229
000 common human SNPs with a LOD > 1%.229
In addition to PCR-based discrimination, the unique RNA cleavage activity has also enabled dynamic DNA machineries that are sensitive to SNVs. For example, Salaita and colleagues reported a high-speed DNA-based rolling motor powered by RNase H (Fig. 15d).230 The motor consisted of a DNA-coated microparticle that hybridized to a surface modified with complementary RNA. Upon the addition of RNase H, the selective hydrolysis of hybrid RNA with full complementary to DNA drove the movement of the motor particle. The cleavage of thousands of anchoring strands co-hybridized on a single particle through multivalent hybridization triggered the rolling of the microparticle. By visually tracking the moving trajectory of the particle, the authors found that the particle travelled ∼60% slower when a single nucleotide mismatch existed in the DNA–RNA duplex, allowing the conversion of the SNV discrimination into a direct visual readout.
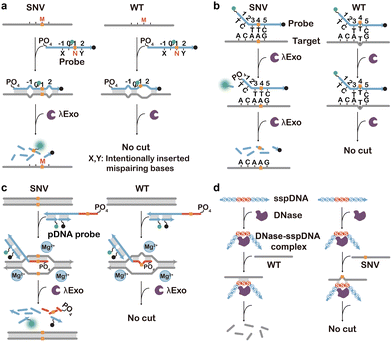 | ||
| Fig. 16 (a) λ-exonuclease-mediated selective cleavage of phosphorated sequences via the rational introduction of synthetic mismatches.231 (b) SNV-specific cleavage mediated by the noncanonical phosphorylation-independent activity of λ-exonuclease.234 (c) Selective recognition and unwinding of SNV-containing dsDNA using λ-exonuclease and phosphorated guide DNA (pDNA) probes, triggering subsequent cleavage of the pDNA probe for fluorescence signal generation.236 (d) DNase-mediated SNV-specific cleavage guided by a single-stranded phosphoreothioated DNA (sspDNA).237 | ||
While λ-exo was considered to execute exonuclease activity in the presence of 5′-PO4, Zhao and colleagues found that dsDNA with a 5′ nonphosphorylated two-nucleotide-protruding end could also be digested by λ-exo with high efficiency.233 More interestingly, chemical modifications, such as fluorescent dyes, at the 5′-end could further accelerate the enzymatic digestion by λ-exo. Such a noncanonical substrate feature made it easier for designing fluorescent DNA probes, as it did not rely on internal fluorescent labels (Fig. 16b).234 In a later study, the authors further demonstrated that this DNA terminal structure-mediated enzymatic reaction was highly sensitive to mismatches near the 5′-end, thus enabling the ultrasensitive discrimination of SNVs in circulating cell-free DNA with the LOD as low as 0.02% for BRAF V600E, NRAS Q61R, and three types of EGFR SNVs in the plasma cancer patients. As λ-exo could be used to convert 5′-PO4 PCR amplicons into ssDNA and then execute SNV-specific cleavage of fluorescently labelled DNA probes, it has also been employed to design one-pot enzymatic reactions for amplified detection of ultralow abundant SNVs in clinical samples.235
Recently, Su and colleagues revealed another nontrivial enzymatic property of λ-exo using single-molecule resonance energy transfer analysis, where λ-exo was found to facilitate 5′-PO4 DNA probes (pDNA) to bind and unwind dsDNA or DNA-RNA duplexes that had complementary sequences with pDNA (Fig. 16c).236 In the presence of Mg2+, pDNA was rapidly digested by λ-exo, allowing the design of fluorogenic pDNA probes and signal amplification methods for dsDNA analysis. Moreover, a single mismatch at the 5′-end or near the middle of the complementary region of pDNA was found to prevent pDNA binding and subsequent enzymatic cleavage. By integrating fluorescently labelled pDNA probes with recombinase polymerase amplification (RPA), direct and multiplexed analysis of RPA amplicons was achieved, allowing the simultaneous detection of D614G and N501Y mutations in the SARS-CoV-2 S gene with high sensitivity and specificity.
4. CRISPR-enabled tools for SNV discrimination
Awarded the 2020 Nobel Prize in Chemistry, CRISPR-Cas technology has emerged as one of the most exciting and rapidly growing fields in chemistry and biology, achieving numerous groundbreaking advancements spanning from gene editing to molecular diagnosis.239–242 As both gene editing and diagnostic applications impose exacting demands on the sequence specificity, intense research efforts have been made to engineer CRISPR systems with enhanced sequence selectivity. Such efforts position CRISPR as an idea toolbox for the detection and enrichment of SNVs. Moreover, CRISPR Cas machineries possess both target-specific endonuclease activity for cis-cleavage of the target and exonuclease activity for trans-cleavage of nonspecific DNA or RNA reporters with multiple turnovers. These unique features make CRISPR Cas highly flexible tools for designing diverse assays and methods for SNV discrimination.374.1. Cas9-enabled tools for SNV discrimination
As one of the first CRISPR systems engineered for genome editing, CRISPR-Cas9 has also gained extensive attention in CRISPR diagnosis. A classic CRISPR-Cas9 system, such as the widely used Streptococcus pyogenes Cas9 (spCas9), consists of three components, including Cas9 protein, crRNA, and tracrRNA. The crRNA contains a 20-nt sequence complementary to the target DNA. Upon target binding, crRNA and tracrRNA further recruit the Cas9 via the alpha-helical recognition (REC) lobe of the protein effector and meanwhile activate its endonuclease (NUC) lobe that degrades the dsDNA target 3-nt away from the 5′-side of the protospacer adjacent motif (PAM).243The short PAM domain (5′-NGG for spCas9) located at the 3′-downstream of the target dsDNA sequence is essential for docking Cas9 onto the target. Single nucleotide mismatches on the PAM domain will significantly disrupt the activity of CRISPR Cas9, which served as a fundamental principle for designing the first generation of Cas9-based probes for SNV discrimination.244–247 For example, in the seminal work conducted by Pardee et al., Cas9 cleavage was combined with nucleic acid sequence-based amplification (NASBA)-mediated RNA amplification to discriminate between the African and American Zika strains (Fig. 17a).243 Because of the single nucleotide difference in the trigger regions of the two strains, a PAM domain only existed in the American strain sequence, which leads to the cleavage by the Cas9 and truncated RNA amplicon during NASBA amplification. Conversely, the African strain sequence did not contain the PAM site and was not cleaved by Cas9, resulting in the full-length RNA amplicon. By further integrating this Cas9-enabled SNV detection system with a cell-free gene-expression system on paper, the authors demonstrated a paper-based diagnostic assay for rapid Zika detection with naked-eye readouts. The PAM-dependent SNV recognition was also integrated with PCR to develop the CUT (CRISPR-mediated ultrasensitive detection of target DNA)-PCR capable of eliminating PAM-containing WT sequences before PCR amplification.248 As CUT-PCR required the oncogenic SNV to destroy the PAM sequence to avoid cis-cleavage, spCas9 and Cpf1 (a type V Cas9 from Francisella novicida) were combined to recognize the 5′-NGG-3′ and 5′-TTN-3′ PAMs, respectively, which covered ∼80% of oncogenic SNVs in the COSMIC database. In addition to PCR, PAM-dependent recognition and enrichment of SNVs were also integrated with downstream isothermal amplification, such as RPA, for the ultrasensitive detection of rare SNV alleles.249–251
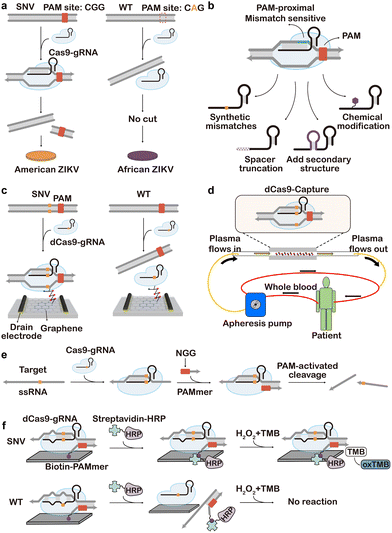 | ||
| Fig. 17 Cas9-enabled assays for SNV discrimination. (a) SNV-specific cis-cleavage of target DNA by destroying PAM via SNV-based mismatch.243 (b) Engineering gRNA to enhance the selectivity for SNV discrimination via strategies, including synthetic mismatch, spacer truncation, adding secondary structure, and chemical modification.259 (c) Amplification-free detection of SNVs using a graphene field-effect transistor immobilized with a dCas9-gRNA complex.282 (d) dCas9-based enrichment of ctDNA into the apheresis machine.281 Modified with permission, Copyright (2022) American Chemical Society. (e) Principle of Cas9 for RNA targeting via a complementary PAM-containing short DNA (PAMmer). (f) PAMmer-based selective recognition of SNV in ssRNA, leading to SNV-specific capture of HRP to generate colorimetric readout.283 | ||
A major drawback of PAM-dependent SNV recognition was the limited sequence combinations even when multiple Cas9 systems were combined. To address this challenge, efforts have been made to investigate and enhance the binding-specificity between the target sequence and the crRNA. By observing the DNA recognition and rejection during interactions with crRNA and spCas9 in real-time using single-molecule FRET, Ha and colleagues found that the dissociation rate between DNA and the Cas9–RNA complex greatly increased from <0.006 s−1 to >−2 s−1 upon introducing mismatches proximal to the PAM.252 This finding demonstrated that it was possible to directly harness Cas9 to discriminate SNVs located within 8–11 nt proximal to the PAM site (so-called seed sequences) (Fig. 17b). This PAM-independent mutation recognition and cleavage principle of spCas9 has been integrated with many up- or downstream amplification systems, such as PCR,253 RCA,254 and EXPAR,255 for the highly sensitive detection and enrichment of SNVs.
As a single mutation on the seed sequences may not be sufficient to completely inhibit the Cas9 activity, gRNAs were further engineered for improving the sequence selectivity to SNVs, including incorporation of more mismatches between the spacer and target sequences,244 truncation of the crRNA by 2–3 nt at the 5′ end,256,257 chemical modifications, such as 2′-fribose modification, to the 5′ end of crRNA,258 and adding secondary structures in the spacer of crRNA (Fig. 17b).259
Besides the widely used spCas9, Cas9 orthologs with enhanced sequence specificity were also discovered and engineered over the past years. For example, using single-molecule FRET, Chen et al. demonstrated that spCas9 was trapped in an inactive state when bound to mismatched targets, which led to the engineering of a hyper-accurate Cas9 variant with much enhanced sequence specificity without compromising on-target activity.260 The Cas9 ortholog from Francisella novicida (FnCas9) was also found to be highly sensitive to mismatches in the sgRNA-DA duplex, as FnCas9 showed negligible binding affinity to substrates that harbored mismatches.261–263 Based on this finding, Maiti and colleagues developed a FnCas9 editor linked uniform detection assay (FELUDA) for accurate identification of SNVs in sickle cell disease and SARS-CoV-2 viral infections via a fluorescence-based lateral flow readout.264
Rather than its conventional role as a target-specific endonuclease that degrades dsDNA to produce a blunt dsDNA break, it is also possible to engineer Cas9 into nickase (nCas9) to produce nicks in dsDNA or nuclease-deficient Cas9 (dCas9) as sequence-specific binders for dsDNA targets through protein engineering.265–268 Specifically, dCas9 can be engineered by introducing H840A and D10A mutations into the NUC lobe, and the introduction of either mutation will lead to the generation of nCas9. Both dCas9 and nCas9 mutants preserve the high sequence selectivity either for PAM-dependent or sgRNA-dependent target recognition and thus have been widely used as engineered chemical tools for SNV discrimination in various assay formats. For example, Zhou et al. integrated strand displacement amplification with nCas9 to develop CRISPR-Cas9-triggered nicking endonuclease-mediated strand displacement amplification (CRISDA) assay.269 Unlike conventional nicking endonuclease-mediated strand displacement amplification where cleavage only occurred at limited nicking cleavage sequences, nCas9 could act on any target DNA containing a PAM sequence. Moreover, the authors further demonstrated that CRISDA was highly sensitive to SNVs on the PAM sequence or first two nucleotides on the seed sequence, allowing discrimination of SNVs related to breast cancer with high sensitivity and specificity. Despite the ultrahigh sensitivity and SNV specificity, CRISDA required complex reaction reagents in addition to nCas9, such as nicking endonuclease, single-stranded DNA binding protein, and PNA probes. Ye and colleagues simplified the assay design by only using nCas9 as the nicking endonuclease and Klenow polymerase for strand displacement amplification at the nicks.270 This system was found to be sensitive to SNVs throughout the PAM and seed sequences and capable of discriminating KRAS SNVs in genomic DNA samples collected from human colon cancer cell lines.249
Besides gFET, similar dCas9-enabled detection platforms with electrochemical,271–274 electrochemiluminescence,275 and impedance readouts276 have also been created for the rapid and sensitive detection of SNVs related to cancer and drug-resistance. Efforts have also been made to immobilize dCas9 on magnetic beads and nanoparticles via click chemistry or streptavidin–biotin conjugation to enable varying spectroscopic readouts for SNV discrimination, such as surface-enhanced Raman spectroscopy277–279 and surface plasmon resonance.280 Besides in vitro assays, dCas9 has also been proposed to enrich ctDNA via an apheresis machine. Apheresis machine is a device that draws out whole blood, separate blood components, and then infuses the blood back into the human body. Downs and Sukumar proposed to integrate dCas9-based enrichment of ctDNA into the apheresis machine by first immobilizing dCas9-gRNA complexes onto magnetic beads via biotin–streptavidin conjugation and then magnetic capturing (Fig. 17d).281 Using BRAF T1799A as a model SNV target, the authors demonstrated efficient capture of the target SNV in unaltered flowing plasma and enriched SNVs by 1.8 to 3.3-fold. Although this work was only demonstrated using a model set-up, it shows an interesting direction for directly enriching ctDNA from cancer patients using CRISPR technology.
As dCas9 can directly recognize SNVs in dsDNA without subsequent cleavage, it has also been used extensively as a highly specific binder for the capture and enrichment of SNV sequences. Immobilization of dCas9 on a solid support, such as electrodes, was one of the most straightforward ways to enable dCas9 based SNV sensing. For example, Aran and colleagues immobilized dCas9 on the surface of a graphene field-effect transistor (gFET) via a chemical linker, 1-pyrenebutanoic acid. The immobilized dCas9 formed a binding complex with a gRNA for capturing SNV associated sickle cell disease or amyotrophic lateral sclerosis. Because of the high sensitivity of gFET, unamplified DNA samples could be analyzed directly during the incubation on the graphene surface (Fig. 17c).282 In the presence of the SNV target, complete hybridization occurred, leaving the target DNA on the gFET surface via the Cas–gRNA complex. For WT targets containing a single nucleotide mismatch, the Cas–gRNA complex had low affinity for the DNA, leading to the dissociation from gFET surfaces. Once removing nonspecifically bound DNA through a rinsing step, electrical measurements were conducted to obtain SNV-specific changes in conductance, source–drain current, transconductance and effective gate potential.
Although CRISPR-Cas9 has evolved to recognize dsDNA, Doudna and colleagues demonstrated that programmable targeting and cleavage of RNAs with Cas9 was possible by partially hybridizing the target RNAs with PAM-presenting oligonucleotides (PAMmers) (Fig. 17e).283 Yeo and colleagues further showed that the dCas9 retained the high binding affinity to RNAs in the presence of PAMmers with no cleavage.284 These pioneering works formulate the design principle to engineer dCas9 for discriminating SNVs in RNA sequences. As an example, using the RNA-binding ability of dCas9, Moon et al. developed a colorimetric assay for the detection of SARS-CoV-2 mutants and drug-resistant pH1N1.285 In this assay, the dCas9–gRNA complex was immobilized on a well microplate, which captured viral RNAs in the presence of biotin-modified PAMmers (Fig. 17f). A synthetic mismatch was incorporated into the gRNA to enhance the sequence selectivity for SNV recognition. By further supplying this system with streptavidin-labeled horseradish peroxidase (HRP) and 3,3′,5,5′-tetramethylbenzidine (TMB), the authors achieved the colorimetric detection of RNAs with SNVs 2-nt adjacent to the PAMmers.
4.2. Cas12-enabled tools for SNV discrimination
The Cas12, including Cas12a, Cas12b, Cas12f (also known as Cas14), and other members, recognizes and cleaves dsDNA in a PAM-dependent manner, producing sticky ends.286 They also recognize and cleave ssDNA in a PAM-free manner. By forming the binding complex among Cas12, crRNA, and the target DNA, the catalytic site of Cas12 is transformed into an active configuration which not only cleaves the target DNA (cis-cleavage) but also nearby nontarget ssDNAs in an indiscriminative manner (trans-cleavage).287 Because trans-cleavage is a multiple turnover process, this feature makes Cas12 an ideal signal amplifier for diverse biosensing and diagnostic applications.288–290 Cas12a is one of the most widely used Cas12 family members for CRISPR biosensing and diagnosis. It consists of a REC lobe for crRNA binding and a NUC lobe for PAM recognition and target-specific cleavage. Upon PAM recognition, Cas12a unwinds dsDNA, and the spacer of crRNA hybridizes to the target strand, forming an R-loop structure together with the non-target strand. The stability of the R-loop was found to be critical to the specificity of Cas12a and was strongly affected by the mismatches between the target DNA and crRNA.291 More importantly, such influences were imposed on both cis- and trans-cleavage activities of Cas12. These observations serve as the fundamental basis for harnessing Cas12a and other Cas12 family proteins for SNV recognition and subsequent signal amplification.When deploying Cas12 for discriminating SNVs in dsDNA, the canonical PAM domain (e.g., 5′-TTTV-3′ for LbCas12a and AsCas12a) is essential to mediate Cas12 recognition and cleavage.292,293 When the sequence of a target SNV overlapped with the PAM sequence, it can be readily discriminated by selectively turning on or off the trans-cleavage activity of Cas12.294 For example, Rodrigo and colleagues demonstrated the CRISPR-Cas12a-based detection of SARS-CoV-2 harboring an E484K G to A mutation (Fig. 18a).295 Upon reverse-transcription, the amplicon of the SNV was 5′-TTTA-3′ capable of serving as the PAM domain for Cas12a docking and subsequent trans-cleavage of the fluorogenic ssDNA reporter. By contrast, a 2-time reduced fluorescence signal was generated because of lack of PAM due to the single nucleotide mismatch on the WT sequence (5′-TTCA-3′). To further expand this strategy to variant-specific SARS-CoV-2 SNVs, Hu and colleagues further designed a CRISPR-Cas12a assay by detecting unique SNVs for Alpha and Delta variants.296 Specifically, SNV S982A was chosen for the Alpha variant, corresponding to the change of a PAM sequence (5′-TTTC-3′) in the WT to a non-canonical PAM (5′-TTGC-3′) in the mutant. SNV D950N was chosen for the Delta variant, corresponding to a PAM sequence (5′-TTTG-3′) in the anti-sense strand of the mutant over that of the WT (5′-CTTG-3′). By further integrating the trans-cleaved fluorogenic DNA reporters with a cellphone-based fluorescence reader, the authors achieved highly sensitive and specific detection of SARS-CoV-2 mutants with clinical sensitivity and specificity at 88.2% and 92.6%, respectively.
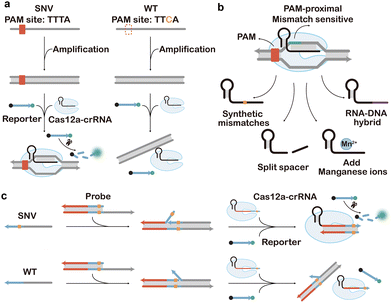 | ||
| Fig. 18 Cas12-enabled assays for SNV discrimination. (a) SNV-specific trans-cleavage of target DNA by destroying PAM via SNV-based mismatch.295 (b) Engineering crRNA to enhance the selectivity of Cas12 for SNV discrimination via strategies, including synthetic mismatch, split crRNA, using Mn2+ as the bivalent metal ion, and DNA–RNA hybrid crRNA. (c) PAM-independent SNV discrimination by coupling toehold-mediated strand displacement with a PAM-free duplex activator with a sticky end.328 | ||
As SNVs do not always lead to changes in PAM sequences or the changes may lead to sub-optimal PAM that can be tolerant by Cas12, tremendous efforts have been made to develop PAM-independent strategies to enable Cas12-based SNV discrimination. Several mechanistic studies revealed that the activity of Cas12a was sensitive to mismatches between the target and crRNA within 6 nt proximal to the PAM domain (Fig. 18b).288,297–299 Strategies have been developed to further enhance the specificity by engineering crRNA within this seed sequence, including introducing additional mismatches to the spacer,296,300–307 truncating the crRNA,256,308–310 engineering the spacer to contain secondary structures,259,311–313 splitting crRNA,314–316 incorporating chemical modifications,312,317 and use of DNA chimeric crRNA318–320 (Fig. 18b). For instant, two seminal Cas12a-based CRISPR diagnostic systems, DNA endonuclease-targeted CRISPR trans reporter (DETECTR) and one-hour low-cost multipurpose highly efficient system (HOLMES), achieved highly specific SNV detection by introducing additional mismatches into the seed sequence of crRNA.288–290 Similar engineering approaches have also been employed by cascaded Cas12 systems, such as CRISPR-Cas-only amplification network (CONAN) and autocatalytic Cas12a circular DNA amplification reaction (AutoCAR).321,322 In such systems, multiple cycled Cas12 recognition and trans-cleavage were shown to further enhance the assay specificity to SNVs compared to the single cycle Cas12 systems.
Another unique characteristic of Cas12 was the ability to recognize ssDNA in a PAM-independent manner, whereas the recognition of dsDNA requires a PAM sequence. This characteristic makes it possible to achieve highly specific activation of Cas12 by incorporating SNV-specific strand displacement reactions.323–326 For example, Wu et al. designed a PAM-free target duplex DNA with a stick end, which was recognized by crRNA via toehold-exchange rather than direct binding.327 As toehold-exchange was highly sensitive to mismatches on the invading strand, this system was found to be ultraspecific to SNVs. To further enable target specific detection, a branch-migration probe was designed capable of converting ssDNA targets into DNA-crRNA-Cas12a binding complexes through two consecutive toehold-exchange reactions (Fig. 18c). With the high specificity, this system achieved the sensitive detection of low-abundant EGFR L858R with a LOD at 0.05%.328
Like Cas9 systems, the choice of Cas12 orthologs is also critical to ensure the assay specificity for SNVs, as enzymes, such as LbCas12a, AsCas12a, and FnCas12a, exhibit varying sensitivity to mismatches with different types and positions. Some Cas12 enzymes, such as CasDx1 and Cas12f (also known as Cas14), were demonstrated to outperform widely used LbCas12a and AsCas12a for discriminating SNVs.301,329,330 For example, Chen and colleagues demonstrated that Cas12f was exceptionally well-suited for discriminating challenging SNVs, such as BRAF V600E because of its PAM-independent and low mutation tolerance characteristics.301 Buffer conditions were also found to influence assay sensitivity and specificity.325,331,332 For example, Mn2+ was shown to increase the detection sensitivity of Cas12a trans-cleavage assays by 13-fold and enhance the specificity for SNV discrimination without the need for introducing additional mismatches (Fig. 18b).333,334 Denaturing chemicals, such as DMSO and glycerol, were also shown to enhance the specificity of Cas12a mainly by reducing the Tm of the target dsDNA.335–337
While most CRISPR-Cas12 assays employ end-point measurements of Cas12 trans-cleavage activity after target-specific activation of Cas12, such measurements could be limited in accuracy and robustness due to the arbitrary experimental choices for SNV discrimination. To address this issue, Santiago and colleagues harnessed Michaelis–Menten parameters, such as substrate affinity (KM) and apparent catalytic efficiency (kcat/KM) of Cas12 trans-cleavage, to discriminate SNVs from WTs.338 They found that the apparent catalytic efficiency was 130-fold greater for fully matched targets than sequences containing single nucleotide mismatches, whereas only a 4.8-fold difference was achieved for the end-point measurement. In a subsequent study, they further achieved accurate discrimination of EGFR SNVs in tumor-extracted amplicons by measuring the kinetics of Cas12 trans-cleavage as the readout.339
The unique recognition ability towards both dsDNA and ssDNA streamlined with target-specific signal amplification positioned Cas12 as a unique molecular unit to be integrated with diverse nucleic acid amplification reactions for further enhancing the sensitivity and specificity for SNV discrimination. For example, allele-specific PCR and RPA reactions have been integrated with Cas12 recognition and amplification for the ultrasensitive detection of SNVs related to cancer and drug-resistance pathogens.340–343 Ligation-based SNV recognition and subsequent RCA reactions have also been integrated with Cas12 for in situ signal amplification for clinically relevant SNVs. Numerous microfluidic and signal readouts have also been integrated with Cas12 to achieve flexible sample treatment and downstream detection of SNVs towards point-of-care (POC) diagnostics.344–346
4.3. Cas13-enabled tools for SNV discrimination
The Cas13 family, including Cas13a, Cas13b, Cas13c, and Cas13d, is a class of RNA-guided ribonucleases. Cas13 has several distinct features from other RNA-guided Cas effectors, including (1) the activator of Cas13 is ssRNA rather than DNA, (2) Cas13 cleaves ssRNA, and (3) the target recognition of Cas13 prefers a protospacer flanking site (PFS) rather than PAM on the 3′-end of the ssRNA target.303,347,348 Structurally, binding of an RNA target complementary to the crRNA triggers Cas13 to cleave RNA nonspecifically by activating the enzyme's two higher eukaryote and prokaryote nucleotide (HEPN) binding domains to form a single RNase active site.349 The activated Cas13 is a general nuclease, capable of cleaving either the target RNA (cis-cleavage) or encountered nonspecific RNA (trans-cleavage). The trans-cleavage activity of Cas13 exhibits high turnover numbers, offering an excellent signal amplification mechanism for RNA detection and diagnostic applications.The sequence specificity of Cas13 was determined by both complementarity between target RNA and crRNA and the activation of HEPN-nuclease. Using library-based, high-throughput RNA binding assays and biochemical experiments to investigate the effects of mismatches on Cas13a from Leptotrichia buccalis (LbuCas13a), Tambe et al. revealed that mismatches in the middle seed region (position 9–12) of the crRNA spacer had the largest defect in binding affinity, whereas mismatches in positions 5–8 of the crRNA spacer were bound with high affinity but failed to promote HEPN-nuclease activity (Fig. 19a).349 In a subsequent study, Vargas et al. further demonstrated that certain positions such as 7, 8, 12, 13, and 19 of the crRNA spacer were particularly sensitive to mismatches and this effect could be enhanced by shortening the crRNA spacer or engineering Cas13a variants with altered allostery communication pathways (e.g., LbuCas13aN378A and LbuCas13aR973A).350 By combining the optimal Cas13a variant and crRNA design strategies, the authors have achieved superior discrimination of SARS-CoV-2 SNVs over wild-type LbuCas13a. Using a microchamber-based digital detection platform, Shinoda et al. investigated the specificity of Cas13a from Leptotrchia trevisanii (LtrCas13a) for detecting mutant SNVs of SARS-CoV-2.351,352 They designed a pair of crRNAs for each of the SNV and WT sequences and found that the optimal position on the crRNA spacer was 7 for N501 (WT) and Y501 (mutant), 5 for E484 (WT) and K484 (mutant), and 7 for L452 (WT) and 8 for R452 (mutant), evidenced by the 2-fold decrease in the number of positive chambers when activated by mismatched crRNA. Using the three crRNA pairs, the authors achieved the precise discrimination of B.1.1.7 (N501Y), B.1.351(N501Y + E484K), and B.1.617 (L452R) SARS-CoV-2 variants by monitoring the ratios between positive chambers generated by WTs and SNVs, respectively.352 Besides digital readouts, the signal ratio generated by detecting the same sample using a pair of SNV-specific crRNA and WT-specific crRNA has been demonstrated to be a generalizable strategy for SNV discrimination, enabling sensitive discrimination of SARS-CoV-2 variants in a concentration-independent manner.347,353
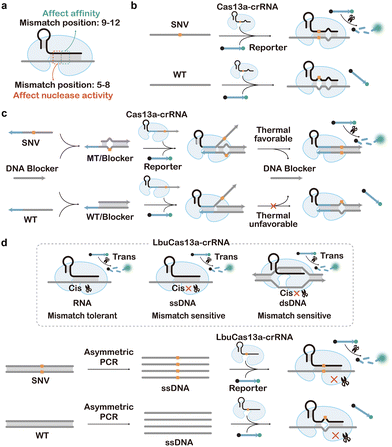 | ||
| Fig. 19 Cas13-enabled assays for SNV discrimination. (a) Effect of mismatches between target RNA and crRNA on the binding and nuclease activity of Cas13.349 (b) Engineering crRNA with synthetic mismatch to enhance the selectivity of Cas13 for SNV discrimination.303 (c) SNV discrimination by activating the trans-cleavage of Cas13 using toehold-mediated strand displacement.364 (d) SNV discrimination based on DNA activated LbuCas13a demonstrated high SNV specificity over RNA targets.360 | ||
Despite efforts being made to engineer Cas13 proteins and optimize crRNA designs, it remains difficult to discriminate SNVs due to the tolerance of mismatches between target RNA and crRNA at high target RNA concentrations. Efforts to further improve the specificity of Cas13 include the incorporation of synthetic mismatches,289,303,350,354–358 truncating the spacer of crRNA,350,359,360 and engineering crRNA with secondary structures.361,362 The incorporation of one or more synthetic mismatch into the crRNA is one of the simplest yet most effective strategies to enhance the sequence specificity of Cas13 assays (Fig. 19b). The first reported use of synthetic mismatch was made by Zhang and colleagues to enable the SNV specificity of specific high-sensitivity enzymatic reporter unlocking (SHERLOCK).303 The synthetic mismatches incorporated in position 5 of the crRNA spacer while maintaining the target SNV at the position 3 was found to maximize the assay specificity, allowing discrimination of closely related strains of Zika virus (ZIKV) and flavivirus dengue (DENV). This strategy also enabled SHERLOCK to analyze low-frequency cancer mutations, including EGFR L858R and BRAF V600E, in cfDNA at levels as low as 0.1%. The strategy of incorporating synthetic mismatches was also employed by SHERLOCK version 2 (SHERLOCKv2), a multiplexed CRISPR diagnostic assay with orthogonal CRISPR enzymes, for the detection of SNVs in ZIKV, DENV, and cancer liquid biopsy samples via lateral flow readouts.289 Casati et al. further developed an accurate detection method for evolving SARS-CoV-2 through SHERLOCK optimization (SDESSO).363 In this design, the target SNV was placed at the position 7 of the crRNA spacer with three synthetic mismatches incorporated in positions 3, 5, and 24. This design led to successful discrimination between Alpha and Omicron variants in clinical samples.
Like Cas12, the specificity of Cas13 may also be enhanced via strand displacement reactions. For example, Hu et al. designed a short DNA blocker that prehybridized to the ssRNA target with a toehold domain exposed on the RNA, so that target-recognition by the Cas13a-crRNA complex was achieved through a toehold-mediated strand displacement (Fig. 19c).364 For the SNV target, a mismatch existed between the blocker and target RNA and the full complementarity was achieved between target RNA and crRNA. Therefore, both target recognition and subsequent trans-cleavage of Cas13a were favourable, resulting in high detection signals. By contrast, the blocker hybridized with the WT RNA sequence with full complementarity and led to a single mismatch when forming the RNA–crRNA duplex. As both target activation and subsequent Cas13 trans-cleavage were hampered by the mismatch, this assay demonstrated high specificity for SNVs with a maximal DF value over 1000 and the detection of low-abundant SNVs at frequencies as low as 0.1%.
Since its first discovery, Cas13a has been perceived as an RNA-specific nuclease for RNA editing and detection. Recently, Wu et al. discovered that LbuCas13a could be directly activated by DNA without the restrictions of PAM or PFS, which elicited strong trans-cleavage activity but with no degradation of the DNA target (Fig. 19d).360 More interestingly, LbuCas13a activation was achieved for both ssDNA and dsDNA in a PAM-free manner, and in both cases, the detection system demonstrated high SNV specificity without the need for synthetic mismatches. Further investigation revealed that the DNA-targeting ability of LbuCas13a was highly sensitive to truncation at the 3′-end over the 5′-end, and a minimal of 24 nucleotides were required to ensure successful activation. High specificity of DNA-targeting LbuCas13a was also found for both ssDNA and dsDNA at the positions 4 to 20 with a 19-fold increase in SNV discrimination specificity over its RNA-targeting counterpart, likely due to the weaker binding affinities of DNA–RNA binding compared to RNA–RNA binding. Based on this exciting finding, the authors developed the superior universal rapid enhanced specificity test with the LbuCas13a (SUREST) platform, enabling highly specific and sensitive detection of both human SNVs at allele frequency as low as 0.01% and SARS-CoV-2 variants. Because LbuCas13a was more sensitive to the ssDNA target over the dsDNA target, the authors also integrated this Cas13-based DNA targeting system with λ-exonuclease-based conversion of double-stranded RPA amplicons to ssDNA.365 To also achieve simultaneous detection of the presence of a target DNA and its SNV, Zhong et al. combined the DNA targeting LbuCas13a for highly specific SNV detection with AsCas12a for targeting a conserved region of the DNA target.366 This strategy, termed specific and precise mutation recognition with Cas12a/Cas13a (SPARC), could effectively distinguish between genuine SNVs and false-negative results due to target absence.
4.4. Ago-enabled tools for SNV discrimination
Argonaute (Ago) proteins are key players in all small-RNA-guided gene-silencing processes.367–370 In 2014, Swarts et al. demonstrated that the Ago of the bacterium Thermus thermophilus (TtAgo) loaded with 13 to 25-nt 5′-PO4-DNA guides cleaved complementary DNA strands, paving the way for harnessing Ago as a CRISPR-Cas-alternative for DNA interference and molecular diagnosis.371 Unlike Cas9 or Cas12 endonucleases, the Ago family does not depend on specific PAM motifs, thereby offering broader sequence compatibility. Agos programmed with DNA guides are highly sensitive to mismatches. Even a single nucleotide mismatch between the gDNA and the target region impedes cleavage activity, thereby enabling the sensitive detection of SNVs.372,373 The use of DNA guides is also highly advantageous due to their greater stability and lower cost.PfAgo-mediated nucleic acid detection (PAND) is the first Ago-based test developed to detect ∼0.1% mutant alleles.374 PAND employs Pyrococcus furiosus Ago (PfAgo) in a two-step cleavage manner. First, PfAgo processes three input gDNAs to generate a 16-nt new guide DNA (ngDNA) containing the mutant allele. This ngDNA then directs PfAgo to cleave fully matched molecular beacons specific to the mutation, enabling fluorescence-based detection. The Ago-directed specific target enrichment and detection (A-Star) technique, another Ago-based assay for specific SNV enrichment and detection, also utilized thermostable PfAgo in a single tube.375 By introducing a synthetic mismatch into the gDNA, A-star effectively cleaved WT ssDNA containing two mismatches, whereas the SNV ssDNA with a single mismatch was tolerated by the PfAgo. Besides PfAgo, TtAgo has also been employed for SNV discrimination. For example, Kong et al. integrated TtAgo with gFET to achieve a “test-and-go” method for SNV discrimination.376 In this assay, the TtAgo–gDNA complex was immobilized on gFET with a monolayer graphene channel bridging the drain and source electrodes. Once the gDNA recognized the target sequence including both RNA and ssDNA to form a TtAgo–gDNA-target ternary complex, the doping effect shifted the Dirac point to generate the detection signal. This assay was highly sensitive to SNVs located in the seed region of gDNA and could achieve amplification-free detection of SNVs within 5 min.
Collectively, CRISPR-enabled tools are currently revolutionizing nucleic acid testing and SNV detection. The key advantage of CRISPR Cas and Ago machineries over other enzymes is the unique combination of high sequence specificity guided by the CRISPR RNA or guide DNA and the need for only a few conserved nucleotides. The broad target sets (dsDNA, ssDNA, and RNA) and multiple cleavage or binding modes, such as cis-cleavage, trans-cleavage, nicking cleavage, and binding-only, further expand the assay flexibility. To help guide choosing CRISPR systems for specific assay design, we summarize the key features of each type of Cas protein in Table 1, which includes PAM requirements, target sets, cleavage modes, specificity, and potential for multiplexing.
| Name | Enzyme | PAM requirements | Target set | Cleavage mode | Multiplexity |
|---|---|---|---|---|---|
| CUT-PCR248 | Cas9 | Yes | dsDNA | cis-Cleavage | Multiplex |
| CRISDA269 | Yes | dsDNA | cis-Cleavage | Single | |
| CRISPR-SNP-Chip282 | Yes | dsDNA | cis-Cleavage | Single | |
| HOLMES290 | Cas12 | Yes | dsDNA | trans-Cleavage | Single |
| CONAN321 | Yes | dsDNA | trans-Cleavage | Single | |
| THMSD325 | No | ssDNA | trans-Cleavage | Single | |
| SHERLOCK303 | Cas13 | No | RNA | trans-Cleavage | 4-Plex |
| SHINEv.2347 | No | RNA | trans-Cleavage | 3-Plex | |
| SECND364 | No | RNA | trans-Cleavage | Single | |
| SUREST360 | No | RNA, ssDNA, and dsDNA | trans-Cleavage | Single |
5. Emerging applications of SNV discrimination tools to clinical diagnosis
One of the ultimate goals of developing chemical tools for discriminating SNVs is to enable their clinical applications in real-world settings. Currently, there are two main drivers for the technological innovation and translation of SNV discrimination tools from the laboratory to the clinic. One driver is the ongoing need of the point-of-care (POC) medical diagnostics in resource-limited settings, which requires SNV tests that are not only accurate but also affordable and accessible.377–379 Particularly, rapid and on-demand management of emerging infectious diseases, such as TB380–382 and COVID,383–385 relies on field-deployable SNV detection tools at remote areas of low-income countries and regions for tracing rapidly evolving mutants and identifying drug resistance. The other driver is the precision diagnosis and personalized treatment of devastating diseases, such as cancer.386–389 In these clinical scenarios, panels of SNVs need to be analyzed from tissue, blood, or urine samples. Although the workhorse for such tasks has shifted from PCR-based techniques to NGS and the third-generation sequencing, chemical tools for enriching low-abundant SNVs are increasingly needed to address the intrinsic challenges of varying sequencing techniques. Here, we focus on two emerging applications of SNV discrimination tools, including POC diagnosis of infectious diseases and precision diagnosis of cancer.5.1. SNV discrimination tools for POC diagnosis of infectious diseases
Infectious diseases pose a severe threat to human lives and global public health.390,391 Detection of SNVs in infectious diseases is critical for tracking pathogen variation and drug resistance. In 2003, the World Health Organization (WHO) published ASSURED (affordable, sensitive, specific, user-friendly, rapid, equipment-free, delivered) criteria for guiding the design of POC diagnostic tests of infectious diseases.392 In 2019, the real-time connectivity and ease of specimen collection were added to provide real-time quality control for testing and overcoming the difficulties in specimen collection and processing.393 The REASSURED principle sets a guideline for developing and translating chemical tools of SNV discrimination into real clinical tests in POC settings. According to this principle, a field-deployable SNV test shall not only be sensitive and specific but also streamline sample processing, nucleic acid extraction, amplification, and signal readout in a rapid and user-friendly manner with minimal equipment need. Toward this need, extensive efforts have been made to advance SNV discrimination technologies with innovations ranging from field-deployable reagents to user-friendly readouts and to fully integrated devices with sample-in answer-out capabilities. | ||
| Fig. 20 Sample processing and reagent storage strategies for delivering SNV discrimination assays in POC settings. (a) HUDSON that combined heating and chemical denaturation for sample processing.354 (b) A modified HUDSON method in SHINEv1 with a shorter incubation time.394 (c) A further modified HUDSON method in SHINEv2 with a shorter incubation time at ambient temperature.347 (d) Processing respiratory samples for MTB and drug resistance detection via chemical denaturation and heating.395 Reprinted with permission, Copyright (2025) American Association for the Advancement of Science. (e) Long-term storage and cold-chain free transportation of reagents of cell-free protein expression-based sensing of nucleic acids and SNVs via freeze-dried paper discs.243 Modified with permission, Copyright (2016) Elsevier. | ||
Besides viral infections, Hu and colleagues also developed a respiratory sample processing procedure for analyzing MTB infection and drug-resistance in sputum and saliva samples (Fig. 20d).395 Dithiothreitol (DTT) that dissociates disulfide bonds was chosen as a reducing reagent to reduce the viscosity of the specimens and a 15-min 90 °C incubation was selected for DNase and MTB inactivation and MTB lysis. This sample processing procedure was also found to be highly compatible with RPA-CRISPR assay for subsequent nucleic acid amplification and SNV discrimination.
To enable SNV discrimination in POC setting, all reagents need to be deliverable to end-users, which is another highly important criterion in the REASSURED principle yet is often overlooked in assay design. Lyophilization can facilitate the transport and storage of diagnostic reagents by eliminating the need for the cold chain and increasing shelf life. By developing SHINEv.2, Myhrvold et al. explored the lyophilization of Cas-13-based CRISPR diagnostic reagents, including that for discriminating SARS-CoV-2 SNVs.347 They found that direct lyophilization of the Cas13 diagnostic reagent led to a profound drop in assay performance. By adding non-reducing disaccharides, such as sucrose and mannitol, as stabilizers and removing destabilizing components, such as polyethylene glycol (PEG) and KCl, the ultimate lyophilized reagents retained the high analytical performance. Meanwhile, lyophilized reagents were found to be unaffected after 5 months in storage, which greatly simplified distribution logistics.
An alternative approach for long-term storage and cold-chain-free transportation is to freeze dry the diagnostic reagent onto paper, which enables the inexpensive, sterile, and abiotic distribution of fragile biological reagents for the clinic and global health. The freeze-dried, paper-based biomolecular platform was first reported by Pardee et al. in 2014, where a toehold-switch-regulated cell-free protein expression system was freeze dried into paper, enabling a low-cost, deliverable diagnostic platform for the detection of antibiotic resistance genes.396 In 2016, Pardee et al. further integrated the freeze-dried, paper-based, cell-free protein expression system with Cas9-based SNV discrimination (assay design is detailed in Section 4.1), allowing the rapid prototyping of paper-based biosensors for portable and low-cost diagnosis of ZIKA (Fig. 20e).243 The freeze-dried, cell-free protein expression system was also integrated with toehold-exchange, enabling the cold-chain-free delivery of SNIPR technology (assay design is detailed in Section 2.4) for SNV discrimination in POC settings.130,397
While reagents for both CRISPR and cell-free protein expression-based diagnostic assays contain thermally unstable proteins and RNAs, all-DNA-based assay reagents can be more stable and reduce the difficulty for long-term storage and transportation. Functional nucleic acids, such as RNA-cleaving DNAzymes, can be programmed to offer both target-recognition and catalytic signal amplification capabilities and are ideal platforms for designing SNV discrimination assays deliverable for POC diagnosis. For example, Yang et al. reported a multicomponent-DNAzyme-based detection platform for the rapid and sensitive detection of SARS-CoV-2 SNVs in patient-derived clinical samples.398 In combination with RPA and T7 transcription, this RNA-encoded viral nucleic acid analytic reporter (REVEALR) allowed rapid identification of WT, alpha, gamma, epsilon, delta, and omicron with 100% accuracy.398–400 Gao et al. demonstrated that the DNAzyme-base CRISPR-like diagnostic system was more stable than CRISPR Cas systems against adverse conditions, such as elevated temperatures and freeze–thaw cycles.401
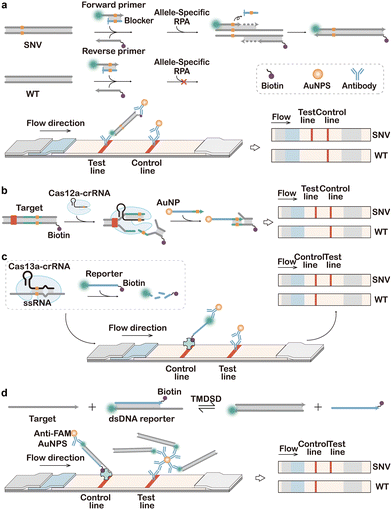 | ||
| Fig. 21 SNV discrimination via lateral flow assays (LFA). (a) Allele-specific RPA coupled with LFA for visual detection of SNVs.405 (b) SNV discrimination via the cis-cleavage of Cas12a and strand invasion to generate an SNV-specific LFA readout.406 (c) LFA-based discrimination of SNVs via the mutation-specific trans-cleavage activity of Cas13a.289 (d) LFA-based discrimination of SNVs by separating chemical labels via toehold-exchange.407 | ||
Besides allele-specific isothermal nucleic acid amplification, the dual-labelled reaction product and subsequent LFA analysis could also be achieved using SNV-specific ligation reactions, enabling rapid and accurate detection of rifampicin-resistant MTB. The cis-cleavage activity of Cas12a has also been employed to facilitate the LFA-based visual analysis of SNVs. For example, Lin et al. harnessed biotin-modified primers to generate biotinylated RPA amplicons (Fig. 21b).406 Cas12a was then employed to cleave the amplicons into truncated amplicons with sticky ends, providing a toehold for subsequent strand invasion hybridization with an invading strand immobilized on AuNPs. The successful invading event in the presence of the target SNV led to the production of a biotin-AuNP dual-labelled product that was visually analyzed via LFA. Because both Cas12a recognition and strand invasion were SNV-specific, this Cas12a cis-cleavage-mediated LFA method demonstrated a considerably higher specificity for SNV discrimination compared to assays making use of Cas12a trans-cleavage activity or dCas9-based LFA.
In addition to generating dual-labelled nucleic acid products for classic LFA designs, it is also possible to disassemble chemical labels to enable LFA-based SNV discrimination. The disassembly design first appeared in the seminal work of SHERLOCKv2, where Gootenberg et al. designed a short dual-labelled RNA reporter with FAM and biotin, respectively (Fig. 21c).289 In the presence of the target SNV, the trans-cleavage activity of Cas13 was induced to degrade the RNA reporter and thus disassemble the two chemical labels. Different from classic LFA where reagents flow from the test line to control line, abundant intact reporter accumulated anti-FAM antibody-AuNP conjugates at the first line on the LFA strip as the control line, preventing the binding of the antibody-AuNP conjugates to protein A on the second test line. The cleavage of the reporter reduced accumulation at the control line and resulted in the signal on the test line. Since its first introduction, this strategy has been employed extensively by CRISPR Cas12 and Cas13 assays for diverse LFA-based nucleic acid detection and SNV discrimination.354,408–410 Besides enzymatic cleavage, Li and colleagues also introduced a toehold-exchange-based strategy for disassembling chemical labels, allowing highly sensitive and specific discrimination of SNVs in buccal swab samples via the coupling of RPA (Fig. 21d).407
As LFA is qualitative, efforts have also been made to develop naked-eye-based quantitative readouts for improving SNV discrimination in POC settings. For example, SNIPR and SNV-specific cell-free protein expression systems translated the SNV recognition into the production of β-galactosidase via toehold-switch and transcription–translation reactions hosted on paper discs.130 β-galactosidase cleaved a yellow substrate, chlorophenol red-β-galactopyranoside, to produce a purple product that had strong absorbance at a wavelength of 575 nm. This color change was then quantified using portable color readers243 or a 3D-printed portable microplate reader397 to achieve quantitative colorimetric readouts for the detection of drug-resistant SNVs in malaria and HIV. Rather than cell-free enzyme expression, Zhang et al. reported an alternative enzyme-based colorimetric detection of SARS-CoV-2 variants at single nucleotide resolution (Fig. 22a).411 In this approach SNV recognition was achieved through a toehold-exchange, where the reverse toehold contained non-canonical C–Ag–C base pairs. SNV-specific strand displacement between the target RNA and reporter probe released Ag(I) ions that were found to effectively inhibit urease for producing NH4+ from urea. As the production of NH4+ increased the pH of the solution, it led to a color change of a sensitive pH indicator, phenol red, from red to yellow. As successful toehold-exchange inhibited the color change, the red color indicated the presence of SNVs, whereas WTs led to the yellow color. By further quantifying the color change using a smartphone, the authors achieved the ultrasensitive discrimination of SARS-CoV-2 variants without the need for pre-amplification.
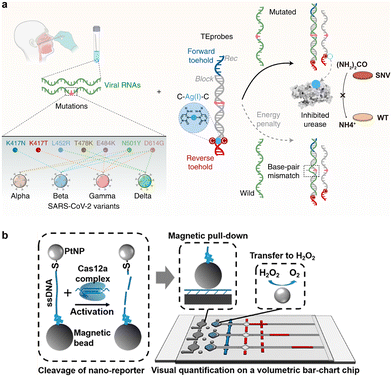 | ||
| Fig. 22 (a) Colorimetric SNV discrimination via toehold-exchange and the mutation-specific modulation of urease activity.411 Reprinted with permission, Copyright (2022) Springer. (b) Distance-based discrimination of SNVs using a volumetric bar-chart chip that converted CRISPR-Cas12a-based SNV detection into the migration distance of red ink bars.412 Reprinted with permission, Copyright (2019) American Chemical Society. | ||
Distance is another quantitative readout that can be readily perceived by human eyes without the need for any equipment. To enable a distance-based readout for SNV discrimination, Shao et al. developed a volumetric bar-chart chip that generated distance changes in response to CRISPR-Cas12a-based SNV detection and a platinum nanoreporter.412 In the presence of a target SNV, Cas12a with synthetic mismatches incorporated in the crRNA was activated and trans-cleaved a ssDNA link used to immobilize platinum nanoparticles (PtNPs) on magnetic beads. Upon releasing, PtNPs entered the bar-chart chip and catalyzed H2O2 decomposition to generate oxygen. The advancements of the red ink bars resulting from the generated oxygen gas offered a distance-based readout for quantifying SNVs (Fig. 22b). Although this distance-based SNV discrimination method had not been demonstrated in real clinical samples, it directed an interesting avenue for achieving a visual yet digital readout for POC SNV discrimination via the integration of advanced microfluidic techniques with CRISPR diagnostic assays.
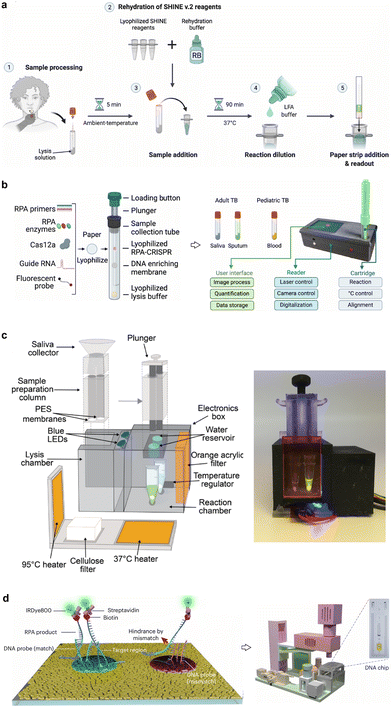 | ||
| Fig. 23 (a) Schematic illustration of the field-deployable LFA-based test for SARS-CoV-2 and its variants using CRISPR-Cas13-based SHINEv.2.347 Reprinted with permission, Copyright (2022) Springer. (b) An inexpensive, handheld, battery-powered, lab-in-tube TB assay detecting MTB DNA and drug resistant SNVs, where RPA and Cas12a were combined for signal amplification and highly specific SNV recognition.395 Reprinted with permission, Copyright (2025) American Association for the Advancement of Science. (c) A minimally instrumented SHERLOCK platform that streamlines all steps of the CRISPR Cas13-based test for analyzing SARS-CoV-2 RNA and SNVs.415 Reprinted with permission, Copyright (2021) American Association for the Advancement of Science. (d) Multiplexed detection of SARS-CoV-2 variants via a fluorescence-enhanced microarray equipped in a portable and automated device.416 Reprinted with permission, Copyright (2023) Springer. | ||
As early as 2010, the WHO has recommended the implementation of the GeneXpert MTB/RIF test from Cephei for screening MTB infections and drug resistance in low- and middle-income countries, as this system automates all operational steps ranging from nucleic acid extraction to real-time PCR and MB-based SNV discrimination into single equipment, enabling sample-in answer-out testing.413,414 Nevertheless, the broad use of GeneXpert or its latest version Xpert Ultra as a true POC test has been limited due to the requirements for stable electrical power supply, temperature control, cartridge supply, cost, lack of quality assurance and maintenance support. With the advancement of addictive manufacturing, such as 3D printing, prototyping of battery-powered low-cost portable diagnostic devices has become ever simpler and faster. For example, Hu and colleagues recently developed an inexpensive, handheld, battery-powered, lab-in-tube (LIT)-TB assay detecting MTB DNA and drug resistant SNVs in patient serum, saliva, and sputum samples with a sample-to-answer time of less than 1 hour (Fig. 23b).381 This system integrated the abovementioned sputum or saliva specimen liquification and MTB lysis, RPA, and CRISPR Cas12a detection steps into a single consumable tube that was processed and analyzed by a portable assay device. In this LIT approach, a respiratory sample was collected in the assay tube that was then transferred to the incubator port of the portable device for sample processing. The LIT plunger was then depressed to load the lysate onto the DNA capture membrane and initiated the RPA and CRISPR Cas12a-based reaction. SNV discrimination was also achieved by introducing synthetic mismatches into crRNA. Upon completion, the tube was transferred to the readout port of the device to detect and analyze the resulting fluorescent signals via a camera equipped with a fluorescence imaging filter to read the assay and a Raspberry Pi microprocessor to integrate fluorescence imaging and automated analysis. With excellent performance in clinical validation against patients with TB infection and drug resistance, the LIT test demonstrated excellent potential for analyzing respiratory specimens at the POC in remote or resource-limited settings without access to Xpert to expand TB diagnostic efforts.
Efforts have also been made to develop minimally instrumented nucleic acid and SNV detection platforms for detecting SARS-CoV-2 variants.411,415 For example, Collins and colleagues described the development of a low-cost, self-contained, POC diagnostic called minimally instrumented SHERLOCK (miSHERLOCK) capable of concurrent detection of SARS-CoV-2 and B.1.1.7, B.1.351, or P.1 variants (Fig. 23c).415 The miSHERLOCK platform integrated instrument-free, built-in sample preparation from saliva with an optimized one-pot SHERLOCK assay. Specifically, RNAs in the saliva sample were concentrated onto a 4-mm polyethersulfone membrane that was then physically transferred into room temperature stable reagents for SHERLOCK reaction. With battery-powered incubation, and simple visual and mobile phone-enabled output interpretation, miSHERLOCK enabled highly sensitive and specific detection of SARS-CoV-2 RNA and SNVs with a LOD of 1000 copies per mL. Besides singlet discrimination assay, multiplexed discrimination of SARS-CoV-2 variants has also been achieved via fluorescence-enhanced microarray technology in a portable and automated device (Fig. 23d). A plasmonic gold (pGOLD) chip with surface plasmonic resonance and local-electric-field enhancement effects was employed for enhancing near-infrared fluorescence signals of DNA on microarrays.416–420 A microfluid chip with one-axis camshaft design was also fabricated to fully automate heat-based sample lysis, asymmetric RPA to generate ssDNA amplicons, and on-chip hybridization and fluorescence enhancement. By probing 12 targets in three mutational hot spots (G142-Y145, K417, and L452) in the S gene, this platform provided high-throughput, rapid, and multiplexed nucleic acid detection with single-molecule sensitivity and SNV discrimination.
5.2. SNV enrichment tools for cancer diagnosis
With ever sharpened tools for discriminating SNVs at ultralow abundance, increasing SNV-based tests have been implemented in all stages of the clinical management of cancer from early screening, to companion tests for guiding personalized treatment, and to the monitoring of minimal residual disease (MRD) for evaluating recurrence risk and informing follow-up treatment. While PCR-based tests, such as the FDA-approved Cobas EGFR mutation test for non-small-cell lung cancer (NSCLC),421 are suitable for detecting small panels of SNVs, the high genetic diversity of tumor often requires the simultaneous detection of hundreds of SNVs via high-throughput sequencing.422–427 Nevertheless, the ultralow abundant nature of tumor-derived SNVs requires an extremely high sequencing depth, that is, many reads per genomic locus. The high sequencing depth not only adds to upfront cost but also limits the total number of distinct loci. This intrinsic trade-off between sequencing breadth and depth makes high-throughput sequencing prohibitively expensive to clinical diagnosis of cancer where large numbers of distinct, low-abundant SNVs exist. Targeted enrichment of low-abundant SNVs ahead of sequencing has been approved to be one of the most efficient approaches for increasing clinical accuracy and meanwhile reducing the upfront cost. Therefore, this section will focus on the design principle of varying chemical tools for targeted SNV enrichment with an emphasis on their applications in cancer diagnosis.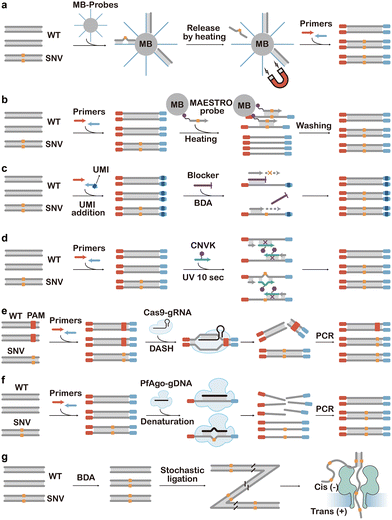 | ||
| Fig. 24 SNV enrichment strategies. (a) Hybridization-based enrichment by selective release of SNVs via heating denaturation.431 (b) Hybridization-based enrichment via shortened hybridization probes.428 (c) PCR-based enrichment by molecular barcoding and blocker displacement amplification.424 (d) PCR-based enrichment by depleting WT sequences via UV crosslinking.442 (e) Enrichment by depleting WT sequences via CRISPR Cas9-based cis-cleavage.443 (f) Enrichment by depleting WT sequences via PfAgo-based cleavage.447 (g) PCR-based enrichment for nanopore sequencing, where stochastic ligation was used to create long amplicons.452 | ||
To date, hybridization-based enrichment has emerged as one of the most widely used principles by commercial vendors for manufacturing targeted SNV enrichment reagents, such as the NimbleGen SeqCap kits by Roche.432–434 Nevertheless, most enrichment strategies often employ long hybridization probes that often capture SNVs and WTs with similar efficiency. To improve the selectivity of capture probes for patient-specific SNVs, Adalsteinsson and colleagues reported a minor-allele-enriched sequencing through recognition oligonucleotides (MAESTRO) method that harnessed short hybridization probes with enhanced SNV specificity (Fig. 24b).428 Remarkably, MAESTRO simultaneously enriched thousands of SNVs with a median ∼1000-fold enrichment from 0.1% VAF to nearly pure mutant DNA, enabling the accurate sequencing of low-abundant SNVs with ∼100-fold fewer reads per locus. When employed for MRD monitoring for patients with breast cancer enrolled in a clinical trial of pre-operative therapy, MAESTRO uncovered more SNVs per patient in cfDNA compared with conventional enrichment strategies. Further validation using 9 melanoma patients in 98 plasma samples revealed that pooling MAESTRO probes improved the sensitivity of MRD by 10- to 100-fold over existing tests.435
Based on the principle of BDA,439 Zhang and colleagues developed multiplex BDA (mBDA),423 a multiplex PCR target-enrichment method that allowed for robust NGS detection and quantification of SNVs with VAF as low as 0.02% using a sequencing depth of only 250×. The mBDA method harnessed rationally designed BDA blockers that perfectly bound to the WT sequence with an overlapping sequence to the forward primer. To initiate primer extension, the forward primer had to displace the blocker to bind to the DNA template, where only the SNV template bearing a mismatch with the blocker could be efficiently displaced. Consequently, the SNV templates were amplified with a notably higher yield per PCR cycle and accumulated through the course of many PCR cycles, leading to a 1000-fold higher efficiency. The authors demonstrated that mBDA scaled well to multiplex panels with a 300-fold median enrichment in an 80-plex panel. Clinical potential of mBDA was further validated with a 16-plex NGS panel covering 145 SNVs across 9 genes involved in melanoma. To further enable calibration-free NGS quantitation of SNVs below 0.01% VAF, Zhang and colleagues integrated molecular barcoding with BDA technology for SNV enrichment (Fig. 24c).424 To do so, molecular identifiers (UMIs) allowing the accurate counting of input molecules were incorporated into the target gene fragment through a pre-amplification step. Upon mBDA enrichment and NGS sequencing, SNVs within targeted genes were simultaneously enriched and accurately quantified. With this quantitative BDA technology, the authors have successfully developed and clinically validated a 20-gene acute myeloid leukemia (AML) panel with VAF down to 0.001% for MRD analysis, as well as two cancer panels including a pan-cancer panel and a specific melanoma panel for tumor tissue and cfDNA samples. Recently, Si et al. further improved BDA technology by extending the overlapping region between the blocker and the forward primer by 2-fold, so that more SNVs could be covered within this elongated enrichment region.440 By operating at higher annealing temperatures, this long BDA (LBDA) technology detected different SNVs down to 0.5% VAF in one reaction with a median enrichment fold of 1000 on 21 SNV templates, enabling a single plex assay covering 81 hotspot SNVs in oncogenic genes.
Besides selective blockage of WTs and activation of SNVs through strand displacement, chemically modified blocker probes were also employed to selectively cross-link and deplete WT sequences.441 For example, Leong et al. described UV-mediated cross-linking minor allele enrichment (UVME) that harnessed ultraviolet irradiation at 365 nm to crosslink DNA probes incorporating a UV-sensitive 3-cyanovinylcarbazole nucleotide modification (Fig. 24d).442 This chemically modified probe was designed to be fully complementary to WT sequences and crosslinked preferentially to the sense strand of WT templates. The crosslinking inhibited further amplification of WTs and thus enriched SNV sequences. The UVME-PCR program included a regular PCR stage for 10–20 cycles to pre-amplify the target DNA, followed by the UV irradiation for 10 s at annealing temperature during each subsequent PCR cycle to induce photo-crosslinking and inhibition of WT amplification, thereby resulting in SNV enrichment up to 800-fold. As a light-activable technique, this strategy was highly flexible, allowing efficient SNV enrichment in a single tube during PCR. Nevertheless, the use of chemical modification may add the assay complexity and upfront cost, especially when large panels of SNVs need to be enriched in clinical samples.
As the need for PAM sequences limited the coverage of Cas9-based SNV enrichment methods, efforts have also been made to develop enzyme-assisted enrichment strategies that are free from PAM. For example, NaME-PrO making use of the thermostable DSN was employed to deplete WT sequences in a sequence-independent manner.444 Nevertheless, this strategy relied on the fine tuning of DNA probes and reaction temperatures to ensure correct formation of duplexes and thus was difficult to expand to large panels of SNVs. Ago enzymes, on the other hand, can be guided using gDNA and offer improved selectivity and flexibility. A-Star was an enrichment strategy that utilized the thermostable PfAgo with gDNAs to selectively bind and cleave PCR amplicons of WT sequences.445 In combination with Sanger sequencing, A-Star achieved the detection of SNVs at 0.01% VAF with 75-fold enrichment of multiple oncogenes in solid tumor tissues and blood samples (Fig. 24e). Nucleic acid enrichment via DNA guided argonaute from Thermus thermophilus (NAVIGATER) is another TtAgo-based strategy that could be used pre-PCR with up to 60-fold enrichment for cancer SNVs, enabling the detection at 0.01% VAF in blood samples upon Sanger sequencing.446 DNase-assisted SNV discrimination tools have also been engineered for enriching large panels of SNVs for cancer diagnosis (Fig. 24f).447 Guiding by the phosphorothioated DNA, DNase selectively cleaved the fully matched WT sequences, enabling 10-plex mutation enrichment. When coupled with NGS, it allowed for high-throughput detection of multiple cancer SNVs at 0.01% VAF in blood samples with a reduction in sequencing depths up to 100-fold. Despite the high efficiency, this DNase-based enrichment strategy required the conversion of dsDNA into ssDNA, which might add the complexity upon integrating into the NGS workflow.
Because PCR-based methods may lead to target duplication, template switching, and loss of epigenetic modifications, several amplification-free strategies have been developed to enrich SNVs for subsequent nanopore sequencing. For example, Cas9-assisted targeting of chromosome segments (CATCH) were introduced, where Cas9 was employed to create targeted fragmentation for nanopore sequencing.427,454 To further improve the recovery rate and specificity to SNVs, Gilpatrick et al. developed nanopore Cas9-targeted sequencing (nCATS), an enrichment strategy that selectively ligated sequencing adapters to fresh cut sites created by Cas9-gRNA complexes.455 With this innovation, nCATS could simultaneously detect SNVs, structural variations, and methylation. The clinical applicability of nCATS was demonstrated for the detection of SNVs in the BRF, KRAS, and TP53 genes in both tumor and normal breast tissue samples, achieving coverage depths of 70×.
6. Conclusion and outlook
Because of their essential roles in biological evolution and pathogenesis, SNVs are an important class of nucleic acid biomarkers for disease diagnosis and precision medicine. The high sequence and structural similarity between an SNV and its WT counterpart impose a formidable challenge in molecular recognition and assay specificity. In this review, we provide a comprehensive overview of three main classes of chemical tools for SNV recognition and discrimination. A timeline of key milestones and representative techniques is presented in Fig. 1 to facilitate the tracing of the critical development of this field. Key features including the SNV discrimination principle and analytical performance of each technique are summarized in Table 2. We emphasize on uncovering the thermodynamic, kinetic, and enzymatic basis for probe and assay designs. Gaining such insights is critical for choosing and optimizing tools best suited for specific conditions and applications. For example, by understanding that the high entropy penalty is the key thermodynamic factor for the narrow temperature window for SNV discrimination, one may choose or design entropy-compensating hybridization probes (e.g., an S-probe or an MB with a long stem domain) when precise control of temperature is unavailable.114 It also explains why structured hybridization probes generally work better at elevated temperatures, whereas toehold-exchange probes are better suited for use at ambient temperatures.97,100,121,126,456 As another example, the specificity of nucleic acid probes for guiding enzyme-based recognition (e.g., gRNA, crRNA, and pDNA) obeys both fundamental hybridization thermodynamics/kinetics and enzymology of the protein effectors, serving as the basis for guiding their designs and uses.244,256,298,299,302,359,361,457| Chemical tools | Name | Discrimination principle | Key parameters | Enzyme | VAFs |
|---|---|---|---|---|---|
| Linear probe | Linear probe67 | Free energy difference | Probe length, temperature, denaturant, LNA and PNA | — | 5%–10% |
| Structured hybridization | Molecular beacon113 | Free energy difference | Loop length, temperature | — | 1%–5% |
| Triple-helix probe106 | Free energy difference | Probe length, temperature, LNA and PNA modification | — | — | |
| S-probe114 | Free energy difference | Loop length, temperature | — | ∼1% | |
| Multicomponent hybridization | Hetero-multivalent particles125 | Multiple probes act cooperatively to increase specificity | Probe length, temperature | — | — |
| Toehold-exchange reaction | Toehold-exchange probe137 | Free energy difference | Probe length, temperature | — | ∼5% |
| M-probe129 | Free energy difference | Probe length, temperature | — | ∼5% | |
| Anti-toehold probe128 | Free energy difference | Probe length, temperature | — | — | |
| SNV discrimination in dsDNA | Double-stranded toehold-exchange probe137 | Free energy difference | Forward and reverse toeholds | — | ∼5% |
| DEG138 | Free energy difference in the assembly process | Helper probe number, length, binding region | — | 1% | |
| PANDA139 | — | 0.1% | |||
| Competitive hybridization | Molecular sink146 | Free energy difference/kinetics difference | Sink design | — | 0.01%–0.1% |
| Proofreading reaction network147 | Kinetics difference | Network design | — | — | |
| PCR-based discrimination of SNVs | LightCycler152 | Differences in the melting curve | Inserting dyes, LNA and PNA modification | DNA polymerase | 1%–5% |
| ARMS-PCR150 | Primers bind more stably to SNV templates | Primer modification, WT-blockers, and the length of the blocker | DNA polymerase | 1% | |
| COLD-PCR174 | Slight Tm differences between targets | Critical denaturation temperature, synthetic reference sequence | DNA polymerase | 0.1%–0.5% | |
| dPCR188 | Partitioning a sample into thousands to millions of independent reactions | Allele-specific primers, SNV-specific MGB-modified hybridization probes | Polymerase | 0.01%–0.1% | |
| Ligation-based tools | MLPA196 | Specific ligation of DNA ligase to SNV | Padlock probe, target-specific dNTP, and competitive probe | DNA ligase | ∼0.1% |
| Endonuclease-assisted tools | RSM218/FLAG221 | Specific cleavage of WTs | Types of endonucleases, artificial introduction of a restriction site | Msp I enzyme/Endonuclease PspGI | ∼1% |
| Duplex-specific-nuclease-assisted tools | NaME-PrO226 | Specific cleavage of WTs | Renaturation temperature | DSN | 0.01%–0.1% |
| Ribonuclease H-assisted tools | rhPCR228 | SNV-specific RNA bases in blocking cleavable nucleotides | Temperature, mutation location | RNase H2 | 0.1% |
| λ-exonuclease-assisted tools | λ-exo-pDNA236 | Selective cleavage of DNA probes for WT targets | Modification of DNA probes, metal ions | λ-exo | 0.5% |
| Deoxyribonuclease-assisted tools | sgDNAse238 | Selective degradation of sspDNA and DNase1 | Length of sspDNA, cleavage temperature | DNAase I | 0.01% |
| Cas9-enabled tools | CUT-PCR248 | Mutation in PAM or seed sequences | Mutation location | spCas9 | 0.01% |
| FELUDA264 | Mutation in the sgRNA-DNA duplex | sgRNA design | FnCas9 | — | |
| CRISDA269 | PAM-or sgRNA-dependent target recognition | Mutation location, PNA modification | nCas9 | — | |
| CRISPR-SNP-Chip282 | PAM-dependent of dCas9 | gRNA design | dCas9 | — | |
| Cas12-enabled tools | HOLMES290 | Mismatch in PAM or seed sequences | gRNA design, mutation location | LbCas12a | — |
| CONAN321 | Mutation location | LbCas12a | 0.1% | ||
| THMSD325 | Synthetic mismatch in gRNA | gRNA design | LbaCas12a | 0.05%–0.1% | |
| Cas13-enabled tools | SHERLOCK303 | Synthetic mismatches in the crRNA:target duplex | crRNA design, Cas13 variants | LwCas13a | 0.1% |
| SHINEv.2347 | crRNA design | LwCas13a | 0.1% | ||
| SECND364 | Specificity of strand exchange and cleavage of Cas 13 | DNA blocker design | Cas13 | 0.01%–0.1% | |
| SUREST360 | High specificity of LbuCas13a | crRNA length, Mutation position | LbuCas13a | 0.01% | |
| Ago-enabled tools | PAND374 | Using PfAgo in a two-step cleavage manner | gDNA design | PfAgo | 0.1% |
| A-star375 | Introducing mismatch and diphosphorylation modification directly | gDNA design, annealing temperature | PfAgo | 0.01% |
Advantages and limitations also exist for each class of SNV recognition principles. For example, hybridization probes generally work best for SNVs located in the middle of the probe and become ineffective for SNVs located at the end.458 By contrast, ligation-based strategies are most effective when SNVs are located close to the end of the probe, as only those mismatches can significantly influence the subsequent ligation reaction.39,195,204 Similarly, most nuclease-based and CRISPR-based SNV recognition strategies are highly sensitive to SNV locations, which may limit their applications.225,226,231,243,244 Therefore, current principles for SNV recognition are highly complementary and may be combined for addressing challenges brought about by complex nucleic acid targets, such as those containing synonymous SNPs or multiple hotspot SNVs.289,398,415,423,439,459
In this review, we also identified two emerging clinical scenarios that drive continuous innovations to further sharpen our tools for SNV recognition and discrimination, including POC diagnosis of infectious diseases and molecular tests for cancer diagnosis and prognosis. The increasing need for on-site monitoring of rapidly evolving and drug-resistant viral and bacterial pathogens under resource-limited conditions drives the extensive integration of various SNV recognition strategies with field-deployable reagents and equipment-free or minimally instrumented readouts. Advances in addictive manufacturing and smartphone-based terminals make it ever faster to prototype fully integrated devices that streamline all steps of SNV tests in a sample-in answer-out manner. Precision cancer diagnosis, on the other hand, drives the transformation of various SNV recognition mechanisms into enrichment tools for the simultaneous capturing of large panels of oncogenic SNVs for subsequent high-throughput sequencing analysis. These SNV enrichment tools, many of which have been commercialized, play essential roles in overcoming intrinsic fidelity limitations of sequencing techniques, balancing sequencing depth and breath, and reducing the turnaround time and upfront costs. Such SNV enrichment tools may also be expanded to address clinical challenges beyond cancer diagnosis, such as non-invasive prenatal testing and monitoring donor-derived cfDNA for organ transplant patients.
Despite advancements over the past few decades, the development and applications of SNV recognition and discrimination tools face several challenges. Confronting these challenges presents new opportunities for future research and development. First, there is still lack of strategies capable of breaking the thermodynamic limit of selective hybridization. The intrinsic trade-off between binding affinity and sequence selectivity imposes a fundamental limit to hybridization processes involved in nearly all SNV discrimination mechanisms, ranging from hybridization-based recognition to enzyme-assisted selective catalysis and to gRNA and crRNA designs in CRISPR technologies. Current strategies focusing on finding the affinity hotspot achieving the optimal balance between assay sensitivity and specificity cannot meet clinical needs where both high sensitivity and specificity are required. A representative example of this is the detection of SNVs in fine-needle aspiration (FNA) biopsy that is commonly used to facilitate the confirmatory of thyroid and breast cancers.460 The low total sample input (requires high sensitivity) and low SNV abundance (requires high specificity) often lead to false negative test results and thus require new discrimination mechanisms that simultaneously enhance sensitivity and specificity. Although several kinetic and enzymatic strategies have been developed possessing signal amplification capacities, strategies such as noncovalent DNA catalysis, nucleic acid amplification, and trans-cleavage of Cas12 and Cas13, often indiscriminatively amplify both SNV-specific and spurious detection signals and thus with reduced specificity in the presence of high concentrations of WT sequences.138,139,461
Second, the specificity of enzyme-assisted assays or SNV enrichment methods is also limited by the intrinsic activities of enzymes. Currently, many enzymatic systems are optimized for specific SNVs but are generally difficult to expand for the simultaneous detection or enrichment of multiple SNVs that are frequently encountered in real clinical settings. Despite numerous efforts having been made to study the specificity and off-target effects of enzymes, particularly CRISPR Cas proteins, our understanding of enzymatic tolerance of mismatch-containing sequences remains limited. Combined experimental high-throughput screening and in silico analysis are critically needed to accumulate high-quality data to elucidate the structure–activity relationships, which in turn can help guide the precise crRNA design and protein engineering.462–466 A recent example of this is the successful engineering of an advanced-fidelity FnCas9 variant, FnCas9-AF2, via high throughput screening and genome-wide analysis, leading to enhanced specificity of FnCas9 across multiple target sites and mismatch conditions.467
Third, when employed for multiplexed SNV detection or enrichment, the design of large panels of nucleic acid probes for direct hybridization or guiding enzymatic catalysis remains challenging. Conventional software tools, such as OligoAnalyzer Toolkit (IDT), Primer Express (ThermoFisher), NCBI Primer-BLAST, and Beacon Designer focus primarily on primer and probe designs for conventional PCR-based tests, which are of limited functionality and throughput for more challenging tasks. Artificial intelligence (AI) is currently revolutionizing the way how we understand and design chemical tools for diverse applications. AI-facilitated design and development of SNV discrimination probes and tools direct a promising area of future research. Excitingly, machine learning, a subfield of AI harnessing data to train machines to make predictions, has already enabled several sequence-to-function models, language models, and generative models capable of directing the generation of high-performance crRNA sequences for improving CRISPR-based nucleic acid detection.468–473
Finally, it remains challenging to systematically compare various types of SNV detection tools due to the lack of standardized parameters and protocol for assay characterization. The DF value is one of the most commonly used parameters to characterize the assay specificity. However, as the DF value is the ratio of the detection signal, hybridization yield, or occasionally the initial rate between SNVs and WTs at an equal concentration, the choice of target concentration may significantly influence the test results. Moreover, the DF value is not applicable for SNV and WT mixtures that are encountered for many clinical samples. In such scenarios, minimal detectable abundance of SNV or VAF is often used to characterize the detection method. Nevertheless, this parameter can be largely influenced by many experimental conditions, such as the type of SNV and choice of the target concentration. To better reflect the nature of the analytical method, Chen et al. introduced the concentration equivalence denoting the excess of WT needed to yield the same level of hybridization yield (typically 50%) as that of the intended SNV.137 Wang et al. further established the theoretical basis of the concentration equivalence and defined the quantitative expression as the robustness factor (RF).138 The RF value is a concentration-independent characteristic for evaluating a given method against low abundant SNVs, as it compares the two calibration curves (one for SNVs and the other for WTs) rather than detection signals at a certain concentration. However, the calculation of RF value relies on an arbitrarily chosen reaction yield (typically lower than 50%) and no algorithm has been reported so far for directly quantifying the difference in calibration curves. Standardization of assay evaluation with well-defined analytical characteristics and protocols (e.g., the choice of SNVs and experimental conditions) may ease the comparison across varying SNV detection methods and accelerate clinical translation. With innovations in advancing SNV recognition mechanisms, AI-facilitated probe designs, assay standardization, as well as device and equipment fabrication, we foresee the development of next-generation SNV discrimination and enrichment tools enabling accurate, accessible, and affordable diagnostics for cancer, infectious diseases, and beyond.
Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results and no new data were generated or analysed in this review.Acknowledgements
We thank the National Key R&D Program of China (No. 2024YFA1209401), the National Natural Science Foundation of China (22474082), the Sichuan Science and Technology Program (No. 2025NSFTD0001 and No. 2025NSFJQ0019), and the Institutional Research Fund from Sichuan University (2021SCUNL105).Notes and references
- A. Auton, G. R. Abecasis, D. M. Altshuler, R. M. Durbin, G. R. Abecasis, D. R. Bentley, A. Chakravarti, A. G. Clark, P. Donnelly and E. E. Eichler, et al., A global reference for human genetic variation, Nature, 2015, 526, 68–74 CrossRef PubMed.
- T. A. Cooper, L. Wan and G. Dreyfuss, RNA and Disease, Cell, 2009, 136, 777–793 CrossRef CAS PubMed.
- Y. Wan, K. Qu, Q. C. Zhang, R. A. Flynn, O. Manor, Z. Ouyang, J. Zhang, R. C. Spitale, M. P. Snyder and E. Segal, et al., Landscape and variation of RNA secondary structure across the human transcriptome, Nature, 2014, 505, 706–709 CrossRef CAS.
- M. Steri, V. Orrù, M. L. Idda, M. Pitzalis, M. Pala, I. Zara, C. Sidore, V. Faà, M. Floris and M. Deiana, et al., Overexpression of the Cytokine BAFF and Autoimmunity Risk, N. Engl. J. Med., 2017, 376, 1615–1626 CrossRef CAS PubMed.
- F. Robert and J. Pelletier, Exploring the Impact of Single-Nucleotide Polymorphisms on Translation, Front. Genet., 2018, 9, 507 CrossRef CAS.
- G. J. Goodall and V. O. Wickramasinghe, RNA in cancer, Nat. Rev. Cancer, 2021, 21, 22–36 CrossRef CAS PubMed.
- F. Lim, J. J. Solvason, G. E. Ryan, S. H. Le, G. A. Jindal, P. Steffen, S. K. Jandu and E. K. Farley, Affinity-optimizing enhancer variants disrupt development, Nature, 2024, 626, 151–159 CrossRef CAS.
- M. M. Scotti and M. S. Swanson, RNA mis-splicing in disease, Nat. Rev. Genet., 2016, 17, 19–32 CrossRef CAS.
- K. S. Manning and T. A. Cooper, The roles of RNA processing in translating genotype to phenotype, Nat. Rev. Mol. Cell Biol., 2017, 18, 102–114 CrossRef CAS PubMed.
- K. Jaganathan, S. Kyriazopoulou Panagiotopoulou, J. F. McRae, S. F. Darbandi, D. Knowles, Y. I. Li, J. A. Kosmicki, J. Arbelaez, W. Cui and G. B. Schwartz, et al., Predicting Splicing from Primary Sequence with Deep Learning, Cell, 2019, 176, 535–548 CrossRef CAS.
- A. A. Komar, SNPs, Silent But Not Invisible, Science, 2007, 315, 466–467 CrossRef CAS PubMed.
- R. C. Hunt, V. L. Simhadri, M. Iandoli, Z. E. Sauna and C. Kimchi-Sarfaty, Exposing synonymous mutations, Trends Genet., 2014, 30, 308–321 CrossRef CAS.
- M. Davyt, N. Bharti and Z. Ignatova, Effect of mRNA/tRNA mutations on translation speed: Implications for human diseases, J. Biol. Chem., 2023, 299, 105089 CrossRef CAS PubMed.
- M. T. Maurano, R. Humbert, E. Rynes, R. E. Thurman, E. Haugen, H. Wang, A. P. Reynolds, R. Sandstrom, H. Qu and J. Brody, et al., Systematic Localization of Common Disease-Associated Variation in Regulatory DNA, Science, 2012, 337, 1190–1195 CrossRef CAS PubMed.
- J. F. Degner, A. A. Pai, R. Pique-Regi, J.-B. Veyrieras, D. J. Gaffney, J. K. Pickrell, S. De Leon, K. Michelini, N. Lewellen and G. E. Crawford, et al., DNase I sensitivity QTLs are a major determinant of human expression variation, Nature, 2012, 482, 390–394 CrossRef CAS PubMed.
- M. J. Landrum, J. M. Lee, G. R. Riley, W. Jang, W. S. Rubinstein, D. M. Church and D. R. Maglott, ClinVar: public archive of relationships among sequence variation and human phenotype, Nucleic Acids Res., 2014, 42, D980–D985 CrossRef CAS.
- S. S. Nishizaki and A. P. Boyle, Mining the Unknown: Assigning Function to Noncoding Single Nucleotide Polymorphisms, Trends Genet., 2017, 33, 34–45 CrossRef CAS.
- H. J. Kim, B. Keam, S. Im, H. S. Ham, D. Oh, J. Kim, W. S. Han, T. Kim, I. A. Park and Y. J. Bang, Use of MDR1/ABCB1 single nucleotide polymorphism (SNP) as a prognostic factor for breast cancer patients receiving docetaxel + doxorubicin neoadjuvant chemotherapy, J. Clin. Oncol., 2008, 26, 569 Search PubMed.
- E. Caiola, M. Broggini and M. Marabese, Genetic markers for prediction of treatment outcomes in ovarian cancer, Pharmacogenomics J., 2014, 14, 401–410 CrossRef CAS.
- A. N. Yadon, K. Maharaj, J. H. Adamson, Y.-P. Lai, J. C. Sacchettini, T. R. Ioerger, E. J. Rubin and A. S. Pym, A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide, Nat. Commun., 2017, 8, 588 CrossRef PubMed.
- C. J. Meehan, G. A. Goig, T. A. Kohl, L. Verboven, A. Dippenaar, M. Ezewudo, M. R. Farhat, J. L. Guthrie, K. Laukens and P. Miotto, et al., Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues, Nat. Rev. Microbiol., 2019, 17, 533–545 CrossRef CAS PubMed.
- K. T. Skinner, A. M. Palkar and A. L. Hong, Genetics of ABCB1 in Cancer, Cancers, 2023, 15, 4236 CrossRef CAS PubMed.
- H. A. Yu, M. E. Arcila, N. Rekhtman, C. S. Sima, M. F. Zakowski, W. Pao, M. G. Kris, V. A. Miller, M. Ladanyi and G. J. Riely, Analysis of Tumor Specimens at the Time of Acquired Resistance to EGFR-TKI Therapy in 155 Patients with EGFR-Mutant Lung Cancers, Clin. Cancer Res, 2013, 19, 2240–2247 CrossRef CAS PubMed.
- K. Michailidou, S. Lindström, J. Dennis, J. Beesley, S. Hui, S. Kar, A. Lemaçon, P. Soucy, D. Glubb and A. Rostamianfar, et al., Association analysis identifies 65 new breast cancer risk loci, Nature, 2017, 551, 92–94 CrossRef CAS PubMed.
- F. Bertucci, C. K. Y. Ng, A. Patsouris, N. Droin, S. Piscuoglio, N. Carbuccia, J. C. Soria, A. T. Dien, Y. Adnani and M. Kamal, et al., Genomic characterization of metastatic breast cancers, Nature, 2019, 569, 560–564 CrossRef CAS.
- D. Gupta, S. Sturtevant, B. Vieira, Y. Nakamura, S. Krishnamoorthy and M. Demers, Characterization of a Genetically Engineered HUDEP2 Cell Line Harboring a Sickle Cell Disease Mutation As a Potential Research Tool for Preclinical Sickle Cell Disease Drug Discovery, Blood, 2019, 134, 3559 CrossRef.
- L. Huang, Z. Guo, F. Wang and L. Fu, KRAS mutation: from undruggable to druggable in cancer, Signal Transduct. Target. Ther., 2021, 6, 386 CrossRef CAS.
- K.-T. Lee, D.-P. Chen, Z.-J. Loh, W.-P. Chung, C.-Y. Wang, P.-S. Chen, C. H. A. Cheung, C.-P. Chang and H.-P. Hsu, Benign polymorphisms in the BRCA genes with linkage disequilibrium is associated with cancer characteristics, Cancer Sci., 2024, 115, 3973–3985 CrossRef CAS.
- W. Wang, H. Zhou, L. Cai and T. Yang, Association between the rifampicin resistance mutations and rifabutin susceptibility in Mycobacterium tuberculosis: A meta-analysis, J. Glob. Antimicrob. Resist., 2025, 40, 53–61 CrossRef CAS.
- J. A. Plante, Y. Liu, J. Liu, H. Xia, B. A. Johnson, K. G. Lokugamage, X. Zhang, A. E. Muruato, J. Zou and C. R. Fontes-Garfias, et al., Spike mutation D614G alters SARS-CoV-2 fitness, Nature, 2021, 592, 116–121 CrossRef CAS.
- A. Ivey, R. K. Hills, M. A. Simpson, J. V. Jovanovic, A. Gilkes, A. Grech, Y. Patel, N. Bhudia, H. Farah and J. Mason, et al., Assessment of Minimal Residual Disease in Standard-Risk AML, N. Engl. J. Med., 2016, 374, 422–433 CrossRef CAS.
- J. Phallen, M. Sausen, V. Adleff, A. Leal, C. Hruban, J. White, V. Anagnostou, J. Fiksel, S. Cristiano and E. Papp, et al., Direct detection of early-stage cancers using circulating tumor DNA, Sci. Transl. Med., 2017, 9, eaan2415 CrossRef.
- L. Keller, Y. Belloum, H. Wikman and K. Pantel, Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond, Br. J. Cancer, 2021, 124, 345–358 CrossRef PubMed.
- P. Song, L. R. Wu, Y. H. Yan, J. X. Zhang, T. Chu, L. N. Kwong, A. A. Patel and D. Y. Zhang, Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics, Nat. Biomed. Eng., 2022, 6, 232–245 CrossRef CAS PubMed.
- I. Landa and M. E. Cabanillas, Genomic alterations in thyroid cancer: biological and clinical insights, Nat. Rev. Endocrinol., 2024, 20, 93–110 CrossRef PubMed.
- K. Pantel and C. Alix-Panabières, Minimal residual disease as a target for liquid biopsy in patients with solid tumours, Nat. Rev. Clin. Oncol., 2025, 22, 65–77 CrossRef.
- K. a V. Kohabir, E. A. Sistermans and R. M. F. Wolthuis, Recent advances in CRISPR-based single-nucleotide fidelity diagnostics, Commun. Med., 2025, 5, 252 CrossRef CAS PubMed.
- A. Abi and A. Safavi, Targeted Detection of Single-Nucleotide Variations: Progress and Promise, ACS Sens., 2019, 4, 792–807 CrossRef CAS PubMed.
- K. Zhang, R. Deng, H. Gao, X. Teng and J. Li, Lighting up single-nucleotide variation in situ in single cells and tissues, Chem. Soc. Rev., 2020, 49, 1932–1954 RSC.
- D. Khodakov, C. Wang and D. Y. Zhang, Diagnostics based on nucleic acid sequence variant profiling: PCR, hybridization, and NGS approaches, Adv. Drug Delivery Rev., 2016, 105, 3–19 CrossRef CAS.
- E. Xiong, P. Liu, R. Deng, K. Zhang, R. Yang and J. Li, Recent advances in enzyme-free and enzyme-mediated single-nucleotide variation assay in vitro, Nat. Sci. Rev., 2024, 11, nwae118 CrossRef CAS.
- F. Darbeheshti, F. Yu and G. M. Makrigiorgos, Pre-PCR Mutation-Enrichment Methods for Liquid Biopsy Applications, Cancers, 2022, 14, 3143 CrossRef CAS PubMed.
- A. E. Rodda, B. J. Parker, A. Spencer and S. R. Corrie, Extending Circulating Tumor DNA Analysis to Ultralow Abundance Mutations: Techniques and Challenges, ACS Sens., 2018, 3, 540–560 CrossRef CAS PubMed.
- M. Li, F. Yin, L. Song, X. Mao, F. Li, C. Fan, X. Zuo and Q. Xia, Nucleic Acid Tests for Clinical Translation, Chem. Rev., 2021, 121, 10469–10558 CrossRef CAS.
- C. Guo, R. Ding, Z. Zhao, J. Guo and F. Li, Enrichment Strategies for Low-Abundant Single Nucleotide Mutations, Chem. – Eur. J., 2025, 31, e202402872 CrossRef CAS.
- D. Y. Zhang, S. X. Chen and P. Yin, Optimizing the specificity of nucleic acid hybridization, Nat. Chem., 2012, 4, 208–214 CrossRef CAS PubMed.
- J. C. M. Wan, C. Massie, J. Garcia-Corbacho, F. Mouliere, J. D. Brenton, C. Caldas, S. Pacey, R. Baird and N. Rosenfeld, Liquid biopsies come of age: towards implementation of circulating tumour DNA, Nat. Rev. Cancer, 2017, 17, 223–238 CrossRef CAS PubMed.
- Z. E. Sauna and C. Kimchi-Sarfaty, Understanding the contribution of synonymous mutations to human disease, Nat. Rev. Genet., 2011, 12, 683–691 CrossRef CAS PubMed.
- Y. Sharma, M. Miladi, S. Dukare, K. Boulay, M. Caudron-Herger, M. Groß, R. Backofen and S. Diederichs, A pan-cancer analysis of synonymous mutations, Nat. Commun., 2019, 10, 2569 CrossRef PubMed.
- R. Sajwan, L. Wang, O. Casar-Borota, K. Karakostis, S. Chen, R. Fahraeus and X. Gu, and S. Vadivel Gnanasundram, A cancer-associated TP53 synonymous mutation induces synthesis of the p53 isoform p53/47, Br. J. Cancer, 2025, 1–6 Search PubMed.
- Y. Lan, Z. Xia, Q. Shao, P. Lin, J. Lu, X. Xiao, M. Zheng, D. Chen, Y. Dou and Q. Xie, Synonymous mutations promote tumorigenesis by disrupting m6A-dependent mRNA metabolism, Cell, 2025, 188, 1828–1841 CrossRef CAS PubMed.
- S. Wakita, H. Yamaguchi, I. Omori, K. Terada, T. Ueda, E. Manabe, S. Kurosawa, S. Iida, T. Ibaraki and Y. Sato, et al., Mutations of the epigenetics-modifying gene (DNMT3a, TET2, IDH1/2) at diagnosis may induce FLT3-ITD at relapse in de novo acute myeloid leukemia, Leukemia, 2013, 27, 1044–1052 CrossRef CAS.
- J. Tie, Y. Wang, C. Tomasetti, L. Li, S. Springer, I. Kinde, N. Silliman, M. Tacey, H.-L. Wong and M. Christie, et al., Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer, Sci. Transl. Med., 2016, 8, 346ra92 Search PubMed.
- M. Jamal-Hanjani, G. A. Wilson, S. Horswell, R. Mitter, O. Sakarya, T. Constantin, R. Salari, E. Kirkizlar, S. Sigurjonsson and R. Pelham, et al., Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer, Ann. Oncol., 2016, 27, 862–867 CrossRef CAS PubMed.
- C. Abbosh, N. J. Birkbak, G. A. Wilson, M. Jamal-Hanjani, T. Constantin, R. Salari, J. Le Quesne, D. A. Moore, S. Veeriah and R. Rosenthal, et al., Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution, Nature, 2017, 545, 446–451 CrossRef CAS PubMed.
- J. B. Dunlap, J. Leonard, M. Rosenberg, R. Cook, R. Press, G. Fan, P. W. Raess, B. J. Druker and E. Traer, The combination of NPM1, DNMT3A, and IDH1/2 mutations leads to inferior overall survival in AML, Am. J. Hematol., 2019, 94, 913–920 CrossRef CAS.
- R. C. Coombes, K. Page, R. Salari, R. K. Hastings, A. Armstrong, S. Ahmed, S. Ali, S. Cleator, L. Kenny and J. Stebbing, et al., Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence, Clin. Cancer Res, 2019, 25, 4255–4263 CrossRef CAS.
- N. R. Faria, T. A. Mellan, C. Whittaker, I. M. Claro, D. da, S. Candido, S. Mishra, M. A. E. Crispim, F. C. S. Sales, I. Hawryluk and J. T. McCrone, et al., Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil, Science, 2021, 372, 815–821 CrossRef CAS PubMed.
- H. Tegally, E. Wilkinson, R. J. Lessells, J. Giandhari, S. Pillay, N. Msomi, K. Mlisana, J. N. Bhiman, A. von Gottberg and S. Walaza, et al., Sixteen novel lineages of SARS-CoV-2 in South Africa, Nat. Med., 2021, 27, 440–446 CrossRef CAS.
- R. Johnson, J. R. Sharma, P. Ramharack, N. Mangwana, C. Kinnear, A. Viraragavan, B. Glanzmann, J. Louw, N. Abdelatif and T. Reddy, et al., Tracking the circulating SARS-CoV-2 variant of concern in South Africa using wastewater-based epidemiology, Sci. Rep., 2022, 12, 1182 CrossRef CAS PubMed.
- E. B. Hodcroft, M. Zuber, S. Nadeau, T. G. Vaughan, K. H. D. Crawford, C. L. Althaus, M. L. Reichmuth, J. E. Bowen, A. C. Walls and D. Corti, et al., Spread of a SARS-CoV-2 variant through Europe in the summer of 2020, Nature, 2021, 595, 707–712 CrossRef CAS.
- R. Chen, Y. Liu, D. Luo, L. Si, B. Huang, J. Wang, X. Li, F. Cheng, D. Xu and C. Duan, Hepatitis B virus mutation pattern rtA181S+T184I+M204I may contribute to multidrug resistance in clinical practice: Analysis of a large cohort of Chinese patients, Antiviral Res., 2020, 180, 104852 CrossRef CAS.
- E. Sanchez-Padilla, M. Merker, P. Beckert, F. Jochims, T. Dlamini, P. Kahn, M. Bonnet and S. Niemann, Detection of Drug-Resistant Tuberculosis by Xpert MTB/RIF in Swaziland, N. Engl. J. Med., 2015, 372, 1181–1182 CrossRef CAS.
- A. M. Femino, F. S. Fay, K. Fogarty and R. H. Singer, Visualization of Single RNA Transcripts in Situ, Science, 1998, 280, 585–590 CrossRef CAS.
- X. Pichon, M. Lagha, F. Mueller and E. Bertrand, A Growing Toolbox to Image Gene Expression in Single Cells: Sensitive Approaches for Demanding Challenges, Mol. Cell, 2018, 71, 468–480 CrossRef CAS.
- V. Marx, Method of the Year: spatially resolved transcriptomics, Nat. Methods, 2021, 18, 9–14 CrossRef CAS PubMed.
- R. B. Wallace, J. Shaffer, R. F. Murphy, J. Bonner, T. Hirose and K. Itakura, Hybridization of synthetic oligodeoxyribonucleotides to Φ X 174 DNA: the effect of single base pair mismatch, Nucleic Acids Res., 1979, 6, 3543–3558 CrossRef CAS.
- B. J. Conner, A. A. Reyes, C. Morin, K. Itakura, R. L. Teplitz and R. B. Wallace, Detection of sickle cell beta S-globin allele by hybridization with synthetic oligonucleotides, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 278–282 CrossRef CAS.
- S. G. Fischer and L. S. Lerman, DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 1579–1583 CrossRef CAS.
- R. M. Myers, S. G. Fischer, T. Maniatis and L. S. Lerman, Modification of the melting properties of duplex DNA by attachment of a GC-rich DNA sequence as determined by denaturing gradient gel electrophoresis, Nucleic Acids Res., 1985, 13, 3111–3129 CrossRef CAS.
- R. M. Myers, N. Lumelsky, L. S. Lerman and T. Maniatis, Detection of single base substitutions in total genomic DNA, Nature, 1985, 313, 495–498 CrossRef CAS PubMed.
- A.-L. Børresen, E. Hovig and A. Brøgger, Detection of base mutations in genomic DNA using denaturing gradient gel electrophoresis (DGGE) followed by transfer and hybridization with gene-specific probes, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1988, 202, 77–83 CrossRef.
- V. V. Demidov and M. D. Frank-Kamenetskii, Two sides of the coin: affinity and specificity of nucleic acid interactions, Trends Biochem. Sci., 2004, 29, 62–71 CrossRef CAS.
- D. D. Nedorezova, M. V. Dubovichenko, E. P. Belyaeva, E. D. Grigorieva, A. V. Peresadina and D. M. Kolpashchikov, Specificity of oligonucleotide gene therapy (OGT) agents, Theranostics, 2022, 12, 7132–7157 CrossRef CAS.
- D. Tulpan, M. Andronescu, S. B. Chang, M. R. Shortreed, A. Condon, H. H. Hoos and L. M. Smith, Thermodynamically based DNA strand design, Nucleic Acids Res., 2005, 33, 4951–4964 CrossRef CAS.
- H. Koltai and C. Weingarten-Baror, Specificity of DNA microarray hybridization: characterization, effectors and approaches for data correction, Nucleic Acids Res., 2008, 36, 2395–2405 CrossRef CAS.
- S. Itzkovitz and A. van Oudenaarden, Validating transcripts with probes and imaging technology, Nat. Methods, 2011, 8, S12–S19 CrossRef CAS PubMed.
- J. Bjørheim and P. O. Ekstrøm, Review of denaturant capillary electrophoresis in DNA variation analysis, Electrophoresis, 2005, 26, 2520–2530 CrossRef.
- W. Reisner, N. B. Larsen, A. Silahtaroglu, A. Kristensen, N. Tommerup, J. O. Tegenfeldt and H. Flyvbjerg, Single-molecule denaturation mapping of DNA in nanofluidic channels, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 13294–13299 CrossRef CAS.
- H. Ørum, M. H. Jakobsen, T. Koch, J. Vuust and M. B. Borre, Detection of the Factor V Leiden Mutation by Direct Allele-specific Hybridization of PCR Amplicons to Photoimmobilized Locked Nucleic Acids, Clin. Chem., 1999, 45, 1898–1905 Search PubMed.
- M. Petersen and J. Wengel, LNA: a versatile tool for therapeutics and genomics, Trends Biotechnol., 2003, 21, 74–81 CrossRef CAS.
- M. P. Johnson, L. M. Haupt and L. R. Griffiths, Locked nucleic acid (LNA) single nucleotide polymorphism (SNP) genotype analysis and validation using real-time PCR, Nucleic Acids Res., 2004, 32, e55 CrossRef.
- Y. You, B. G. Moreira, M. A. Behlke and R. Owczarzy, Design of LNA probes that improve mismatch discrimination, Nucleic Acids Res., 2006, 34, e60 CrossRef.
- J. M. Obliosca, S. Y. Cheng, Y.-A. Chen, M. F. Llanos, Y.-L. Liu, D. M. Imphean, D. R. Bell, J. T. Petty, P. Ren and H.-C. Yeh, LNA Thymidine Monomer Enables Differentiation of the Four Single-Nucleotide Variants by Melting Temperature, J. Am. Chem. Soc., 2017, 139, 7110–7116 CrossRef CAS.
- H. Ørum, P. E. Nielsen, M. Egholm, R. H. Berg, O. Buchardt and C. Stanley, Single base pair mutation analysis by PNA directed PCR clamping, Nucleic Acids Res., 1993, 21, 5332–5336 CrossRef.
- P. L. Ross, K. Lee and P. Belgrader, Discrimination of Single-Nucleotide Polymorphisms in Human DNA Using Peptide Nucleic Acid Probes Detected by MALDI-TOF Mass Spectrometry, Anal. Chem., 1997, 69, 4197–4202 CrossRef CAS.
- A. Lomakin and M. D. Frank-Kamenetskii, A theoretical analysis of specificity of nucleic acid interactions with oligonucleotides and peptide nucleic acids (PNAs), J. Mol. Biol., 1998, 276, 57–70 CrossRef CAS PubMed.
- M. Komiyama, S. Ye, Liang, Y. Yamamoto, T. Tomita, J.-M. Zhou and H. Aburatani, PNA for One-Base Differentiating Protection of DNA from Nuclease and Its Use for SNPs Detection, J. Am. Chem. Soc., 2003, 125, 3758–3762 CrossRef CAS PubMed.
- G. He, S. Rapireddy, R. Bahal, B. Sahu and D. H. Ly, Strand Invasion of Extended, Mixed-Sequence B-DNA by γPNAs, J. Am. Chem. Soc., 2009, 131, 12088–12090 CrossRef CAS PubMed.
- L. Zhang, R. Parvin, S. Lin, M. Chen, R. Zheng, Q. Fan and F. Ye, Peptide Nucleic Acid Clamp-Assisted Photothermal Multiplexed Digital PCR for Identifying SARS-CoV-2 Variants of Concern, Adv. Sci., 2024, 11, 2306088 CrossRef CAS.
- Z. Guo, Q. Liu and L. M. Smith, Enhanced discrimination of single nucleotide polymorphisms by artificial mismatch hybridization, Nat. Biotechnol., 1997, 15, 331–335 CrossRef CAS PubMed.
- A. G. Frutos, S. Pal, M. Quesada and J. Lahiri, Method for Detection of Single-Base Mismatches Using Bimolecular Beacons, J. Am. Chem. Soc., 2002, 124, 2396–2397 CrossRef CAS PubMed.
- I. Lee, S. S. Ajay, H. Chen, A. Maruyama, N. Wang, M. G. McInnis and B. D. Athey, Discriminating single-base difference miRNA expressions using microarray Probe Design Guru (ProDeG), Nucleic Acids Res., 2008, 36, e27 CrossRef.
- I. I. Cisse, H. Kim and T. Ha, A rule of seven in Watson-Crick base-pairing of mismatched sequences, Nat. Struct. Mol. Biol., 2012, 19, 623–627 CrossRef CAS PubMed.
- H. E. Moser and P. B. Dervan, Sequence-Specific Cleavage of Double Helical DNA by Triple Helix Formation, Science, 1987, 238, 645–650 CrossRef CAS.
- R. W. Roberts and D. M. Crothers, Specificity and stringency in DNA triplex formation, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 9397–9401 CrossRef CAS.
- G. Bonnet, S. Tyagi, A. Libchaber and F. R. Kramer, Thermodynamic basis of the enhanced specificity of structured DNA probes, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 6171–6176 CrossRef CAS PubMed.
- S. Tyagi and F. R. Kramer, Molecular Beacons: Probes that Fluoresce upon Hybridization, Nat. Biotechnol., 1996, 14, 303–308 CrossRef CAS PubMed.
- L. G. Kostrikis, S. Tyagi, M. M. Mhlanga, D. D. Ho and F. R. Kramer, Spectral Genotyping of Human Alleles, Science, 1998, 279, 1228–1229 CrossRef CAS.
- A. S. Piatek, S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer and D. Alland, Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis, Nat. Biotechnol., 1998, 16, 359–363 CrossRef CAS.
- S. Tyagi, D. P. Bratu and F. R. Kramer, Multicolor molecular beacons for allele discrimination, Nat. Biotechnol., 1998, 16, 49–53 CrossRef CAS PubMed.
- S. Raja, J. Ching, L. Xi, S. J. Hughes, R. Chang, W. Wong, W. McMillan, W. E. Gooding, K. S. McCarty Jr and M. Chestney, et al., Technology for Automated, Rapid, and Quantitative PCR or Reverse Transcription-PCR Clinical Testing, Clin. Chem., 2005, 51, 882–890 CrossRef CAS.
- S. A. E. Marras, S. Tyagi and F. R. Kramer, Real-time assays with molecular beacons and other fluorescent nucleic acid hybridization probes, Clin. Chim. Acta, 2006, 363, 48–60 CrossRef CAS.
- S. Chakravorty, B. Aladegbami, M. Burday, M. Levi, S. A. E. Marras, D. Shah, H. H. El-Hajj, F. R. Kramer and D. Alland, Rapid Universal Identification of Bacterial Pathogens from Clinical Cultures by Using a Novel Sloppy Molecular Beacon Melting Temperature Signature Technique, J. Clin. Microbiol., 2010, 48, 258–267 CrossRef CAS.
- T. Bentin, H. J. Larsen and P. E. Nielsen, Combined Triplex/Duplex Invasion of Double-Stranded DNA by “Tail-Clamp” Peptide Nucleic Acid, Biochemistry, 2003, 42, 13987–13995 CrossRef CAS.
- T. N. Grossmann, L. Röglin and O. Seitz, Triplex Molecular Beacons as Modular Probes for DNA Detection, Angew. Chem., Int. Ed., 2007, 46, 5223–5225 CrossRef CAS.
- O. Seitz, Solid-Phase Synthesis of Doubly Labeled Peptide Nucleic Acids as Probes for the Real-Time Detection of Hybridization, Angew. Chem., Int. Ed., 2000, 39, 3249–3252 CrossRef CAS.
- H. Kuhn, V. V. Demidov, J. M. Coull, M. J. Fiandaca, B. D. Gildea and M. D. Frank-Kamenetskii, Hybridization of DNA and PNA Molecular Beacons to Single-Stranded and Double-Stranded DNA Targets, J. Am. Chem. Soc., 2002, 124, 1097–1103 CrossRef CAS.
- L. Wang, C. J. Yang, C. D. Medley, S. A. Benner and W. Tan, Locked Nucleic Acid Molecular Beacons, J. Am. Chem. Soc., 2005, 127, 15664–15665 CrossRef CAS.
- B. Guo, Y. Sheng, K. Zhou, Q. Liu, L. Liu and H.-C. Wu, Analyte-Triggered DNA-Probe Release from a Triplex Molecular Beacon for Nanopore Sensing, Angew. Chem., Int. Ed., 2018, 57, 3602–3606 CrossRef CAS.
- Y. Xiao, X. Lou, T. Uzawa, K. J. I. Plakos, K. W. Plaxco and H. T. Soh, An Electrochemical Sensor for Single Nucleotide Polymorphism Detection in Serum Based on a Triple-Stem DNA Probe, J. Am. Chem. Soc., 2009, 131, 15311–15316 CrossRef CAS PubMed.
- Y. Xiao, K. J. I. Plakos, X. Lou, R. J. White, J. Qian, K. W. Plaxco and H. T. Soh, Fluorescence Detection of Single-Nucleotide Polymorphisms with a Single, Self-Complementary, Triple-Stem DNA Probe, Angew. Chem., Int. Ed., 2009, 48, 4354–4358 CrossRef CAS PubMed.
- C. Guo, H. Deng, Q. Yang, D. Huang, C. Shen, G. A. Wang and F. Li, Coding Intrinsic Disorder into DNA Hybridization Probes Enables Discrimination of Single Nucleotide Variants over Wide and Tunable Temperature Ranges, Angew. Chem., 2023, 135, e202314386 CrossRef.
- E. M. Cornett, E. A. Campbell, G. Gulenay, E. Peterson, N. Bhaskar and D. M. Kolpashchikov, Molecular Logic Gates for DNA Analysis: Detection of Rifampin Resistance in M. tuberculosis DNA, Angew. Chem., Int. Ed., 2012, 51, 9075–9077 CrossRef CAS.
- D. M. Kolpashchikov, A Binary DNA Probe for Highly Specific Nucleic Acid Recognition, J. Am. Chem. Soc., 2006, 128, 10625–10628 CrossRef CAS PubMed.
- D. M. Kolpashchikov, Split DNA Enzyme for Visual Single Nucleotide Polymorphism Typing, J. Am. Chem. Soc., 2008, 130, 2934–2935 CrossRef CAS PubMed.
- J. Grimes, Y. V. Gerasimova and D. M. Kolpashchikov, Real-Time SNP Analysis in Secondary-Structure-Folded Nucleic Acids, Angew. Chem., Int. Ed., 2010, 49, 8950–8953 CrossRef CAS.
- D. M. Kolpashchikov, Binary Probes for Nucleic Acid Analysis, Chem. Rev., 2010, 110, 4709–4723 CrossRef CAS PubMed.
- M. Stancescu, T. A. Fedotova, J. Hooyberghs, A. Balaeff and D. M. Kolpashchikov, Nonequilibrium Hybridization Enables Discrimination of a Point Mutation within 5–40 °C, J. Am. Chem. Soc., 2016, 138, 13465–13468 CrossRef CAS.
- R. J. Karadeema, M. Stancescu, T. P. Steidl, S. C. Bertot and D. M. Kolpashchikov, The owl sensor: a ‘fragile’ DNA nanostructure for the analysis of single nucleotide variations, Nanoscale, 2018, 10, 10116–10122 RSC.
- D. M. Kolpashchikov, Evolution of Hybridization Probes to DNA Machines and Robots, Acc. Chem. Res., 2019, 52, 1949–1956 CrossRef CAS PubMed.
- B. L. Mueller, M. J. Liberman and D. M. Kolpashchikov, OWL2: a molecular beacon-based nanostructure for highly selective detection of single-nucleotide variations in folded nucleic acids, Nanoscale, 2023, 15, 5735–5742 RSC.
- D. M. Kolpashchikov, A Binary Deoxyribozyme for Nucleic Acid Analysis, ChemBioChem, 2007, 8, 2039–2042 CrossRef CAS.
- B. R. Deal, R. Ma, V. P.-Y. Ma, H. Su, J. T. Kindt and K. Salaita, Engineering DNA-Functionalized Nanostructures to Bind Nucleic Acid Targets Heteromultivalently with Enhanced Avidity, J. Am. Chem. Soc., 2020, 142, 9653–9660 CrossRef CAS PubMed.
- B. R. Deal, R. Ma, S. Narum, H. Ogasawara, Y. Duan, J. T. Kindt and K. Salaita, Heteromultivalency enables enhanced detection of nucleic acid mutations, Nat. Chem., 2024, 16, 229–238 CrossRef CAS PubMed.
- D. Y. Zhang and E. Winfree, Control of DNA Strand Displacement Kinetics Using Toehold Exchange, J. Am. Chem. Soc., 2009, 131, 17303–17314 CrossRef CAS PubMed.
- L. R. Wu, J. S. Wang, J. Z. Fang, E. R. Evans, A. Pinto, I. Pekker, R. Boykin, C. Ngouenet, P. J. Webster and J. Beechem, et al., Continuously tunable nucleic acid hybridization probes, Nat. Methods, 2015, 12, 1191–1196 CrossRef CAS.
- L. Wu, G. A. Wang and F. Li, Plug-and-Play Module for Reversible and Continuous Control of DNA Strand Displacement Kinetics, J. Am. Chem. Soc., 2024, 146, 6516–6521 CrossRef CAS.
- J. S. Wang, Y. H. Yan and D. Y. Zhang, Modular probes for enriching and detecting complex nucleic acid sequences, Nat. Chem., 2017, 9, 1222–1228 CrossRef CAS.
- F. Hong, D. Ma, K. Wu, L. A. Mina, R. C. Luiten, Y. Liu, H. Yan and A. A. Green, Precise and Programmable Detection of Mutations Using Ultraspecific Riboregulators, Cell, 2020, 180, 1018–1032 CrossRef CAS PubMed.
- Y. Li, G. A. Wang, S. D. Mason, X. Yang, Z. Yu, Y. Tang and F. Li, Simulation-guided engineering of an enzyme-powered three dimensional DNA nanomachine for discriminating single nucleotide variants, Chem. Sci., 2018, 9, 6434–6439 RSC.
- Y. Li, S. C. Meng, Y. Wang, C. M. Platnich, M. K. Earle, E. Mylona, P. Naydenova, S. Baker, J. Zhu and U. F. Keyser, Nanopore detection of single-nucleotide RNA mutations and modifications with programmable nanolatches, Nat. Nanotechnol., 2025, 1–9 Search PubMed.
- M. Lyu, L. Kong, Z. Yang, Y. Wu, C. E. McGhee and Y. Lu, PNA-Assisted DNAzymes to Cleave Double-Stranded DNA for Genetic Engineering with High Sequence Fidelity, J. Am. Chem. Soc., 2021, 143, 9724–9728 CrossRef CAS PubMed.
- N. O. Bukanov, V. V. Demidov, P. E. Nielsen and M. D. Frank-Kamenetskii, PD-loop: A complex of duplex DNA with an oligonucleotide, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 5516–5520 CrossRef CAS.
- J. Das, I. Ivanov, E. H. Sargent and S. O. Kelley, DNA Clutch Probes for Circulating Tumor DNA Analysis, J. Am. Chem. Soc., 2016, 138, 11009–11016 CrossRef CAS.
- D. A. Khodakov, A. S. Khodakova, A. Linacre and A. V. Ellis, Toehold-Mediated Nonenzymatic DNA Strand Displacement As a Platform for DNA Genotyping, J. Am. Chem. Soc., 2013, 135, 5612–5619 CrossRef CAS.
- S. X. Chen, D. Y. Zhang and G. Seelig, Conditionally fluorescent molecular probes for detecting single base changes in double-stranded DNA, Nat. Chem., 2013, 5, 782–789 CrossRef CAS.
- G. A. Wang, X. Xie, H. Mansour, F. Chen, G. Matamoros, A. L. Sanchez, C. Fan and F. Li, Expanding detection windows for discriminating single nucleotide variants using rationally designed DNA equalizer probes, Nat. Commun., 2020, 11, 5473 CrossRef CAS PubMed.
- D. Huang, H. Deng, J. Zhou, G. A. Wang, Q. Lei, C. Guo, W. Peng, P. Liang, C. Shen and B. Ying, et al., Mismatch-Guided Deoxyribonucleic Acid Assembly Enables Ultrasensitive and Multiplex Detection of Low-Allele-Fraction Variants in Clinical Samples, J. Am. Chem. Soc., 2023, 145, 20412–20421 CrossRef CAS PubMed.
- J. S. Wang and D. Y. Zhang, Simulation-guided DNA probe design for consistently ultraspecific hybridization, Nat. Chem., 2015, 7, 545–553 CrossRef CAS PubMed.
- G. Gilliland, S. Perrin, K. Blanchard and H. F. Bunn, Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 2725–2729 CrossRef CAS PubMed.
- P. D. Siebert and J. W. Larrick, Competitive PCR, Nature, 1992, 359, 557–558 CrossRef CAS PubMed.
- P. R. Prestwood, M. Yang, G. V. Lewis, S. Balaratnam, K. Yazdani and J. S. Schneekloth, Competitive Microarray Screening Reveals Functional Ligands for the DHX15 RNA G-Quadruplex, ACS Med. Chem. Lett., 2024, 15, 814–821 CrossRef CAS PubMed.
- X. Chen, N. Liu, L. Liu, W. Chen, N. Chen, M. Lin, J. Xu, X. Zhou, H. Wang and M. Zhao, et al., Thermodynamics and kinetics guided probe design for uniformly sensitive and specific DNA hybridization without optimization, Nat. Commun., 2019, 10, 4675 CrossRef PubMed.
- Y. Tan, D. Huang, G. A. Wang, C. Shen, H. Deng and F. Li, Concentration-Bias-Free Discrimination of Single Nucleotide Variants Using Isothermal Nucleic Acid Amplification and Mismatch-Guided DNA Assembly, Anal. Chem., 2025, 97, 1917–1924 CrossRef CAS PubMed.
- S. X. Chen and G. Seelig, An Engineered Kinetic Amplification Mechanism for Single Nucleotide Variant Discrimination by DNA Hybridization Probes, J. Am. Chem. Soc., 2016, 138, 5076–5086 CrossRef CAS PubMed.
- R. Mukherjee, A. Sengar, J. Cabello-García and T. E. Ouldridge, Kinetic Proofreading Can Enhance Specificity in a Nonenzymatic DNA Strand Displacement Network, J. Am. Chem. Soc., 2024, 146, 18916–18926 CrossRef CAS.
- J. J. Hopfield, Kinetic Proofreading: A New Mechanism for Reducing Errors in Biosynthetic Processes Requiring High Specificity, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 4135–4139 CrossRef CAS.
- C. Petersen, A. Johnson-Buck and N. G. Walter, Iterative Kinetic Proofreading for High-Specificity DNA Sequence Discrimination, Biophys. J., 2021, 120, 270a CrossRef.
- C. R. Newton, A. Graham, L. E. Heptinstall, S. J. Powell, C. Summers, N. Kalsheker, J. C. Smith and A. F. Markham, Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS), Nucleic Acids Res., 1989, 17, 2503–2516 CrossRef CAS.
- F. Darbeheshti and G. M. Makrigiorgos, Enzymatic Methods for Mutation Detection in Cancer Samples and Liquid Biopsies, Int. J. Mol. Sci., 2023, 24, 923 CrossRef CAS PubMed.
- C. T. Wittwer, K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry and U. J. Balis, The LightCyclerTM: A Microvolume Multisample Fluorimeter with Rapid Temperature Control, BioTechniques, 1997, 22, 176–181 CrossRef CAS.
- C. T. Wittwer, G. H. Reed, C. N. Gundry, J. G. Vandersteen and R. J. Pryor, High-Resolution Genotyping by Amplicon Melting Analysis Using LCGreen, Clin. Chem., 2003, 49, 853–860 CrossRef CAS PubMed.
- C. T. Wittwer, A. C. Hemmert, J. O. Kent and N. A. Rejali, DNA melting analysis, Mol. Aspects Med., 2024, 97, 101268 CrossRef CAS.
- L. S. Chou, C. Meadows, C. T. Wittwer and E. Lyon, Unlabeled Oligonucleotide Probes Modified with Locked Nucleic Acids for Improved Mismatch Discrimination in Genotyping by Melting Analysis, BioTechniques, 2005, 39, 644–650 CrossRef CAS PubMed.
- S. O. Sundberg, C. T. Wittwer, R. M. Howell, J. Huuskonen, R. J. Pryor, J. S. Farrar, H. M. Stiles, R. A. Palais and I. T. Knight, Microfluidic Genotyping by Rapid Serial PCR and High-Speed Melting Analysis, Clin. Chem., 2014, 60, 1306–1313 CrossRef CAS PubMed.
- J.-Y. Jong, Multiplex molecular diagnostics, https://www.mlo-online.com/home/article/13005002/multiplex-molecular-diagnostics-shifting-the-paradigm, (accessed 9 June 2025).
- Q. Huang, D. Chen, C. Du, Q. Liu, S. Lin, L. Liang, Y. Xu, Y. Liao and Q. Li, Highly multiplex PCR assays by coupling the 5′-flap endonuclease activity of Taq DNA polymerase and molecular beacon reporters, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2110672119 CrossRef CAS.
- G. Fu, A. Miles and L. Alphey, Multiplex Detection and SNP Genotyping in a Single Fluorescence Channel, PLoS One, 2012, 7, e30340 CrossRef CAS.
- D. Gonzalez de Castro, B. Angulo, B. Gomez, D. Mair, R. Martinez, A. Suarez-Gauthier, F. Shieh, M. Velez, V. H. Brophy and H. J. Lawrence, et al., A comparison of three methods for detecting KRAS mutations in formalin-fixed colorectal cancer specimens, Br. J. Cancer, 2012, 107, 345–351 CrossRef CAS.
- B. Angulo, E. Conde, A. Suárez-Gauthier, C. Plaza, R. Martínez, P. Redondo, E. Izquierdo, B. Rubio-Viqueira, L. Paz-Ares and M. Hidalgo, et al., A Comparison of EGFR Mutation Testing Methods in Lung Carcinoma: Direct Sequencing, Real-time PCR and Immunohistochemistry, PLoS One, 2012, 7, e43842 CrossRef CAS PubMed.
- J. B. de Kok, E. T. G. Wiegerinck, B. A. J. Giesendorf and D. W. Swinkels, Rapid genotyping of single nucleotide polymorphisms using novel minor groove binding DNA oligonucleotides (MGB probes), Hum. Mutat., 2002, 19, 554–559 CrossRef CAS PubMed.
- R. Barbano, B. Pasculli, M. Coco, A. Fontana, M. Copetti, M. Rendina, V. M. Valori, P. Graziano, E. Maiello and V. M. Fazio, et al., Competitive allele-specific TaqMan PCR (Cast-PCR) is a sensitive, specific and fast method for BRAF V600 mutation detection in Melanoma patients, Sci. Rep., 2015, 5, 18592 CrossRef CAS.
- J. Stadler, J. Eder, B. Pratscher, S. Brandt, D. Schneller, R. Müllegger, C. Vogl, F. Trautinger, G. Brem and J. P. Burgstaller, SNPase-ARMS qPCR: Ultrasensitive Mutation-Based Detection of Cell-Free Tumor DNA in Melanoma Patients, PLoS One, 2015, 10, e0142273 CrossRef.
- Q. Liu and S. S. Sommer, Detection of extremely rare alleles by bidirectional pyrophosphorolysis-activated polymerization allele-specific amplification (Bi-PAP-A): measurement of mutation load in mammalian tissues, BioTechniques, 2004, 36, 156–166 CrossRef CAS PubMed.
- Q. Liu and S. S. Sommer, Pyrophosphorolysis-Activated Polymerization (PAP): Application to Allele-Specific Amplification, BioTechniques, 2000, 29, 1072–1083 CrossRef CAS PubMed.
- H. Ørum, P. E. Nielsen, M. Egholm, R. H. Berg, O. Buchardt and C. Stanley, Single base pair mutation analysis by PNA directed PCR clamping, Nucleic Acids Res., 1993, 21, 5332–5336 CrossRef.
- H. Ørum, P. E. Nielsen, M. Egholm, R. H. Berg, O. Buchardt and C. Stanley, Single base pair mutation analysis by PNA directed PCR clamping, Nucleic Acids Res., 1993, 21, 5332–5336 CrossRef PubMed.
- Y. Nagai, H. Miyazawa, Huqun, T. Tanaka, K. Udagawa, M. Kato, S. Fukuyama, A. Yokote, K. Kobayashi and M. Kanazawa, et al., Genetic Heterogeneity of the Epidermal Growth Factor Receptor in Non–Small Cell Lung Cancer Cell Lines Revealed by a Rapid and Sensitive Detection System, the Peptide Nucleic Acid-Locked Nucleic Acid PCR Clamp, Cancer Res., 2005, 65, 7276–7282 CrossRef CAS PubMed.
- S.-T. Lee, J.-Y. Kim, M.-J. Kown, S. W. Kim, J. H. Chung, M.-J. Ahn, Y. L. Oh, J.-W. Kim and C.-S. Ki, Mutant Enrichment with 3′-Modified Oligonucleotides: A Practical PCR Method for Detecting Trace Mutant DNAs, J. Mol. Diagn., 2011, 13, 657–668 CrossRef CAS.
- L. R. Wu, S. X. Chen, Y. Wu, A. A. Patel and D. Y. Zhang, Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification, Nat. Biomed. Eng., 2017, 1, 714–723 CrossRef CAS.
- Y. Si, X. Wang, X. Su, Z. Weng, Q. Hu, Q. Li, C. Fan, D. Y. Zhang, Y. Wang and S. Luo, et al., Extended Enrichment for Ultrasensitive Detection of Low-Frequency Mutations by Long Blocker Displacement Amplification, Angew. Chem., Int. Ed., 2024, 63, e202400551 CrossRef CAS PubMed.
- J. Li, L. Wang, H. Mamon, M. H. Kulke, R. Berbeco and G. M. Makrigiorgos, Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing, Nat. Med., 2008, 14, 579–584 CrossRef CAS.
- C. A. Milbury, J. Li and G. M. Makrigiorgos, Ice -COLD-PCR enables rapid amplification and robust enrichment for low-abundance unknown DNA mutations, Nucleic Acids Res., 2011, 39, e2 CrossRef.
- E. Castellanos-Rizaldos, P. Liu, C. A. Milbury, M. Guha, A. Brisci, L. Cremonesi, M. Ferrari, H. Mamon and G. M. Makrigiorgos, Temperature-Tolerant COLD-PCR Reduces Temperature Stringency and Enables Robust Mutation Enrichment, Clin. Chem., 2012, 58, 1130–1138 CrossRef CAS.
- B. Vogelstein and K. W. Kinzler, Digital PCR, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 9236–9241 CrossRef CAS.
- C. M. Hindson, J. R. Chevillet, H. A. Briggs, E. N. Gallichotte, I. K. Ruf, B. J. Hindson, R. L. Vessella and M. Tewari, Absolute quantification by droplet digital PCR versus analog real-time PCR, Nat. Methods, 2013, 10, 1003–1005 CrossRef CAS.
- K. A. Heyries, C. Tropini, M. VanInsberghe, C. Doolin, O. I. Petriv, A. Singhal, K. Leung, C. B. Hughesman and C. L. Hansen, Megapixel digital PCR, Nat. Methods, 2011, 8, 649–651 CrossRef CAS PubMed.
- M. Baker, Digital PCR hits its stride, Nat. Methods, 2012, 9, 541–544 CrossRef CAS.
- A. Trouchet, G. Gines, L. Benhaim and V. Taly, Digital PCR: from early developments to its future application in clinics, Lab. Chip, 2025, 25, 3921–3961 RSC.
- S. Hussung, M. Follo, R. F. U. Klar, S. Michalczyk, K. Fritsch, F. Nollmann, J. Hipp, J. Duyster, F. Scherer and N. Von Bubnoff, et al., Development and Clinical Validation of Discriminatory Multitarget Digital Droplet PCR Assays for the Detection of Hot Spot KRAS and NRAS Mutations in Cell-Free DNA, J. Mol. Diagn., 2020, 22, 943–956 CrossRef CAS.
- M. Mitake, S. Hirano and T. Kishino, Imprinting analysis by droplet digital PCR coupled with locked nucleic acid TaqMan probes, Epigenetics, 2021, 16, 729–740 CrossRef.
- C. Tan, Y. Yan, N. Guo, F. Wang, S. Wang, L. Zhu, Y. Wang, Y. Ma and Y. Guo, Single-Tube Multiplex Digital Polymerase Chain Reaction Assay for Molecular Diagnosis and Prediction of Severity of Spinal Muscular Atrophy, Anal. Chem., 2022, 94, 3517–3525 CrossRef CAS.
- L. M. Wainman, S. H. Sathyanarayana and J. A. Lefferts, Applications of Digital Polymerase Chain Reaction (dPCR) in Molecular and Clinical Testing, J. Appl. Lab. Med, 2024, 9, 124–137 CrossRef PubMed.
- I. V. Kutyavin, I. A. Afonina, A. Mills, V. V. Gorn, E. A. Lukhtanov, E. S. Belousov, M. J. Singer, D. K. Walburger, S. G. Lokhov and A. A. Gall, et al., 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures, Nucleic Acids Res., 2000, 28, 655–661 CrossRef CAS PubMed.
- Y. Yao, C. Nellåker and H. Karlsson, Evaluation of minor groove binding probe and Taqman probe PCR assays: Influence of mismatches and template complexity on quantification, Mol. Cell. Probes, 2006, 20, 311–316 CrossRef CAS.
- K.-L. Xu, Z.-M. Zhang, Y.-D. Wang, X.-L. Cheng, H.-Y. Jin, F. Wei and S.-C. Ma, MGB probe-based multiplex droplet digital PCR for the interspecific identification of Notopterygii Rhizoma et Radix in herbal materials and preparations, Phytomedicine, 2025, 136, 156325 CrossRef CAS.
- H. Yesilkaya, F. Meacci, S. Niemann, D. Hillemann, S. Rüsch-Gerdes, M. R. Barer, P. W. Andrew and M. R. Oggioni, LONG DRUG Study Group, Evaluation of Molecular-Beacon, TaqMan, and Fluorescence Resonance Energy Transfer Probes for Detection of Antibiotic Resistance-Conferring Single Nucleotide Polymorphisms in Mixed Mycobacterium tuberculosis DNA Extracts, J. Clin. Microbiol., 2006, 44, 3826–3829 CrossRef CAS.
- A. V. Cherepanov and S. de Vries, Kinetics and thermodynamics of nick sealing by T4 DNA ligase, Eur. J. Biochem., 2003, 270, 4315–4325 CrossRef CAS PubMed.
- Y. Li, X. Wang, M. Wang, M. Liu, H. Wang, W. Xia and L. Liu, Advances in ligase-based nucleic acid amplification technology for detecting gene mutations: a review, Mol. Cell. Biochem., 2023, 478, 1621–1631 CrossRef CAS.
- H. Yuan, W. Liu, J. Hu and C. Zhang, Recent advances in single-nucleotide variant assay: From in vitro detection to in vivo imaging, TrAC Trends Anal. Chem., 2024, 180, 117963 CrossRef CAS.
- M. Wiedmann, W. J. Wilson, J. Czajka, J. Luo, F. Barany and C. A. Batt, Ligase chain reaction (LCR)–overview and applications, Genome Res., 1994, 3, S51–S64 CrossRef CAS.
- U. Landegren, R. Kaiser, J. Sanders and L. Hood, A Ligase-Mediated Gene Detection Technique, Science, 1988, 241, 1077–1080 CrossRef CAS.
- M. Nilsson, G. Barbany, D.-O. Antson, K. Gertow and U. Landegren, Enhanced detection and distinction of RNA by enzymatic probe ligation, Nat. Biotechnol., 2000, 18, 791–793 CrossRef CAS PubMed.
- V. Potapov, J. L. Ong, B. W. Langhorst, K. Bilotti, D. Cahoon, B. Canton, T. F. Knight, T. C. Evans Jr and G. J. S. Lohman, A single-molecule sequencing assay for the comprehensive profiling of T4 DNA ligase fidelity and bias during DNA end-joining, Nucleic Acids Res., 2018, 46, e79 CrossRef PubMed.
- J. P. Schouten, Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification, Nucleic Acids Res., 2002, 30, 57e–557 CrossRef.
- S. C. Shin, G. Kim, H.-B. Yang, K. W. Park, B.-C. Kang and H. G. Park, Application of the ASLP technology to a novel platform for rapid and noise-free multiplexed SNP genotyping, Biosens. Bioelectron., 2014, 54, 687–694 CrossRef CAS.
- H.-Q. Wang, W.-Y. Liu, Z. Wu, L.-J. Tang, X.-M. Xu, R.-Q. Yu and J.-H. Jiang, Homogeneous Label-Free Genotyping of Single Nucleotide Polymorphism Using Ligation-Mediated Strand Displacement Amplification with DNAzyme-Based Chemiluminescence Detection, Anal. Chem., 2011, 83, 1883–1889 CrossRef CAS PubMed.
- X. Chen, A. Ying and Z. Gao, Highly sensitive and selective colorimetric genotyping of single-nucleotide polymorphisms based on enzyme-amplified ligation on magnetic beads, Biosens. Bioelectron., 2012, 36, 89–94 CrossRef CAS.
- J. H. Park, H. Jang, Y. K. Jung, Y. L. Jung, I. Shin, D.-Y. Cho and H. G. Park, A mass spectrometry-based multiplex SNP genotyping by utilizing allele-specific ligation and strand displacement amplification, Biosens. Bioelectron., 2017, 91, 122–127 CrossRef CAS PubMed.
- M. Nilsson, H. Malmgren, M. Samiotaki, M. Kwiatkowski, B. P. Chowdhary and U. Landegren, Padlock Probes: Circularizing Oligonucleotides for Localized DNA Detection, Science, 1994, 265, 2085–2088 CrossRef CAS PubMed.
- M. Nilsson, K. Krejci, J. Koch, M. Kwiatkowski, P. Gustavsson and U. Landegren, Padlock probes reveal single-nucleotide differences, parent of origin and in situ distribution of centromeric sequences in human chromosomes 13 and 21, Nat. Genet., 1997, 16, 252–255 CrossRef CAS PubMed.
- P. M. Lizardi, X. Huang, Z. Zhu, P. Bray-Ward, D. C. Thomas and D. C. Ward, Mutation detection and single-molecule counting using isothermal rolling-circle amplification, Nat. Genet., 1998, 19, 225–232 CrossRef CAS.
- C. Larsson, J. Koch, A. Nygren, G. Janssen, A. K. Raap, U. Landegren and M. Nilsson, In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes, Nat. Methods, 2004, 1, 227–232 CrossRef CAS PubMed.
- O. Söderberg, M. Gullberg, M. Jarvius, K. Ridderstråle, K.-J. Leuchowius, J. Jarvius, K. Wester, P. Hydbring, F. Bahram and L.-G. Larsson, et al., Direct observation of individual endogenous protein complexes in situ by proximity ligation, Nat. Methods, 2006, 3, 995–1000 CrossRef.
- C. Larsson, I. Grundberg, O. Söderberg and M. Nilsson, In situ detection and genotyping of individual mRNA molecules, Nat. Methods, 2010, 7, 395–397 CrossRef CAS PubMed.
- R. Deng, K. Zhang, Y. Sun, X. Ren and J. Li, Highly specific imaging of mRNA in single cells by target RNA-initiated rolling circle amplification, Chem. Sci., 2017, 8, 3668–3675 RSC.
- K. Zhang, R. Deng, X. Teng, Y. Li, Y. Sun, X. Ren and J. Li, Direct Visualization of Single-Nucleotide Variation in mtDNA Using a CRISPR/Cas9-Mediated Proximity Ligation Assay, J. Am. Chem. Soc., 2018, 140, 11293–11301 CrossRef CAS PubMed.
- R. R. G. Soares, N. Madaboosi and M. Nilsson, Rolling Circle Amplification in Integrated Microsystems: An Uncut Gem toward Massively Multiplexed Pathogen Diagnostics and Genotyping, Acc. Chem. Res., 2021, 54, 3979–3990 CrossRef CAS.
- K. Abravaya, J. J. Carrino, S. Muldoon and H. H. Lee, Detection of point mutations with a modified ligase chain reaction (Gap-LCR), Nucleic Acids Res., 1995, 23, 675–682 CrossRef CAS PubMed.
- P. Hardenbol, J. Banér, M. Jain, M. Nilsson, E. A. Namsaraev, G. A. Karlin-Neumann, H. Fakhrai-Rad, M. Ronaghi, T. D. Willis and U. Landegren, et al., Multiplexed genotyping with sequence-tagged molecular inversion probes, Nat. Biotechnol., 2003, 21, 673–678 CrossRef CAS PubMed.
- Y. Huang, Y.-L. Zhang, X. Xu, J.-H. Jiang, G.-L. Shen and R.-Q. Yu, Highly Specific and Sensitive Electrochemical Genotyping via Gap Ligation Reaction and Surface Hybridization Detection, J. Am. Chem. Soc., 2009, 131, 2478–2480 CrossRef CAS.
- J. Kim, J. K. Ahn, J. S. Kim, B. R. Choi, J. Cho and H. Lee, Highly selective detection of single nucleotide polymorphism (SNP) using a dumbbell DNA probe with a gap-filling approach, J. Ind. Eng. Chem., 2020, 88, 78–83 CrossRef CAS.
- Y. Wang, M. Cottman and J. D. Schiffman, Molecular inversion probes: a novel microarray technology and its application in cancer research, Cancer Genet., 2012, 205, 341–355 CrossRef CAS PubMed.
- G. W. Shin, B. Chung, G. Y. Jung and G. Y. Jung, Multiplex ligase-based genotyping methods combined with CE, Electrophoresis, 2014, 35, 1004–1016 CrossRef CAS PubMed.
- M. Mignardi, A. Mezger, X. Qian, L. La Fleur, J. Botling, C. Larsson and M. Nilsson, Oligonucleotide gap-fill ligation for mutation detection and sequencing in situ, Nucleic Acids Res., 2015, 43, e151 CrossRef PubMed.
- A. Pingoud, Structure and function of type II restriction endonucleases, Nucleic Acids Res., 2001, 29, 3705–3727 CrossRef CAS.
- J. M. Parry, M. Shamsher and D. O. F. Skibinski, Restriction site mutation analysis, a proposed methodology for the detection and study of DNA base changes following mutagen exposure, Mutagenesis, 1990, 5, 209–212 CrossRef CAS.
- M. S. Sandy, S. M. Chiocca and P. A. Cerutti, Genotypic Analysis of Mutations in Taq I Restriction Recognition Sites by Restriction Fragment Length Polymorphism/Polymerase Chain Reaction, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 890–894 CrossRef CAS.
- R. Ward, N. Hawkins, R. O’Grady, C. Sheehan, T. O’Connor, H. Impey, N. Roberts, C. Fuery and A. Todd, Restriction Endonuclease-Mediated Selective Polymerase Chain Reaction, Am. J. Pathol., 1998, 153, 373–379 CrossRef CAS.
- G. Amicarelli, E. Shehi, G. M. Makrigiorgos and D. Adlerstein, FLAG assay as a novel method for real-time signal generation during PCR: application to detection and genotyping of KRAS codon 12 mutations, Nucleic Acids Res., 2007, 35, e131 CrossRef.
- A. Haliassos, J. C. Chomel, S. Grandjouan, J. Kruh, J. C. Kaplan and A. Kitzis, Detection of minority point mutations by modified PCR technique: a new approach for a sensitive diagnosis of tumor-progression markers, Nucleic Acids Res., 1989, 17, 8093–8099 CrossRef CAS.
- M. Kaur, Ligation of a primer at a mutation: a method to detect low level mutations in DNA, Mutagenesis, 2002, 17, 365–374 CrossRef CAS.
- I. Ivanov, J. A. Tainer and J. A. McCammon, Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 1465–1470 Search PubMed.
- X. Xiao, Y. Liu and M. Zhao, Endonuclease IV discriminates mismatches next to the apurinic/apyrimidinic site in DNA strands: constructing DNA sensing platforms with extremely high selectivity, Chem. Commun., 2013, 49, 2819–2821 Search PubMed.
- C. Song, Y. Liu, R. Fontana, A. Makrigiorgos, H. Mamon, M. H. Kulke and G. M. Makrigiorgos, Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment, Nucleic Acids Res., 2016, 19, e146 Search PubMed.
- I. Ladas, M. Fitarelli-Kiehl, C. Song, V. A. Adalsteinsson, H. A. Parsons, N. U. Lin, N. Wagle and G. M. Makrigiorgos, Multiplexed Elimination of Wild-Type DNA and High-Resolution Melting Prior to Targeted Resequencing of Liquid Biopsies, Clin. Chem., 2017, 63, 1605–1613 CrossRef CAS PubMed.
- J. R. Dobosy, S. D. Rose, K. R. Beltz, S. M. Rupp, K. M. Powers, M. A. Behlke and J. A. Walder, RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers, BMC Biotechnol., 2011, 11, 80 CrossRef CAS PubMed.
- SNP Assays | IDT, https://sg.idtdna.com/pages/products/qpcr-and-pcr/genotyping/rhamp-snp-genotyping/rhamp-snp-assays, (accessed 26 July 2025).
- K. Yehl, A. Mugler, S. Vivek, Y. Liu, Y. Zhang, M. Fan, E. R. Weeks and K. Salaita, High-speed DNA-based rolling motors powered by RNase H, Nat. Nanotechnol., 2016, 11, 184–190 Search PubMed.
- T. Wu, X. Xiao, Z. Zhang and M. Zhao, Enzyme-mediated single-nucleotide variation detection at room temperature with high discrimination factor, Chem. Sci., 2015, 6, 1206–1211, 10.1039/c4sc03375b.
- Y. Yu, L. Ma, L. Li, Y. Deng, L. Xu, H. Liu, L. Xiao and X. Su, Digestion of Dynamic Substrate by Exonuclease Reveals High Single-Mismatch Selectivity, Anal. Chem., 2018, 90, 13655–13662 CrossRef CAS.
- T. Wu, W. Chen, Z. Yang, H. Tan, J. Wang, X. Xiao, M. Li and M. Zhao, DNA terminal structure-mediated enzymatic reaction for ultra-sensitive discrimination of single nucleotide variations in circulating cell-free DNA, Nucleic Acids Res., 2018, 46, e24 CrossRef CAS PubMed.
- T. Wu, Y. Yang, W. Chen, J. Wang, Z. Yang, S. Wang, X. Xiao, M. Li and M. Zhao, Noncanonical substrate preference of lambda exonuclease for 5′-nonphosphate-ended dsDNA and a mismatch-induced acceleration effect on the enzymatic reaction, Nucleic Acids Res., 2018, 46, 3119–3129 CrossRef CAS.
- Z. Yang, W. Chen, J. Wang, M. Shi, R. Zhang, S. Dai, T. Wu and M. Zhao, Programmable One-Pot Enzymatic Reaction for Direct Fluorescence Detection of Ultralow-Abundance Mutations in the DNA Duplex, Anal. Chem., 2021, 93, 7086–7093 CrossRef CAS PubMed.
- S. Fu, J. Li, J. Chen, L. Zhang, J. Liu, H. Liu and X. Su, Bacteriophage λ exonuclease and a 5′-phosphorylated DNA guide allow PAM-independent targeting of double-stranded nucleic acids, Nat. Biotechnol., 2025, 43, 1144–1155 CrossRef CAS.
- X. Xiao, T. Wu, F. Gu and M. Zhao, Generation of artificial sequence-specific nucleases via a preassembled inert-template, Chem. Sci., 2016, 7, 2051–2057 Search PubMed.
- W. Chen, H. Xu, S. Dai, J. Wang, Z. Yang, Y. Jin, M. Zou, X. Xiao, T. Wu and W. Yan, et al., Detection of low-frequency mutations in clinical samples by increasing mutation abundance via the excision of wild-type sequences, Nat. Biomed. Eng., 2023, 7, 1602–1613 CrossRef CAS PubMed.
- M. R. O’Connell, B. L. Oakes, S. H. Sternberg, A. East-Seletsky, M. Kaplan and J. A. Doudna, Programmable RNA recognition and cleavage by CRISPR/Cas9, Nature, 2014, 516, 263–266 CrossRef.
- F. Zhang, Development of CRISPR-Cas systems for genome editing and beyond, Q. Rev. Biophys., 2019, 52, e6 CrossRef.
- J. Y. Wang and J. A. Doudna, CRISPR technology: A decade of genome editing is only the beginning, Science, 2023, 379, eadd8643 CrossRef CAS.
- M. Pacesa, O. Pelea and M. Jinek, Past, present, and future of CRISPR genome editing technologies, Cell, 2024, 187, 1076–1100 CrossRef CAS.
- K. Pardee, A. A. Green, M. K. Takahashi, D. Braff, G. Lambert, J. W. Lee, T. Ferrante, D. Ma, N. Donghia and M. Fan, et al., Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components, Cell, 2016, 165, 1255–1266 CrossRef CAS PubMed.
- P. D. Hsu, D. A. Scott, J. A. Weinstein, F. A. Ran, S. Konermann, V. Agarwala, Y. Li, E. J. Fine, X. Wu and O. Shalem, et al., DNA targeting specificity of RNA-guided Cas9 nucleases, Nat. Biotechnol., 2013, 31, 827–832 CrossRef CAS.
- C. Anders, O. Niewoehner, A. Duerst and M. Jinek, Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease, Nature, 2014, 513, 569–573 CrossRef CAS.
- B. P. Kleinstiver, M. S. Prew, S. Q. Tsai, V. V. Topkar, N. T. Nguyen, Z. Zheng, A. P. W. Gonzales, Z. Li, R. T. Peterson and J.-R. J. Yeh, et al., Engineered CRISPR-Cas9 nucleases with altered PAM specificities, Nature, 2015, 523, 481–485 CrossRef.
- L. Gao, D. B. T. Cox, W. X. Yan, J. C. Manteiga, M. W. Schneider, T. Yamano, H. Nishimasu, O. Nureki, N. Crosetto and F. Zhang, Engineered Cpf1 variants with altered PAM specificities, Nat. Biotechnol., 2017, 35, 789–792 CrossRef CAS.
- S. H. Lee, J. Yu, G.-H. Hwang, S. Kim, H. S. Kim, S. Ye, K. Kim, J. Park, D. Y. Park and Y.-K. Cho, et al., CUT-PCR: CRISPR-mediated, ultrasensitive detection of target DNA using PCR, Oncogene, 2017, 36, 6823–6829 CrossRef CAS.
- T. Wang, Y. Liu, H. Sun, B. Yin and B. Ye, An RNA-Guided Cas9 Nickase-Based Method for Universal Isothermal DNA Amplification, Angew. Chem., Int. Ed., 2019, 58, 5382–5386 CrossRef CAS PubMed.
- M. Huang, X. Zhou, H. Wang and D. Xing, Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection, Anal. Chem., 2018, 90, 2193–2200 CrossRef CAS PubMed.
- W. Zhou, L. Hu, L. Ying, Z. Zhao, P. K. Chu and X.-F. Yu, A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection, Nat. Commun., 2018, 9, 5012 CrossRef PubMed.
- D. Singh, S. H. Sternberg, J. Fei, J. A. Doudna and T. Ha, Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9, Nat. Commun., 2016, 7, 12778 CrossRef CAS PubMed.
- M. Guo, K. Ren, Y. Zhu, Z. Tang, Y. Wang, B. Zhang and Z. Huang, Structural insights into a high fidelity variant of SpCas9, Cell Res., 2019, 29, 183–192 CrossRef CAS PubMed.
- M. G. Mohsen and E. T. Kool, The Discovery of Rolling Circle Amplification and Rolling Circle Transcription, Acc. Chem. Res., 2016, 49, 2540–2550 CrossRef CAS PubMed.
- J. Van Ness, L. K. Van Ness and D. J. Galas, Isothermal reactions for the amplification of oligonucleotides, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4504–4509 CrossRef CAS PubMed.
- Y. Fu, J. D. Sander, D. Reyon, V. M. Cascio and J. K. Joung, Improving CRISPR-Cas nuclease specificity using truncated guide RNAs, Nat. Biotechnol., 2014, 32, 279–284 CrossRef CAS PubMed.
- Y. S. Dagdas, J. S. Chen, S. H. Sternberg, J. A. Doudna and A. Yildiz, A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9, Sci. Adv., 2017, 3, eaao0027 CrossRef PubMed.
- M. Rahdar, M. A. McMahon, T. P. Prakash, E. E. Swayze, C. F. Bennett and D. W. Cleveland, Synthetic CRISPR RNA-Cas9–guided genome editing in human cells, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E7110–E7117 CrossRef CAS.
- D. D. Kocak, E. A. Josephs, V. Bhandarkar, S. S. Adkar, J. B. Kwon and C. A. Gersbach, Increasing the specificity of CRISPR systems with engineered RNA secondary structures, Nat. Biotechnol., 2019, 37, 657–666 CrossRef CAS.
- J. S. Chen, Y. S. Dagdas, B. P. Kleinstiver, M. M. Welch, A. A. Sousa, L. B. Harrington, S. H. Sternberg, J. K. Joung, A. Yildiz and J. A. Doudna, Enhanced proofreading governs CRISPR–Cas9 targeting accuracy, Nature, 2017, 550, 407–410 CrossRef CAS.
- H. Hirano, J. S. Gootenberg, T. Horii, O. O. Abudayyeh, M. Kimura, P. D. Hsu, T. Nakane, R. Ishitani, I. Hatada and F. Zhang, et al., Structure and Engineering of Francisella novicida Cas9, Cell, 2016, 164, 950–961 CrossRef CAS PubMed.
- S. Acharya, A. Mishra, D. Paul, A. H. Ansari, M. Azhar, M. Kumar, R. Rauthan, N. Sharma, M. Aich and D. Sinha, et al., Francisella novicida Cas9 interrogates genomic DNA with very high specificity and can be used for mammalian genome editing, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 20959–20968 CrossRef CAS.
- S. Acharya, A. H. Ansari, P. Kumar Das, S. Hirano, M. Aich, R. Rauthan, S. Mahato, S. Maddileti, S. Sarkar and M. Kumar, et al., PAM-flexible Engineered FnCas9 variants for robust and ultra-precise genome editing and diagnostics, Nat. Commun., 2024, 15, 5471 CrossRef CAS.
- M. Azhar, R. Phutela, M. Kumar, A. H. Ansari, R. Rauthan, S. Gulati, N. Sharma, D. Sinha, S. Sharma and S. Singh, et al., Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis, Biosens. Bioelectron., 2021, 183, 113207 CrossRef CAS PubMed.
- A. C. Komor, Y. B. Kim, M. S. Packer, J. A. Zuris and D. R. Liu, Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage, Nature, 2016, 533, 420–424 CrossRef CAS PubMed.
- Y. B. Kim, A. C. Komor, J. M. Levy, M. S. Packer, K. T. Zhao and D. R. Liu, Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions, Nat. Biotechnol., 2017, 35, 371–376 CrossRef CAS PubMed.
- N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson and D. R. Liu, Programmable base editing of A T to G C in genomic DNA without DNA cleavage, Nature, 2017, 551, 464–471 CrossRef CAS.
- A. V. Anzalone, P. B. Randolph, J. R. Davis, A. A. Sousa, L. W. Koblan, J. M. Levy, P. J. Chen, C. Wilson, G. A. Newby and A. Raguram, et al., Search-and-replace genome editing without double-strand breaks or donor DNA, Nature, 2019, 576, 149–157 CrossRef CAS.
- W. Zhou, L. Hu, L. Ying, Z. Zhao, P. K. Chu and X.-F. Yu, A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection, Nat. Commun., 2018, 9, 5012 CrossRef PubMed.
- T. Wang, Y. Liu, H. Sun, B. Yin and B. Ye, An RNA-Guided Cas9 Nickase-Based Method for Universal Isothermal DNA Amplification, Angew. Chem., Int. Ed., 2019, 58, 5382–5386 CrossRef CAS PubMed.
- T. Liang, X. Qin, Y. Zhang, Y. Yang, Y. Chen, L. Yuan, F. Liu, Z. Chen, X. Li and F. Yang, CRISPR/dCas9-Mediated Specific Molecular Assembly Facilitates Genotyping of Mutant Circulating Tumor DNA, Anal. Chem., 2023, 95, 16305–16314 CrossRef CAS.
- L. Wu, T. Zhou and R. Huang, A universal CRISPR/Cas9-based electrochemiluminescence probe for sensitive and single-base-specific DNA detection, Sens. Actuators B Chem., 2022, 357, 131411 CrossRef CAS.
- S. H. Im, A. I. Robby, H. Choi, J. Y. Chung, Y. S. Kim, S. Y. Park and H. J. Chung, A Wireless, CRISPR-Polymer Dot Electrochemical Sensor for the Diagnosis of Bacterial Pneumonia and Multi-Drug Resistance, ACS Appl. Mater. Interfaces, 2024, 16, 5637–5647 CrossRef CAS.
- Z. O. Uygun, L. Yeniay and F. G
![[i with combining dot above]](https://www.rsc.org/images/entities/char_0069_0307.gif) rg
rg![[i with combining dot above]](https://www.rsc.org/images/entities/char_0069_0307.gif) n Sağın, CRISPR-dCas9 powered impedimetric biosensor for label-free detection of circulating tumor DNAs, Anal. Chim. Acta, 2020, 1121, 35–41 CrossRef CAS PubMed.
n Sağın, CRISPR-dCas9 powered impedimetric biosensor for label-free detection of circulating tumor DNAs, Anal. Chim. Acta, 2020, 1121, 35–41 CrossRef CAS PubMed. - L. Wu, T. Zhou and R. Huang, A universal CRISPR/Cas9-based electrochemiluminescence probe for sensitive and single-base-specific DNA detection, Sens. Actuators B Chem., 2022, 357, 131411 CrossRef CAS.
- Z. O. Uygun, L. Yeniay and F. G
![[i with combining dot above]](https://www.rsc.org/images/entities/char_0069_0307.gif) rg
rg![[i with combining dot above]](https://www.rsc.org/images/entities/char_0069_0307.gif) n Sağın, CRISPR-dCas9 powered impedimetric biosensor for label-free detection of circulating tumor DNAs, Anal. Chim. Acta, 2020, 1121, 35–41 CrossRef CAS.
n Sağın, CRISPR-dCas9 powered impedimetric biosensor for label-free detection of circulating tumor DNAs, Anal. Chim. Acta, 2020, 1121, 35–41 CrossRef CAS. - A. Su, Y. Liu, W. Sun, C. Liang, W. Xu, A. Rodger, J. Piper, Y. Wang and S. Xu, Silver Nanoparticles with Dual-Recognition via CRISPR/dCas9 for SERS Identification of Two KRAS Mutations in Nucleic Acid Targets, ACS Appl. Nano Mater., 2024, 7, 9800–9808 CrossRef CAS.
- A. Su, Y. Liu, W. Sun, C. Liang, W. Xu, A. Rodger, J. Piper, Y. Wang and S. Xu, Silver Nanoparticles with Dual-Recognition via CRISPR/dCas9 for SERS Identification of Two KRAS Mutations in Nucleic Acid Targets, ACS Appl. Nano Mater., 2024, 7, 9800–9808 CrossRef CAS.
- H. Kim, S. Lee, H. W. Seo, B. Kang, J. Moon, K. G. Lee, D. Yong, H. Kang, J. Jung and E.-K. Lim, et al., Clustered Regularly Interspaced Short Palindromic Repeats-Mediated Surface-Enhanced Raman Scattering Assay for Multidrug-Resistant Bacteria, ACS Nano, 2020, 14, 17241–17253 CrossRef CAS PubMed.
- F. Zheng, Z. Chen, J. Li, R. Wu, B. Zhang, G. Nie, Z. Xie and H. Zhang, A Highly Sensitive CRISPR-Empowered Surface Plasmon Resonance Sensor for Diagnosis of Inherited Diseases with Femtomolar-Level Real-Time Quantification, Adv. Sci., 2022, 9, 2105231 CrossRef CAS PubMed.
- B. M. Downs and S. Sukumar, Capturing ctDNA from Unaltered Stationary and Flowing Plasma with dCas9, ACS Appl. Mater. Interfaces, 2022, 14, 24113–24121 CrossRef CAS PubMed.
- S. Balderston, J. J. Taulbee, E. Celaya, K. Fung, A. Jiao, K. Smith, R. Hajian, G. Gasiunas, S. Kutanovas and D. Kim, et al., Discrimination of single-point mutations in unamplified genomic DNA via Cas9 immobilized on a graphene field-effect transistor, Nat. Biomed. Eng., 2021, 5, 713–725 CrossRef CAS PubMed.
- M. R. O’Connell, B. L. Oakes, S. H. Sternberg, A. East-Seletsky, M. Kaplan and J. A. Doudna, Programmable RNA recognition and cleavage by CRISPR/Cas9, Nature, 2014, 516, 263–266 CrossRef.
- D. A. Nelles, M. Y. Fang, M. R. O’Connell, J. L. Xu, S. J. Markmiller, J. A. Doudna and G. W. Yeo, Programmable RNA Tracking in Live Cells with CRISPR/Cas9, Cell, 2016, 165, 488–496 CrossRef CAS PubMed.
- J. Moon, H.-J. Kwon, D. Yong, I.-C. Lee, H. Kim, H. Kang, E.-K. Lim, K.-S. Lee, J. Jung and H. G. Park, et al., Colorimetric Detection of SARS-CoV-2 and Drug-Resistant pH1N1 Using CRISPR/dCas9, ACS Sens., 2020, 5, 4017–4026 CrossRef CAS.
- W. Y. Wu, B. Adiego-Pérez and J. van der Oost, Biology and applications of CRISPR–Cas12 and transposon-associated homologs, Nat. Biotechnol., 2024, 42, 1807–1821 CrossRef CAS PubMed.
- S.-Y. Li, Q.-X. Cheng, J.-K. Liu, X.-Q. Nie, G.-P. Zhao and J. Wang, CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA, Cell Res., 2018, 28, 491–493 CrossRef CAS.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity, Science, 2018, 360, 436–439 CrossRef CAS.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6, Science, 2018, 360, 439–444 CrossRef CAS PubMed.
- L. Li, S. Li, N. Wu, J. Wu, G. Wang, G. Zhao and J. Wang, HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation, ACS Synth. Biol., 2019, 8, 2228–2237 CrossRef CAS.
- D. C. Swarts, J. Van Der Oost and M. Jinek, Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a, Mol. Cell, 2017, 66, 221–233 CrossRef CAS PubMed.
- E. Tóth, É. Varga, P. I. Kulcsár, V. Kocsis-Jutka, S. L. Krausz, A. Nyeste, Z. Welker, K. Huszár, Z. Ligeti and A. Tálas, et al., Improved LbCas12a variants with altered PAM specificities further broaden the genome targeting range of Cas12a nucleases, Nucleic Acids Res., 2020, 48, 3722–3733 CrossRef.
- M. H. Tran, H. Park, C. L. Nobles, P. Karunadharma, L. Pan, G. Zhong, H. Wang, W. He, T. Ou and G. Crynen, et al., A more efficient CRISPR-Cas12a variant derived from Lachnospiraceae bacterium MA2020, Mol. Ther. - Nucleic Acids, 2021, 24, 40–53 CrossRef CAS.
- S.-Y. Li, Q.-X. Cheng, J.-M. Wang, X.-Y. Li, Z.-L. Zhang, S. Gao, R.-B. Cao, G.-P. Zhao and J. Wang, CRISPR-Cas12a-assisted nucleic acid detection, Cell Discovery, 2018, 4, 20 CrossRef PubMed.
- M.-C. Marqués, R. Ruiz, R. Montagud-Martínez, R. Márquez-Costa, S. Albert, P. Domingo-Calap and G. Rodrigo, CRISPR-Cas12a-Based Detection of SARS-CoV-2 Harboring the E484K Mutation, ACS Synth. Biol., 2021, 10, 3595–3599 CrossRef.
- B. Ning, B. M. Youngquist, D. D. Li, C. J. Lyon, A. Zelazny, N. J. Maness, D. Tian and T. Y. Hu, Rapid detection of multiple SARS-CoV-2 variants of concern by PAM-targeting mutations, Cell Rep. Methods, 2022, 2, 100173 CrossRef CAS PubMed.
- I. Fonfara, H. Richter, M. Bratovič, A. Le Rhun and E. Charpentier, The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA, Nature, 2016, 532, 517–521 CrossRef CAS PubMed.
- M. Klein, B. Eslami-Mossallam, D. G. Arroyo and M. Depken, Hybridization Kinetics Explains CRISPR-Cas Off-Targeting Rules, Cell Rep., 2018, 22, 1413–1423 CrossRef CAS.
- I. Strohkendl, F. A. Saifuddin, J. R. Rybarski, I. J. Finkelstein and R. Russell, Kinetic Basis for DNA Target Specificity of CRISPR-Cas12a, Mol. Cell, 2018, 71, 816–824 CrossRef CAS.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity, Science, 2012, 337, 816–821 CrossRef CAS.
- Y. He, S. Shao and J. Chen, High-Fidelity Identification of Single Nucleotide Polymorphism by Type V CRISPR Systems, ACS Sens., 2023, 8, 4478–4483 CrossRef CAS.
- K. A. V. Kohabir, J. Linthorst, L. O. Nooi, R. Brouwer, R. M. F. Wolthuis and E. A. Sistermans, Synthetic mismatches enable specific CRISPR-Cas12a-based detection of genome-wide SNVs tracked by ARTEMIS, Cell Rep. Methods, 2024, 4, 100912 CrossRef CAS.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer and C. A. Freije, et al., Nucleic acid detection with CRISPR-Cas13a/C2c2, Science, 2017, 356, 438–442 CrossRef CAS PubMed.
- X. Huang, F. Zhang, K. Zhu, W. Lin and W. Ma, dsmCRISPR: Dual synthetic mismatches CRISPR/Cas12a-based detection of SARS-CoV-2 D614G mutation, Virus Res., 2021, 304, 198530 CrossRef CAS PubMed.
- C. Zhang, Z. Cai, Z. Zhou, M. Li, W. Hong, W. Zhou, D. Yu, P. Wei, J. He and Y. Wang, et al., CASMART, a one-step CRISPR Cas12a-mediated isothermal amplification for rapid and high-resolution digital detection of rare mutant alleles, Biosens. Bioelectron., 2023, 222, 114956 CrossRef CAS PubMed.
- X. Wang, T. Yang, Y. Zhang, Z. Zeng, Q. Wei, P. Chen, S. Yang, Y. Huang, Y. Zhang and H. Lu, et al., Optimization and Clinical Application Potential of Single Nucleotide Polymorphism Detection Method Based on CRISPR/Cas12a and Recombinase Polymerase Amplification, Anal. Chem., 2024, 96, 17567–17575 CrossRef CAS.
- J. Ai, J. Deng, J. Hu, X. Pu, T. Yuan, Y. Teng, H. Li, B. Chen, J. Du and L. Jiang, et al., PAM-Independent CRISPR-Cas12a System for Specific Assays of Single Nucleotide Variants, JACS Au, 2025, 5, 1392–1401 CrossRef CAS.
- J. Yang, N. Barua, M. N. Rahman, C. Li, N. Lo, K. Y. Yeong, T. F. Tsang, X. Yang, Y.-Y. Cheung and A. K. L. Tsang, et al., Rapid SARS-CoV-2 Variants Enzymatic Detection (SAVED) by CRISPR-Cas12a, Microbiol. Spectr., 2022, 10, e03260-22 CrossRef PubMed.
- P. Gao, M. Yang, Y. Chen, J. Yan, M. Han, H. Deng, K. Qian, J. Yang, Y. Lu and L. Zhou, et al., A spacer design strategy for CRISPR-Cas12f1 with single-nucleotide polymorphism mutation resolution capability and its application in the mutations diagnosis of pathogens, J. Med. Virol., 2023, 95, e29189 CrossRef CAS.
- S. Chen, C. Wu, C. Qian, Y. Pang, K. Guo, T. Wang, L. Bai, F. Qian, Z. Ye and Z. Liu, et al., Ultraspecific One-Pot CRISPR-Based “Green-Yellow-Red” Multiplex Detection Strategy Integrated with Portable Cartridge for Point-of-Care Diagnosis, Anal. Chem., 2024, 96, 3145–3152 CAS.
- S. C. A. Creutzburg, W. Y. Wu, P. Mohanraju, T. Swartjes, F. Alkan, J. Gorodkin, R. H. J. Staals and J. van der Oost, Good guide, bad guide: spacer sequence-dependent cleavage efficiency of Cas12a, Nucleic Acids Res., 2020, 48, 3228–3243 CrossRef CAS PubMed.
- Y. Ke, B. Ghalandari, S. Huang, S. Li, C. Huang, X. Zhi, D. Cui and X. Ding, 2′-O-Methyl modified guide RNA promotes the single nucleotide polymorphism (SNP) discrimination ability of CRISPR–Cas12a systems, Chem. Sci., 2022, 13, 2050–2061 RSC.
- W. Zhang, R. Shi, K. Dong, H. Hu, W. Shu, Y. Mu, B. Yan, L. Li, X. Xiao and H. Wang, The Off-Target Effect of CRISPR-Cas12a System toward Insertions and Deletions between Target DNA and crRNA Sequences, Anal. Chem., 2022, 94, 8596–8604 CrossRef CAS.
- H. Lee Yu, Y. Cao, X. Lu and I.-M. Hsing, Detection of rare variant alleles using the AsCas12a double-stranded DNA trans-cleavage activity, Biosens. Bioelectron., 2021, 189, 113382 CrossRef CAS PubMed.
- Q. Liu, Z. Jiang, S. Li, Y. Li, Y. Wan, Z. Hu, S. Ma, Z. Zou and R. Yang, Nonequilibrium hybridization-driven CRISPR/Cas adapter with extended energetic penalty for discrimination of single-nucleotide variants, Nucleic Acids Res., 2025, 53, gkaf287 CrossRef.
- W. Ren, M. Li, X. Liu, W. You, Q. You, B. Li, H. Ye and R. Zhang, Specific detection of DNA and RNA by the CRISPR-Cas12a system containing spacer split crRNA, Anal. Chim. Acta, 2025, 1367, 344204 CrossRef CAS.
- L. T. Nguyen, B. M. Smith and P. K. Jain, Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection, Nat. Commun., 2020, 11, 4906 CrossRef CAS PubMed.
- H. Kim, W. Lee, Y. Oh, S.-H. Kang, J. K. Hur, H. Lee, W. Song, K.-S. Lim, Y.-H. Park and B.-S. Song, et al., Enhancement of target specificity of CRISPR–Cas12a by using a chimeric DNA–RNA guide, Nucleic Acids Res., 2020, 48, 8601–8616 CrossRef CAS PubMed.
- J. Yang, N. Barua, M. N. Rahman, N. Lo, T. F. Tsang, X. Yang, P. K. S. Chan, L. Zhang and M. Ip, Chimeric crRNA improves CRISPR–Cas12a specificity in the N501Y mutation detection of Alpha, Beta, Gamma, and Mu variants of SARS-CoV-2, PLoS One, 2021, 16, e0261778 CrossRef CAS PubMed.
- H. Kim, W. Lee, C. H. Kim, Y. Oh, L. W. Gwon, H. Lee, W. Song, J. K. Hur, K.-S. Lim and K. J. Jeong, et al., Highly specific chimeric DNA-RNA-guided genome editing with enhanced CRISPR-Cas12a system, Mol. Ther. - Nucleic Acids, 2022, 28, 353–362 CrossRef CAS PubMed.
- K. Shi, S. Xie, R. Tian, S. Wang, Q. Lu, D. Gao, C. Lei, H. Zhu and Z. Nie, A CRISPR-Cas autocatalysis-driven feedback amplification network for supersensitive DNA diagnostics, Sci. Adv., 2021, 7, eabc7802 CrossRef CAS.
- F. Deng, Y. Li, B. Yang, R. Sang, W. Deng, M. Kansara, F. Lin, S. Thavaneswaran, D. M. Thomas and E. M. Goldys, Topological barrier to Cas12a activation by circular DNA nanostructures facilitates autocatalysis and transforms DNA/RNA sensing, Nat. Commun., 2024, 15, 1818 CrossRef CAS PubMed.
- W. Zhang, Y. Mu, K. Dong, L. Zhang, B. Yan, H. Hu, Y. Liao, R. Zhao, W. Shu and Z. Ye, et al., PAM-independent ultra-specific activation of CRISPR-Cas12a via sticky-end dsDNA, Nucleic Acids Res., 2022, 50, 12674–12688 CrossRef CAS.
- Y. Li, F. Li, R. Li, Y. Zhu, Y. Lin, Y. Zhang, D. Sun and Y. Yu, Single nucleotide polymorphism discrimination and genotyping based on cascade strand displacement reaction mediated label-free Cas12a system, Sens. Actuators B Chem., 2025, 423, 136832 CrossRef CAS.
- D. S. S. Marpaung, S. S. Jiang, W.-T. Fang, Y.-C. Liao and M.-C. Chuang, Activated single nucleotide mismatch determination through toehold-embedded hairpin-mediated strand displacement reaction alongside CRISPR-Cas12a for detection of TP53 point mutation, Sens. Actuators B Chem., 2025, 423, 136751 CrossRef CAS.
- J. Mo, Y. Zhao, C. Sun, M. Li, H. Chen, Y. Zhang, X. Chen, X. Guo, Z. Wang and Y. Wang, et al., Strand displacement-driven CRISPR/Cas12a framework enables precise identification of low-abundance point mutations, View, 2025, 20250047 CrossRef CAS.
- Y. Wu, W. Luo, Z. Weng, Y. Guo, H. Yu, R. Zhao, L. Zhang, J. Zhao, D. Bai and X. Zhou, et al., A PAM-free CRISPR/Cas12a ultra-specific activation mode based on toehold-mediated strand displacement and branch migration, Nucleic Acids Res., 2022, 50, 11727–11737 CrossRef CAS PubMed.
- Y. Zhu, Y. Lin, B. Gong, Y. Zhang, G. Su and Y. Yu, Dual toeholds regulated CRISPR-Cas12a sensing platform for ApoE single nucleotide polymorphisms genotyping, Biosens. Bioelectron., 2024, 255, 116255 CrossRef CAS.
- C. L. Fasching, V. Servellita, B. McKay, V. Nagesh, J. P. Broughton, A. Sotomayor-Gonzalez, B. Wang, N. Brazer, K. Reyes and J. Streithorst, et al., COVID-19 Variant Detection with a High-Fidelity CRISPR-Cas12 Enzyme, J. Clin. Microbiol., 2022, 60, e00261-22 CrossRef.
- L. B. Harrington, D. Burstein, J. S. Chen, D. Paez-Espino, E. Ma, I. P. Witte, J. C. Cofsky, N. C. Kyrpides, J. F. Banfield and J. A. Doudna, Programmed DNA destruction by miniature CRISPR-Cas14 enzymes, Science, 2018, 362, 839–842 CrossRef CAS PubMed.
- G. T. Nguyen, M. A. Schelling, A. Raju, K. A. Buscher, A. Sritharan and D. G. Sashital, CRISPR-Cas12a exhibits metal-dependent specificity switching, Nucleic Acids Res., 2024, 52, 9343–9359 CrossRef CAS PubMed.
- H. Son, J. Park, I. Hwang, Y. Jung, S. Bae and S. Lee, Mg2+-dependent conformational rearrangements of CRISPR-Cas12a R-loop complex are mandatory for complete double-stranded DNA cleavage, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2113747118 CrossRef CAS.
- R. Sundaresan, H. P. Parameshwaran, S. D. Yogesha, M. W. Keilbarth and R. Rajan, RNA-Independent DNA Cleavage Activities of Cas9 and Cas12a, Cell Rep., 2017, 21, 3728–3739 CrossRef CAS PubMed.
- B. Li, J. Yan, Y. Zhang, W. Li, C. Zeng, W. Zhao, X. Hou, C. Zhang and Y. Dong, CRISPR-Cas12a Possesses Unconventional DNase Activity that Can Be Inactivated by Synthetic Oligonucleotides, Mol. Ther. Nucleic Acids, 2020, 19, 1043–1052 CrossRef CAS PubMed.
- Z. Li, W. Zhao, S. Ma, Z. Li, Y. Yao and T. Fei, A chemical-enhanced system for CRISPR-Based nucleic acid detection, Biosens. Bioelectron., 2021, 192, 113493 CrossRef CAS.
- M. Lin, H. Yue, T. Tian, E. Xiong, D. Zhu, Y. Jiang and X. Zhou, Glycerol Additive Boosts 100-fold Sensitivity Enhancement for One-Pot RPA-CRISPR/Cas12a Assay, Anal. Chem., 2022, 94, 8277–8284 CrossRef CAS PubMed.
- K. Chen, L. Dai, J. Zhao, M. Deng, L. Song, D. Bai, Y. Wu, X. Zhou, Y. Yang and S. Yang, et al., Temperature-boosted PAM-less activation of CRISPR-Cas12a combined with selective inhibitors enhances detection of SNVs with VAFs below 0.01%, Talanta, 2023, 261, 124674 CrossRef CAS.
- C. Blanluet, D. A. Huyke, A. Ramachandran, A. S. Avaro and J. G. Santiago, Detection and Discrimination of Single Nucleotide Polymorphisms by Quantification of CRISPR-Cas Catalytic Efficiency, Anal. Chem., 2022, 94, 15117–15123 CrossRef CAS.
- C. Blanluet, C. J. Kuo, A. Bhattacharya and J. G. Santiago, Design and Evaluation of a Robust CRISPR Kinetic Assay for Hot-Spot Genotyping, Anal. Chem., 2024, 96, 7444–7451 CrossRef CAS.
- H. Wang, R. Liu, K. Dong, L. Zhang, J. Zhang, X. Zhang, J. Zhang, X. Xiao, W. Zhang and X. Wang, A universal and sensitive gene mutation detection method based on CRISPR-Cas12a, Anal. Chim. Acta, 2023, 1246, 340886 CrossRef CAS.
- X. Liu, H. Yang, J. Liu, K. Liu, L. Jin, Y. Zhang, M. R. Khan, K. Zhong, J. Cao and Q. He, et al., In Situ Cas12a-Based Allele-Specific PCR for Imaging Single-Nucleotide Variations in Foodborne Pathogenic Bacteria, Anal. Chem., 2024, 96, 2032–2040 CrossRef CAS.
- Y. Wu, Y. Chang, Y. Sun, Y. Wang, K. Li, Z. Lu, Q. Liu, F. Wang and L. Wei, A multi-AS-PCR-coupled CRISPR/Cas12a assay for the detection of ten single-base mutations, Anal. Chim. Acta, 2024, 1320, 343027 CrossRef CAS PubMed.
- L. Fang, X. Yang, Y. Li, C. Xue, Z. Li, H. Jiang, X. Li, S. Lu, D. Wang and H. He, et al., SPECIAL: Phosphorothioate dNTP assisted RPA equipped with CRISPR/Cas12a amplifier enables high-specific nucleic acid testing, Biosens. Bioelectron., 2025, 279, 117421 CrossRef CAS PubMed.
- H. Yan, Y. Wen, Z. Tian, N. Hart, S. Han, S. J. Hughes and Y. Zeng, A one-pot isothermal Cas12-based assay for the sensitive detection of microRNAs, Nat. Biomed. Eng., 2023, 7, 1583–1601 CrossRef CAS.
- X. Jin, S. Feng, W. Lv, D. Shen and B. Li, Split activator ligation assay: A CRISPR/Cas12a-enabled biosensor for the genotyping of cancer-associated miRNA single-nucleotide polymorphisms, Sens. Actuators B Chem., 2025, 442, 138134 CrossRef CAS.
- G. Lin, J. Li and K. Zhang, Multiple CRISPR zones-driven ultrasensitive detection of DNA via CRISPR-Cas12a and ligation-rolling circle amplification, Talanta, 2025, 295, 128336 CrossRef CAS PubMed.
- J. Arizti-Sanz, A. Bradley, Y. B. Zhang, C. K. Boehm, C. A. Freije, M. E. Grunberg, T.-S. F. Kosoko-Thoroddsen, N. L. Welch, P. P. Pillai and S. Mantena, et al., Simplified Cas13-based assays for the fast identification of SARS-CoV-2 and its variants, Nat. Biomed. Eng., 2022, 6, 932–943 CrossRef CAS.
- Y. Hu, Y. Chen, J. Xu, X. Wang, S. Luo, B. Mao, Q. Zhou and W. Li, Metagenomic discovery of novel CRISPR-Cas13 systems, Cell Discovery, 2022, 8, 107 CrossRef CAS.
- A. Tambe, A. East-Seletsky, G. J. Knott, J. A. Doudna and M. R. O’Connell, RNA Binding and HEPN-Nuclease Activation Are Decoupled in CRISPR-Cas13a, Cell Rep., 2018, 24, 1025–1036 CrossRef CAS PubMed.
- A. M. Molina Vargas, S. Sinha, R. Osborn, P. R. Arantes, A. Patel, S. Dewhurst, D. J. Hardy, A. Cameron, G. Palermo and M. R. O’Connell, New design strategies for ultra-specific CRISPR-Cas13a-based RNA detection with single-nucleotide mismatch sensitivity, Nucleic Acids Res., 2024, 52, 921–939 CrossRef CAS.
- H. Shinoda, Y. Taguchi, R. Nakagawa, A. Makino, S. Okazaki, M. Nakano, Y. Muramoto, C. Takahashi, I. Takahashi and J. Ando, et al., Amplification-free RNA detection with CRISPR–Cas13, Commun. Biol., 2021, 4, 476 CrossRef CAS.
- H. Shinoda, T. Iida, A. Makino, M. Yoshimura, J. Ishikawa, J. Ando, K. Murai, K. Sugiyama, Y. Muramoto and M. Nakano, et al., Automated amplification-free digital RNA detection platform for rapid and sensitive SARS-CoV-2 diagnosis, Commun. Biol., 2022, 5, 473 CrossRef CAS.
- N. L. Welch, M. Zhu, C. Hua, J. Weller, M. E. Mirhashemi, T. G. Nguyen, S. Mantena, M. R. Bauer, B. M. Shaw and C. M. Ackerman, et al., Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and identification of SARS-CoV-2 variants, Nat. Med., 2022, 28, 1083–1094 CrossRef CAS.
- C. Myhrvold, C. A. Freije, J. S. Gootenberg, O. O. Abudayyeh, H. C. Metsky, A. F. Durbin, M. J. Kellner, A. L. Tan, L. M. Paul and L. A. Parham, et al., Field-deployable viral diagnostics using CRISPR-Cas13, Science, 2018, 360, 444–448 CrossRef CAS.
- C. M. Ackerman, C. Myhrvold, S. G. Thakku, C. A. Freije, H. C. Metsky, D. K. Yang, S. H. Ye, C. K. Boehm, T.-S. F. Kosoko-Thoroddsen and J. Kehe, et al., Massively multiplexed nucleic acid detection with Cas13, Nature, 2020, 582, 277–282 CrossRef CAS PubMed.
- W. Chen, H. Luo, L. Zeng, Y. Pan, J. B. Parr, Y. Jiang, C. H. Cunningham, K. L. Hawley, J. D. Radolf and W. Ke, et al., A suite of PCR-LwCas13a assays for detection and genotyping of Treponema pallidum in clinical samples, Nat. Commun., 2022, 13, 4671 CrossRef CAS.
- Y. Wu, Y. Liu, Y. Chang and M. Liu, Integration of CRISPR/Cas13a and V-Shape PCR for Rapid, Sensitive, and Specific Genotyping of CYP2C19 Gene Polymorphisms, Anal. Chem., 2023, 95, 10127–10135 CrossRef CAS PubMed.
- H. Ma, Y. Tian, D. Kong, M. Guo, C. Dai, Q. Wang, S. Li, Z. Tian, Y. Liu and D. Wei, One-base-mismatch CRISPR-based transistors for single nucleotide resolution assay, Biosens. Bioelectron., 2024, 262, 116548 CrossRef CAS PubMed.
- C. Shembrey, R. Yang, J. Casan, W. Hu, H. Chen, G. J. Singh, T. Sadras, K. Prasad, J. Shortt and R. W. Johnstone, et al., Principles of CRISPR-Cas13 mismatch intolerance enable selective silencing of point-mutated oncogenic RNA with single-base precision, Sci. Adv., 2024, 10, eadl0731 CrossRef CAS PubMed.
- X. Wu, S. Luo, C. Guo, Y. Zhao, J. Zhong, R. Hu, X. Yang, C. Liu, Q. Zhang and S. Zhuang, et al., LbuCas13a directly targets DNA and elicits strong trans-cleavage activity, Nat. Biomed. Eng., 2025 DOI:10.1038/s41551-025-01424-6.
- Y. Ke, S. Huang, B. Ghalandari, S. Li, A. R. Warden, J. Dang, L. Kang, Y. Zhang, Y. Wang and Y. Sun, et al., Hairpin-Spacer crRNA-Enhanced CRISPR/Cas13a System Promotes the Specificity of Single Nucleotide Polymorphism (SNP) Identification, Adv. Sci., 2021, 8, 2003611 CrossRef CAS.
- Y. Li, R. Xu, F. Quan, Y. Wu, Y. Wu, Y. Zhang, Y. Liang, Z. Zhang, H. Gao and R. Deng, et al., Topologically constrained DNA-mediated one-pot CRISPR assay for rapid detection of viral RNA with single nucleotide resolution, eBioMedicine, 2025, 112, 105564 CrossRef CAS.
- B. Casati, J. P. Verdi, A. Hempelmann, M. Kittel, A. G. Klaebisch, B. Meister, S. Welker, S. Asthana, S. Di Giorgio and P. Boskovic, et al., Rapid, adaptable and sensitive Cas13-based COVID-19 diagnostics using ADESSO, Nat. Commun., 2022, 13, 3308 CrossRef CAS.
- H. Hu, H. Xue, K. Dong, Y. Li, P. Liu, H. Wang, L. Li, X. Xiao and H. Chen, Strand displacement-enhanced CRISPR-Cas13a system for ultra-specific detection of RNA single nucleotide variation, Biosens. Bioelectron., 2025, 280, 117445 CrossRef CAS PubMed.
- S. Luo, Y. Chen, Z. Li, X. Zhang and Y. Liu, Single-tube Lambda exonuclease-mediated LbuCas13a detect of ssDNA for single-nucleotide polymorphisms genotyping, Biosens. Bioelectron., 2025, 288, 117760 CrossRef CAS.
- J. Zhong, Y. Chen, Y. Dong, C. Guo and Y. Liu, SPARC: An Orthogonal Cas12a/Cas13a Dual-Channel CRISPR Platform for Reliable SNV Identification and Mutation Confirmation, Anal. Chem., 2025, 97, 14629–14636 CrossRef CAS.
- L. Peters and G. Meister, Argonaute Proteins: Mediators of RNA Silencing, Mol. Cell, 2007, 26, 611–623 CrossRef CAS.
- S. Shabalina and E. Koonin, Origins and evolution of eukaryotic RNA interference, Trends Ecol. Evol., 2008, 23, 578–587 CrossRef PubMed.
- A. Kuzmenko, A. Oguienko, D. Esyunina, D. Yudin, M. Petrova, A. Kudinova, O. Maslova, M. Ninova, S. Ryazansky and D. Leach, et al., DNA targeting and interference by a bacterial Argonaute nuclease, Nature, 2020, 587, 632–637 CrossRef CAS PubMed.
- B. Koopal, A. Potocnik, S. K. Mutte, C. Aparicio-Maldonado, S. Lindhoud, J. J. M. Vervoort, S. J. J. Brouns and D. C. Swarts, Short prokaryotic Argonaute systems trigger cell death upon detection of invading DNA, Cell, 2022, 185, 1471–1486 CrossRef CAS PubMed.
- D. C. Swarts, M. M. Jore, E. R. Westra, Y. Zhu, J. H. Janssen, A. P. Snijders, Y. Wang, D. J. Patel, J. Berenguer and S. J. J. Brouns, et al., DNA-guided DNA interference by a prokaryotic Argonaute, Nature, 2014, 507, 258–261 CrossRef CAS PubMed.
- L. Lisitskaya, A. A. Aravin and A. Kulbachinskiy, DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins, Nat. Commun., 2018, 9, 5165 CrossRef PubMed.
- H. Li, F. Zheng, Z. Yang, F. Cun, K. Wu, W. Chen, B. Yang, J. Kong and H. Chen, Trends in the use of argonaute proteins in molecular diagnosis, TrAC Trends Anal. Chem., 2025, 183, 118081 CrossRef CAS.
- R. He, L. Wang, F. Wang, W. Li, Y. Liu, A. Li, Y. Wang, W. Mao, C. Zhai and L. Ma, Pyrococcus furiosus Argonaute-mediated nucleic acid detection, Chem. Commun., 2019, 55, 13219–13222 RSC.
- Q. Liu, X. Guo, G. Xun, Z. Li, Y. Chong, L. Yang, H. Wang, F. Zhang, S. Luo and L. Cui, et al., Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations, Nucleic Acids Res., 2021, 49, e75 CrossRef CAS.
- D. Kong, S. Zhang, M. Guo, S. Li, Q. Wang, J. Gou, Y. Wu, Y. Chen, Y. Yang and C. Dai, et al., Ultra-Fast Single-Nucleotide-Variation Detection Enabled by Argonaute-Mediated Transistor Platform, Adv. Mater., 2024, 36, 2307366 CrossRef CAS.
- C. Wang, M. Liu, Z. Wang, S. Li, Y. Deng and N. He, Point-of-care diagnostics for infectious diseases: From methods to devices, Nano Today, 2021, 37, 101092 CrossRef CAS PubMed.
- G.-R. Han, A. Goncharov, M. Eryilmaz, S. Ye, B. Palanisamy, R. Ghosh, F. Lisi, E. Rogers, D. Guzman and D. Yigci, et al., Machine learning in point-of-care testing: innovations, challenges, and opportunities, Nat. Commun., 2025, 16, 3165 CrossRef CAS PubMed.
- J. Fu, Q. Zhang, S. Liu, D. Puyat, A. Shah, A. Ebrahimimojarad and S. W. Oh, Rapid Amplification and Detection of Single-Stranded Nucleic Acids for Point-of-Care Diagnosis, Small Methods, 2025, 9, 2401733 CrossRef CAS.
- J. M. Hong, H. Lee, N. V. Menon, C. T. Lim, L. P. Lee and C. W. M. Ong, Point-of-care diagnostic tests for tuberculosis disease, Sci. Transl. Med., 2022, 14, eabj4124 CrossRef CAS.
- B. M. Youngquist, J. Saliba, Y. Kim, T. J. Cutro, C. J. Lyon, J. Olivo, N. Ha, J. Fine, R. Colman and C. Vergara, et al., Rapid tuberculosis diagnosis from respiratory or blood samples by a low cost, portable lab-in-tube assay, Sci. Transl. Med., 2025, 17, eadp6411 CrossRef CAS.
- K. Dheda, M. Ruhwald, G. Theron, J. Peter and W. C. Yam, Point-of-care diagnosis of tuberculosis: Past, present and future, Respirology, 2013, 18, 217–232 CrossRef.
- H. Harpaldas, S. Arumugam, C. C. Rodriguez, B. A. Kumar, V. Shi and S. K. Sia, Point-of-care diagnostics: recent developments in a pandemic age, Lab. Chip, 2021, 21, 4517–4548 RSC.
- E. Valera, A. Jankelow, J. Lim, V. Kindratenko, A. Ganguli, K. White, J. Kumar and R. Bashir, COVID-19 Point-of-Care Diagnostics: Present and Future, ACS Nano, 2021, 15, 7899–7906 CrossRef CAS.
- S. Lee, L. Bi, H. Chen, D. Lin, R. Mei, Y. Wu, L. Chen, S.-W. Joo and J. Choo, Recent advances in point-of-care testing of COVID-19, Chem. Soc. Rev., 2023, 52, 8500–8530 RSC.
- T. Mahmoudi, M. de la Guardia and B. Baradaran, Lateral flow assays towards point-of-care cancer detection: A review of current progress and future trends, TrAC Trends Anal. Chem., 2020, 125, 115842 CrossRef CAS.
- L. Syedmoradi, M. L. Norton and K. Omidfar, Point-of-care cancer diagnostic devices: From academic research to clinical translation, Talanta, 2021, 225, 122002 CrossRef CAS PubMed.
- P. Bhardwaj, B. Arora, S. Saxena, S. Singh, P. Palkar, J. S. Goda and R. Banerjee, Paper-based point of care diagnostics for cancer biomarkers, Sens. Diagn., 2024, 3, 504–535 RSC.
- A. R. Meyer and M. A. Gorin, First point-of-care PSA test for prostate cancer detection, Nat. Rev. Urol., 2019, 16, 331–332 CrossRef PubMed.
- COVID-19 cases | WHO COVID-19 dashboard, https://data.who.int/dashboards/covid19/cases, (accessed 2 August 2025).
- Tuberculosis (TB), https://www.who.int/news-room/fact-sheets/detail/tuberculosis, (accessed 2 August 2025).
- D. Mabey, R. W. Peeling, A. Ustianowski and M. D. Perkins, Diagnostics for the developing world, Nat. Rev. Microbiol., 2004, 2, 231–240 CrossRef CAS PubMed.
- K. J. Land, D. I. Boeras, X.-S. Chen, A. R. Ramsay and R. W. Peeling, REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes, Nat. Microbiol., 2019, 4, 46–54 CrossRef CAS.
- J. Arizti-Sanz, C. A. Freije, A. C. Stanton, B. A. Petros, C. K. Boehm, S. Siddiqui, B. M. Shaw, G. Adams, T.-S. F. Kosoko-Thoroddsen and M. E. Kemball, et al., Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2, Nat. Commun., 2020, 11, 5921 CrossRef CAS PubMed.
- B. M. Youngquist, J. Saliba, Y. Kim, T. J. Cutro, C. J. Lyon, J. Olivo, N. Ha, J. Fine, R. Colman and C. Vergara, et al., Rapid tuberculosis diagnosis from respiratory or blood samples by a low cost, portable lab-in-tube assay, Sci. Transl. Med., 2025, 17, eadp6411 CrossRef CAS PubMed.
- K. Pardee, A. A. Green, T. Ferrante, D. E. Cameron, A. DaleyKeyser, P. Yin and J. J. Collins, Paper-Based Synthetic Gene Networks, Cell, 2014, 159, 940–954 CrossRef CAS PubMed.
- M. Karlikow, S. J. R. Da Silva, Y. Guo, S. Cicek, L. Krokovsky, P. Homme, Y. Xiong, T. Xu, M.-A. Calderón-Peláez and S. Camacho-Ortega, et al., Field validation of the performance of paper-based tests for the detection of the Zika and chikungunya viruses in serum samples, Nat. Biomed. Eng., 2022, 6, 246–256 CrossRef CAS PubMed.
- K. Yang, D. N. Schuder, A. K. Ngor and J. C. Chaput, REVEALR-Based Genotyping of SARS-CoV-2 Variants of Concern in Clinical Samples, J. Am. Chem. Soc., 2022, 144, 11685–11692 CrossRef CAS PubMed.
- K. Yang and J. C. Chaput, REVEALR: A Multicomponent XNAzyme-Based Nucleic Acid Detection System for SARS-CoV-2, J. Am. Chem. Soc., 2021, 143, 8957–8961 CrossRef CAS PubMed.
- D. N. Schuder, N. D. Lu and J. C. Chaput, Revealr-Based Diagnostic Panel for Rapid Detection of Acute Respiratory Infections, ACS Synth. Biol., 2024, 13, 4202–4208 CrossRef CAS.
- L. Gao, K. Yi, Y. Tan, C. Guo, D. Zheng, C. Shen and F. Li, Engineering Gene-Specific DNAzymes for Accessible and Multiplexed Nucleic Acid Testing, JACS Au, 2024, 4, 1664–1672 CrossRef CAS.
- J. Pedreira-Rincón, L. Rivas, J. Comenge, V. Skouridou, D. Camprubí-Ferrer, J. Muñoz, C. K. O’Sullivan, A. Chamorro-Garcia and C. Parolo, A comprehensive review of competitive lateral flow assays over the past decade, Lab. Chip, 2025, 25, 2578–2608 RSC.
- S. Kakkar, P. Gupta, S. P. Singh Yadav, D. Raj, G. Singh, S. Chauhan, M. K. Mishra, E. Martín-Ortega, S. Chiussi and K. Kant, Lateral flow assays: Progress and evolution of recent trends in point-of-care applications, Mater. Today Bio, 2024, 28, 101188 CrossRef CAS.
- V. Restrepo-Cano, P. García-Huertas, A. Caraballo-Guzmán, M. M. Sánchez-Jiménez and G. Torres-Lindarte, Back to Basics: Unraveling the Fundamentals of Lateral Flow Assays, J. Appl. Lab. Med, 2025, 10, 476–492 CrossRef.
- L. Zhang, L. Xu, J. Zhang, W. Wang, Y. Huang, Y. Zhou, X. Yao, Z. Liu and Y. Ge, A point-of-care single nucleotide variation assay based on strand-displacement-triggered recombinase polymerase amplification, Sens. Actuators B Chem., 2024, 402, 135075 CrossRef CAS.
- M. Lin, Z. Qiu, M. Hao, W. Qi, T. Zhang, Y. Shen, H. Xiao, C. Liang, L. Xie and Y. Jiang, et al., Cas12a cis-cleavage mediated lateral flow assay enables multiplex and ultra-specific nucleic acid detection, Nat. Commun., 2025, 16, 5597 CrossRef.
- W. Peng, Y. Tan, C. Shen, Y. Tang and F. Li, Enabling a universal lateral flow readout for DNA strand displacement via disassembling chemical labels, Chem. Commun., 2023, 59, 8803–8805 RSC.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados and A. Sotomayor-Gonzalez, et al., CRISPR–Cas12-based detection of SARS-CoV-2, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS.
- H. De Puig, I. Bosch, J. J. Collins and L. Gehrke, Point-of-Care Devices to Detect Zika and Other Emerging Viruses, Annu. Rev. Biomed. Eng., 2020, 22, 371–386 CrossRef CAS.
- R. Hu, C. Guo, C. Liu, Q. Zhang, X. Zhang, Y. Chen and Y. Liu, From Lab to Home: Ultrasensitive Rapid Detection of SARS-CoV-2 with a Cascade CRISPR/Cas13a-Cas12a System Based Lateral Flow Assay, Anal. Chem., 2024, 96, 14197–14204 CrossRef CAS PubMed.
- T. Zhang, R. Deng, Y. Wang, C. Wu, K. Zhang, C. Wang, N. Gong, R. Ledesma-Amaro, X. Teng and C. Yang, et al., A paper-based assay for the colorimetric detection of SARS-CoV-2 variants at single-nucleotide resolution, Nat. Biomed. Eng., 2022, 6, 957–967 CrossRef CAS.
- N. Shao, X. Han, Y. Song, P. Zhang and L. Qin, CRISPR-Cas12a Coupled with Platinum Nanoreporter for Visual Quantification of SNVs on a Volumetric Bar-Chart Chip, Anal. Chem., 2019, 91, 12384–12391 CrossRef CAS PubMed.
- C. C. Boehme, M. P. Nicol, P. Nabeta, J. S. Michael, E. Gotuzzo, R. Tahirli, M. T. Gler, R. Blakemore, W. Worodria and C. Gray, et al., Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study, The Lancet, 2011, 377, 1495–1505 CrossRef.
- S. D. Lawn and M. P. Nicol, Xpert® MTB/RIF Assay: Development, Evaluation and Implementation of a New Rapid Molecular Diagnostic for Tuberculosis and Rifampicin Resistance, Future Microbiol., 2011, 6, 1067–1082 CrossRef.
- H. De Puig, R. A. Lee, D. Najjar, X. Tan, L. R. Soenksen, N. M. Angenent-Mari, N. M. Donghia, N. E. Weckman, A. Ory and C. F. Ng, et al., Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants, Sci. Adv., 2021, 7, eabh2944 CrossRef PubMed.
- Y. Liu, Y. Yang, G. Wang, D. Wang, P.-L. Shao, J. Tang, T. He, J. Zheng, R. Hu and Y. Liu, et al., Multiplexed discrimination of SARS-CoV-2 variants via plasmonic-enhanced fluorescence in a portable and automated device, Nat. Biomed. Eng., 2023, 7, 1636–1648 CrossRef CAS.
- B. Zhang, R. B. Kumar, H. Dai and B. J. Feldman, A plasmonic chip for biomarker discovery and diagnosis of type 1 diabetes, Nat. Med., 2014, 20, 948–953 CrossRef CAS PubMed.
- B. Zhang, B. A. Pinsky, J. S. Ananta, S. Zhao, S. Arulkumar, H. Wan, M. K. Sahoo, J. Abeynayake, J. J. Waggoner and C. Hopes, et al., Diagnosis of Zika virus infection on a nanotechnology platform, Nat. Med., 2017, 23, 548–550 CrossRef CAS.
- T. Liu, J. Hsiung, S. Zhao, J. Kost, D. Sreedhar, C. V. Hanson, K. Olson, D. Keare, S. T. Chang and K. P. Bliden, et al., Quantification of antibody avidities and accurate detection of SARS-CoV-2 antibodies in serum and saliva on plasmonic substrates, Nat. Biomed. Eng., 2020, 4, 1188–1196 CrossRef CAS.
- T. AbdElFatah, M. Jalali, S. G. Yedire, I. I. Hosseini, C. Del Real Mata, H. Khan, S. V. Hamidi, O. Jeanne, R. Siavash Moakhar and M. McLean, et al., Nanoplasmonic amplification in microfluidics enables accelerated colorimetric quantification of nucleic acid biomarkers from pathogens, Nat. Nanotechnol., 2023, 18, 922–932 CrossRef CAS.
- C. for D. E. and Research, cobas EGFR Mutation Test v2, FDA.
- J. H. Bae, R. Liu, E. Roberts, E. Nguyen, S. Tabrizi, J. Rhoades, T. Blewett, K. Xiong, G. Gydush and D. Shea, et al., Single duplex DNA sequencing with CODEC detects mutations with high sensitivity, Nat. Genet., 2023, 55, 871–879 CrossRef CAS.
- P. Song, S. X. Chen, Y. H. Yan, A. Pinto, L. Y. Cheng, P. Dai, A. A. Patel and D. Y. Zhang, Selective multiplexed enrichment for the detection and quantitation of low-fraction DNA variants via low-depth sequencing, Nat. Biomed. Eng., 2021, 5, 690–701 CrossRef CAS.
- P. Dai, L. R. Wu, S. X. Chen, M. X. Wang, L. Y. Cheng, J. X. Zhang, P. Hao, W. Yao, J. Zarka and G. C. Issa, et al., Calibration-free NGS quantitation of mutations below 0.01% VAF, Nat. Commun., 2021, 12, 1–9 CrossRef.
- J. D. Cohen, C. Douville, J. C. Dudley, B. J. Mog, M. Popoli, J. Ptak, L. Dobbyn, N. Silliman, J. Schaefer and J. Tie, et al., Detection of low-frequency DNA variants by targeted sequencing of the Watson and Crick strands, Nat. Biotechnol., 2021, 39, 1220–1227 CrossRef CAS PubMed.
- T. Gilpatrick, I. Lee, J. E. Graham, E. Raimondeau, R. Bowen, A. Heron, B. Downs, S. Sukumar, F. J. Sedlazeck and W. Timp, Targeted nanopore sequencing with Cas9-guided adapter ligation, Nat. Biotechnol., 2020, 38, 433–438 CrossRef CAS PubMed.
- T. Gabrieli, H. Sharim, D. Fridman, N. Arbib, Y. Michaeli and Y. Ebenstein, Selective nanopore sequencing of human BRCA1 by Cas9-assisted targeting of chromosome segments (CATCH), Nucleic Acids Res., 2018, 46, e87 CrossRef.
- G. Gydush, E. Nguyen, J. H. Bae, T. Blewett, J. Rhoades, S. C. Reed, D. Shea, K. Xiong, R. Liu and F. Yu, et al., Massively parallel enrichment of low-frequency alleles enables duplex sequencing at low depth, Nat. Biomed. Eng., 2022, 6, 257–266 CrossRef CAS PubMed.
- E. Samorodnitsky, B. M. Jewell, R. Hagopian, J. Miya, M. R. Wing, E. Lyon, S. Damodaran, D. Bhatt, J. W. Reeser and J. Datta, et al., Evaluation of Hybridization Capture Versus Amplicon-Based Methods for Whole-Exome Sequencing, Hum. Mutat., 2015, 36, 903–914 CrossRef CAS PubMed.
- D. C. Koboldt, Q. Zhang, D. E. Larson, D. Shen, M. D. McLellan, L. Lin, C. A. Miller, E. R. Mardis, L. Ding and R. K. Wilson, VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing, Genome Res., 2012, 22, 568–576 CrossRef CAS PubMed.
- M. Guha, E. Castellanos-Rizaldos, P. Liu, H. Mamon and G. M. Makrigiorgos, Differential strand separation at critical temperature: A minimally disruptive enrichment method for low-abundance unknown DNA mutations, Nucleic Acids Res., 2013, 41, e50 CrossRef CAS.
- A. M. Newman, S. V. Bratman, J. To, J. F. Wynne, N. C. W. Eclov, L. A. Modlin, C. L. Liu, J. W. Neal, H. A. Wakelee and R. E. Merritt, et al., An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage, Nat. Med., 2014, 20, 548–554 CrossRef CAS.
- M. W. Schmitt, E. J. Fox, M. J. Prindle, K. S. Reid-Bayliss, L. D. True, J. P. Radich and L. A. Loeb, Sequencing small genomic targets with high efficiency and extreme accuracy, Nat. Methods, 2015, 12, 423–425 CrossRef CAS PubMed.
- A. M. Newman, A. F. Lovejoy, D. M. Klass, D. M. Kurtz, J. J. Chabon, F. Scherer, H. Stehr, C. L. Liu, S. V. Bratman and C. Say, et al., Integrated digital error suppression for improved detection of circulating tumor DNA, Nat. Biotechnol., 2016, 34, 547–555 CrossRef CAS PubMed.
- T. Blewett, J. Rhoades, R. Liu, K. Xiong, S. Sridhar, A. Crnjac, J. Cheng, A. R. Lawless, D. T. Frederick and K. T. Flaherty, et al., MAESTRO-Pool Enables Highly Parallel and Specific Mutation-Enrichment Sequencing for Minimal Residual Disease Detection in Cohort Studies, Clin. Chem., 2024, 70, 434–443 CrossRef CAS PubMed.
- J. Li, L. Wang, H. Mamon, M. H. Kulke, R. Berbeco and G. M. Makrigiorgos, Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing, Nat. Med., 2008, 14, 579–584 CrossRef CAS PubMed.
- T. Fujita, S. Nagata and H. Fujii, Oligoribonucleotide-Mediated Blockade of DNA Extension by Taq DNA Polymerases Increases Specificity and Sensitivity for Detecting Single-Nucleotide Differences, Anal. Chem., 2023, 95, 3442–3451 CrossRef CAS PubMed.
- Z. Liu, R. Zhang, X. Jiang, L. Ji, P. Sun, Y. Ji, Y. Zhang, Y. Ding, K. Li and Z. Pu, et al., Highly Sensitive Enrichment of Low-Frequency Variants by Hairpin Competition Amplification, Anal. Chem., 2023, 95, 12015–12023 CrossRef CAS.
- L. R. Wu, S. X. Chen, Y. Wu, A. A. Patel and D. Y. Zhang, Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification, Nat. Biomed. Eng., 2017, 1, 714–723 CrossRef CAS.
- Y. Si, X. Wang, X. Su, Z. Weng, Q. Hu, Q. Li, C. Fan, D. Y. Zhang, Y. Wang and S. Luo, et al., Extended Enrichment for Ultrasensitive Detection of Low-Frequency Mutations by Long Blocker Displacement Amplification, Angew. Chem., Int. Ed., 2024, 63, e202400551 CrossRef CAS PubMed.
- M. B. Wabuyele, H. Farquar, W. Stryjewski, R. P. Hammer, S. A. Soper, Y.-W. Cheng and F. Barany, Approaching Real-Time Molecular Diagnostics: Single-Pair Fluorescence Resonance Energy Transfer (spFRET) Detection for the Analysis of Low Abundant Point Mutations in K-ras Oncogenes, J. Am. Chem. Soc., 2003, 125, 6937–6945 CrossRef CAS PubMed.
- K. W. Leong, F. Yu and G. M. Makrigiorgos, Mutation enrichment in human DNA samples via UV-mediated cross-linking, Nucleic Acids Res., 2022, 50, e32 CrossRef CAS PubMed.
- W. Gu, E. D. Crawford, B. D. O’Donovan, M. R. Wilson, E. D. Chow, H. Retallack and J. L. DeRisi, Depletion of Abundant Sequences by Hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications, Genome Biol., 2016, 17, 41 CrossRef CAS.
- C. Song, Y. Liu, R. Fontana, A. Makrigiorgos, H. Mamon, M. H. Kulke and G. M. Makrigiorgos, Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment, Nucleic Acids Res., 2016, 44, e146 CrossRef PubMed.
- Q. Liu, X. Guo, G. Xun, Z. Li, Y. Chong, L. Yang, H. Wang, F. Zhang, S. Luo and L. Cui, et al., Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations, Nucleic Acids Res., 2021, 49, e75 CrossRef CAS PubMed.
- J. Song, J. W. Hegge, M. G. Mauk, J. Chen, J. E. Till, N. Bhagwat, L. T. Azink, J. Peng, M. Sen and J. Mays, et al., Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics, Nucleic Acids Res., 2020, 48, e19 CrossRef.
- W. Chen, H. Xu, S. Dai, J. Wang, Z. Yang, Y. Jin, M. Zou, X. Xiao, T. Wu and W. Yan, et al., Detection of low-frequency mutations in clinical samples by increasing mutation abundance via the excision of wild-type sequences, Nat. Biomed. Eng., 2023, 7, 1602–1613 CrossRef CAS PubMed.
- E. A. Manrao, I. M. Derrington, A. H. Laszlo, K. W. Langford, M. K. Hopper, N. Gillgren, M. Pavlenok, M. Niederweis and J. H. Gundlach, Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase, Nat. Biotechnol., 2012, 30, 349–353 CrossRef CAS.
- M. Ayub and H. Bayley, Individual RNA Base Recognition in Immobilized Oligonucleotides Using a Protein Nanopore, Nano Lett., 2012, 12, 5637–5643 CrossRef CAS.
- J. J. Kasianowicz, E. Brandin, D. Branton and D. W. Deamer, Characterization of individual polynucleotide molecules using a membrane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) channel, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13770–13773 CrossRef CAS PubMed.
channel, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13770–13773 CrossRef CAS PubMed. - G. M. Cherf, K. R. Lieberman, H. Rashid, C. E. Lam, K. Karplus and M. Akeson, Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision, Nat. Biotechnol., 2012, 30, 344–348 CrossRef CAS PubMed.
- D. Thirunavukarasu, L. Y. Cheng, P. Song, S. X. Chen, M. J. Borad, L. Kwong, P. James, D. J. Turner and D. Y. Zhang, Oncogene Concatenated Enriched Amplicon Nanopore Sequencing for rapid, accurate, and affordable somatic mutation detection, Genome Biol., 2021, 22, 227 CrossRef CAS.
- Y. Wu, J. Guo, W. Li, X. Xiu, D. Thirunavukarasu, Y. Wang, K. Wang, W. Chen, D. Yu Zhang and X. Yang, et al., Enhanced Detection of Novel Low-Frequency Gene Fusions via High-Yield Ligation and Multiplexed Enrichment Sequencing, Angew. Chem., Int. Ed., 2024, 63, e202316484 CrossRef CAS.
- W. Jiang, X. Zhao, T. Gabrieli, C. Lou, Y. Ebenstein and T. F. Zhu, Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters, Nat. Commun., 2015, 6, 8101 CrossRef PubMed.
- A. R. Vandiver, B. Pielstick, T. Gilpatrick, A. N. Hoang, H. J. Vernon, J. Wanagat and W. Timp, Long read mitochondrial genome sequencing using Cas9-guided adaptor ligation, Mitochondrion, 2022, 65, 176–183 CrossRef CAS PubMed.
- W. Tang, W. Zhong, Y. Tan, G. A. Wang, F. Li and Y. Liu, DNA Strand Displacement Reaction: A Powerful Tool for Discriminating Single Nucleotide Variants, Top. Curr. Chem., 2020, 378, 10 CrossRef CAS.
- Y. Yu, L. Ma, L. Li, Y. Deng, L. Xu, H. Liu, L. Xiao and X. Su, Digestion of Dynamic Substrate by Exonuclease Reveals High Single-Mismatch Selectivity, Anal. Chem., 2018, 90, 13655–13662 CrossRef CAS PubMed.
- L. Zhang, J. Chen, M. He and X. Su, Molecular dynamics simulation-guided toehold mediated strand displacement probe for single-nucleotide variants detection, Exploration, 2022, 2, 20210265 CrossRef CAS.
- K. Zhang, L. Rodriguez, L. Y. Cheng, M. Wang and D. Y. Zhang, Single-Tube qPCR Detection and Quantitation of Hotspot Mutations Down to 0.01% Variant Allele Fraction, Anal. Chem., 2022, 94, 934–943 CrossRef CAS.
- Y. E. Nikiforov, L. Yip and M. N. Nikiforova, New Strategies in Diagnosing Cancer in Thyroid Nodules: Impact of Molecular Markers, Clin. Cancer Res, 2013, 19, 2283–2288 CrossRef CAS.
- Y. Zhang, T. Huang, F. Yang, Q. Tan, S. Bu, S. Yu, J. Ye, T. Hang, X. Feng and D. Zhang, Mismatch-guided DNA competitive converter enables expanded detection window for discriminating single nucleotide polymorphisms, Chem. Eng. J., 2025, 512, 162549 CrossRef CAS.
- S. Lee, N. Ding, Y. Sun, T. Yuan, J. Li, Q. Yuan, L. Liu, J. Yang, Q. Wang and A. B. Kolomeisky, et al., Single C-to-T substitution using engineered APOBEC3G-nCas9 base editors with minimum genome- and transcriptome-wide off-target effects, Sci. Adv., 2020, 6, eaba1773 CrossRef CAS.
- J. G. Doench, N. Fusi, M. Sullender, M. Hegde, E. W. Vaimberg, K. F. Donovan, I. Smith, Z. Tothova, C. Wilen and R. Orchard, et al., Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9, Nat. Biotechnol., 2016, 34, 184–191 CrossRef CAS PubMed.
- B. Shen, W. Zhang, J. Zhang, J. Zhou, J. Wang, L. Chen, L. Wang, A. Hodgkins, V. Iyer and X. Huang, et al., Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects, Nat. Methods, 2014, 11, 399–402 CrossRef CAS PubMed.
- E. Zuo, Y. Sun, T. Yuan, B. He, C. Zhou, W. Ying, J. Liu, W. Wei, R. Zeng and Y. Li, et al., A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects, Nat. Methods, 2020, 17, 600–604 CrossRef CAS.
- Y. Liu, C. Zhou, S. Huang, L. Dang, Y. Wei, J. He, Y. Zhou, S. Mao, W. Tao and Y. Zhang, et al., A Cas-embedding strategy for minimizing off-target effects of DNA base editors, Nat. Commun., 2020, 11, 6073 CrossRef CAS PubMed.
- S. Ye, J. Kim, M. Kim, K. Kim, Y. Won, T. Park, S. An, H. Jeong, H. Chung and I. S. Lee, et al., MUTE-Seq: An Ultrasensitive Method for Detecting Low-Frequency Mutations in cfDNA With Engineered Advanced-Fidelity FnCas9, Adv. Mater., 2025, e05208 CrossRef CAS.
- A. T. Riley, J. M. Robson, A. Ulanova and A. A. Green, Generative and predictive neural networks for the design of functional RNA molecules, Nat. Commun., 2025, 16, 4155 CrossRef CAS PubMed.
- F. Wong, C. De La Fuente-Nunez and J. J. Collins, Leveraging artificial intelligence in the fight against infectious diseases, Science, 2023, 381, 164–170 CrossRef CAS PubMed.
- H.-H. Wessels, A. Stirn, A. Méndez-Mancilla, E. J. Kim, S. K. Hart, D. A. Knowles and N. E. Sanjana, Prediction of on-target and off-target activity of CRISPR–Cas13d guide RNAs using deep learning, Nat. Biotechnol., 2024, 42, 628–637 CrossRef CAS.
- S. J. Low, M. O’Neill, W. J. Kerry, N. Wild, M. Krysiak, Y. Nong, F. Azzato, E. Hor, L. Williams and G. Taiaroa, et al., PathoGD: an integrative genomics approach to primer and guide RNA design for CRISPR-based diagnostics, Commun. Biol., 2025, 8, 147 CrossRef CAS.
- S. Mantena, P. P. Pillai, B. A. Petros, N. L. Welch, C. Myhrvold, P. C. Sabeti and H. C. Metsky, Model-directed generation of artificial CRISPR–Cas13a guide RNA sequences improves nucleic acid detection, Nat. Biotechnol., 2025, 43, 1266–1273 CrossRef CAS.
- Z. Yao, W. Li, K. He, H. Wang, Y. Xu, Q. Wu, L. Wang and Y. Nie, Facilitating crRNA Design by Integrating DNA Interaction Features of CRISPR-Cas12a System, Adv. Sci., 2025, 12, 2501269 CrossRef CAS.
Footnote |
| † These authors contributed equally to this review. |
| This journal is © The Royal Society of Chemistry 2026 |