Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Realities of the consortium approach in science: sustainable enzymatic production of C1 chemicals from carbon dioxide
Andrea
Rodil
ab,
Ingemar
von Ossowski
 b,
Mari
Nyyssönen
c,
Yufang
Tian
b,
Marleen
Hallamaa
b,
Mari
Nyyssönen
c,
Yufang
Tian
b,
Marleen
Hallamaa
 a,
Jan
Deska
a,
Jan
Deska
 a,
Malin
Bomberg
c and
Silvan
Scheller
*b
a,
Malin
Bomberg
c and
Silvan
Scheller
*b
aUniversity of Helsinki, Department of Chemistry, A. I. Virtasen aukio 1, 00560 Helsinki, Finland
bAalto University, Department of Bioproducts and Biosystems, School of Chemical Engineering, Kemistintie 1, 02150 Espoo, Finland. E-mail: silvan.schelleraalto.fi
cVTT Technical Research Centre of Finland, Maarintie 3, Espoo, Finland
First published on 28th August 2024
Abstract
Research at the frontiers of science is getting increasingly specialised. At the same time, major global challenges require the cooperation and innovation of different research fields. One solution for enhancing scientific discovery and innovation within this landscape is to form research consortia that bring together expertise from different disciplines. Such multidisciplinary efforts are also highly recognized and increasingly enforced by funding agencies. Within this landscape, we established a research consortium consisting of three partners to explore environmental acid-tolerant formate dehydrogenases as novel biocatalysts for formic acid production from CO2. Taking our ambitious project on biocatalytic CO2 valorisation as a case study, we reflect on the realities of forming a research consortium, highlighting some of the related theoretical and technical issues, as well as its intrinsic positive and valuable nourishing effect on researchers. Finally, we offer some constructive criticism and practical advice to other scientists willing to embark on complex scientific projects through collaborations.
Sustainability spotlightAnthropogenic carbon dioxide emissions have led to a massive increase in its atmospheric levels with severe consequences on climate change and global warming. In this perspective, we reflect on the circular utility of CO2 as an abundant resource to store energy. Primarily, we delve on a metagenomic approach to discover new enzymes able to bind/convert CO2 at low temperature/pH (SDG 15), and to use C1 entities in chemical valorization based on an eco-friendly enzymatic approach (SDG 12, SDG 13). With a strong emphasis on the opportunities and challenges regarding the multidisciplinary approach rather than the scientific details of the underlying complex project, this article aims to encourage other researchers to include partnerships in their projects (SDG 17). |
1. Introduction
One of the main virtues, and problems, of modern science is specialisation. Whilst the scientific community is blessed to have many talented researchers able to solve wide-spread and niche problems alike, the tendency is for those researchers to be highly specialised in their fields. This circumstance is equally a catalyst for the progress of science and the inhibitor that stalls the speed of discovery. Nowadays, our community faces increasingly complex challenges, and these challenges will only be resolved by the joint hard work of highly specialised scientists, together. Funding bodies and governmental programmes begin to see the value of multidisciplinary work, and researchers are constantly being encouraged to form partnerships.One of the most prominent threats to humanity is global warming. The current worldwide carbon dioxide (CO2) emissions from fossil fuel use remain excessive and the natural capacity for photosynthetic CO2 assimilation continues to be outstripped.1–5 Thus, the prospect of CO2 utilisation will not only help achieve more tolerable atmospheric CO2 levels, but it will provide a carbon source that is large enough to substitute fossil carbon sources. In this quest to access CO2 as a carbon source, it is crucial that we get inspiration from nature. The last decade has seen an active development on the field of synthetic biology, with cutting-edge technologies designed to biocatalytically convert CO2 emissions into high value-added chemical products such as formic acid (HCOOH).6,7 Formic acid can be further transformed into high-value chemicals.8,9
Soon, we encountered our first challenge: specialisation. From a chemist's perspective, biocatalytic research is often constrained in terms of commercial availability, considerably limiting the scope of experimentation, and failing to adequately answer key questions or deliver novel solutions and data. Although the situation can be improved by producing a broader collection of enzymatic species, the necessary scientific skills, know-how, and infrastructure do not always coexist within a purely chemistry-based research group. At the same time, from the biologist's perspective, the idea of taking formic acid and being able to obtain other molecules might seem unsurpassable. As a solution, entering a formal collaborative partnership, with researchers from various other academic and technical disciplines, is a proven approach for addressing any theoretical or research shortcomings frequently associated with a highly ambitious and complex project and highly specialised researchers.
In this perspective article, we consider the benefits and limitations of forming a consortium, starting with the fact that funding bodies and research agencies are beginning to see the value of this kind of multidisciplinary work, and researchers are constantly being encouraged to form partnerships. In our view, we understood that taking this approach would carry a high degree of conceptual risk, as our proposed project involved highly ambitious and challenging research with a good deal of scientific uncertainty and unpredictability. Even so, by pursuing this ‘high risk-high gain’ strategy, both researchers and funding agencies alike, certainly expected that the anticipated outcome would clearly outweigh the element of risk and possible failure of the research. Take our project as a case study.
2. Research description and objectives
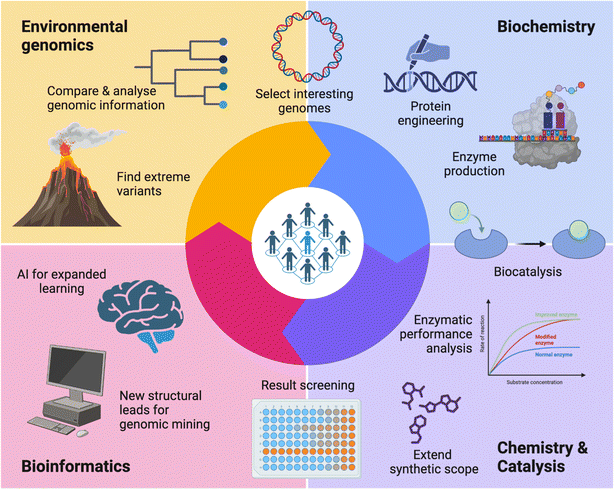
In 2019, the Research Council of Finland (formerly the Academy of Finland) issued a special call aimed at proposals that could strengthen Finnish scientific competence and competitiveness in the utilisation of green chemistry technologies for industrial manufacturing of products from captured CO2 emissions.10 As scientists interested on the topic, we prepared a proposal, and thus, ‘ExtremoForm: extremophile microorganisms as a source of highly productive enzymes for CO2 reduction to formic acid and other C1 fuels and platform chemicals’ was born. With academic and scientific partners from the VTT Technical Research Centre of Finland Ltd, Aalto University, and the University of Helsinki; three disciplines were involved: environmental metagenomics, expression of enzymes and chemical catalysis (Fig. 1).Our main collective aim was to procure a selection of enzymes – formate dehydrogenases (FDHs) – that are acid-tolerant and active in low temperature conditions, and with a strong preference for CO2 reductase catalysis. These novel FDHs represent a sustainable green alternative, because they rely on abundant non-toxic molybdenum and tungsten, instead of late transition metals such as rhodium that are typically used by classical chemical catalysts. Via enzymatic promiscuity, we focused on the direct production of formic acid for the environmentally friendly generation of high value-added chemicals and fuels. Notably, we focus on formic acid, and not formate, to avoid the problem of having to provide a counter ion for each formate produced. In addition, formic acid can be used to address the problematic transport and storage of hydrogen (H2) for the renewable energy industry.
To advance this outcome, our five strategic targets were laid out as follows: (i) to screen and uncover metallo-FDH biocatalysts that efficiently reduce CO2 to formic acid at zero degree temperatures and under extremely acidic conditions (pH < 3), (ii) to attain a fundamental understanding of the molecular mechanisms behind the low temperature activity and high acid stability of the FDH biocatalysts, (iii) to improve the performance of the FDH biocatalysts by using a combination of directed evolution and semi-rational protein engineering and design, (iv) to characterise the properties of the FDH biocatalysts by applying the same conventional methods used for chemical catalysts, and (v) to broaden the reaction specificity of the existing FDH biocatalysts for the formation of alternative products such as formate esters and formamides.
3. Scientific premise and background
3.1 CO2 emissions and formic acid
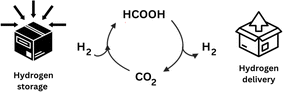
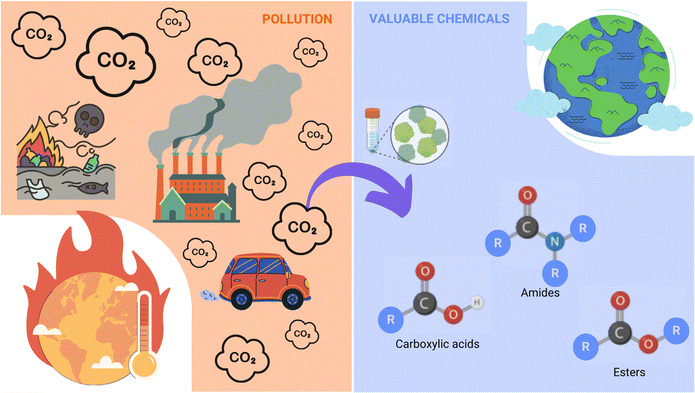
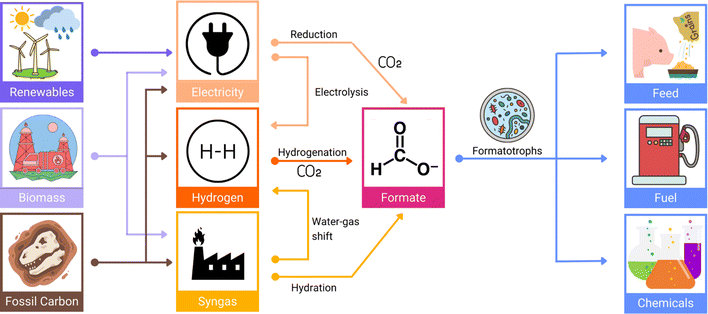
Combustion of hydrocarbon-containing materials has led to a massive increase in atmospheric CO2 emissions, with current global atmospheric CO2 concentrations at 421.47 ppm (March 3rd, 2024), and a steady yearly increase of 2.4 ppm on average since 2012.11 There is a pressing need for the development of more sustainable and clean energy concepts, and although in recent years, most efforts to ease CO2 accumulation have focused on developing new energy efficiency and sequestration technologies,12 the obvious abundance of this greenhouse gas has also come to represent a readily available, cheap, and renewable source of C1-carbon for producing various value-added chemicals such as formic acid.13Molecular hydrogen (H2) is emerging as a carrier of renewable fuel,14 but its necessary compression or liquefaction is highly energetically demanding, resulting in a costly and hazardous procedure.15 Alternatively, formic acid is often touted as a promising H2 carrier, as it achieves a higher volumetric density and is liquid under standard temperatures and pressures, making its handling more economical and much safer (Fig. 2).16
For its role as a liquid H2 carrier, formic acid can be acquired through the catalytic hydrogenation of CO2 emissions.17 In addition to its role as a stabilised form of H2 fuel, formic acid is also viewed as an economically valuable platform feedstock for producing numerous other higher value-added chemicals within the circular bioeconomy (Fig. 3).18
 | ||
| Fig. 3 The processes involved in formic acid (formate) bioeconomy. Formate can be obtained through different methods and from different sources and is ultimately transformed into high-value chemicals affording huge economic benefits. Given its complexity, it is not possible to develop any new work into the formate bioeconomy without including more than one research discipline, allowing for multidisciplinary advancements in a reasonably timely manner. This image has been adapted from Yishai et al. (2016).18 | ||
Achieving formic acid production directly from CO2 and H2 is a rather challenging reaction, as it requires expensive catalysts and harsh conditions to yield high conversion rates.19–22 Thermodynamically, formate formation is favourable at pH = 7 (eqn 1′); but under standard conditions (pH = 0, eqn 1), under which formic acid is produced, the equilibrium favours CO2 formation. This equilibrium, however, is temperature dependent, and low temperatures favour formic acid production. At 0 °C, 50 bar CO2 and 150 bar H2, the maximal formic acid concentration that can be produced is ca. 3 M.23–26
| CO2(g) + H2(g) = HCOOH(aq) ΔG° = +15.3 kJ mol−1 | (1) |
| CO2(g) + H2(g) = HCOO−(aq) + H+ ΔG°′ = −3.5 kJ mol−1 | (1′) |
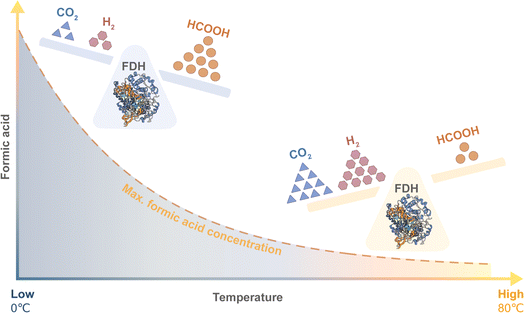
The severity of the reaction conditions can be diminished by using a less energy-demanding biocatalyst: a formate dehydrogenase that is active at low temperature, unlike other catalysts cited in literature.24 So far, there are several reports that the production of formic acid from CO2 can proceed enzymatically through FDH, either as a whole cell biocatalyst or as an immobilised or free enzyme.27–31 However, with the higher expected accumulation of formic acid at lower temperatures, FDH will likely be susceptible to protein denaturation by the ensuing low pH environment and so to maintain its catalytic activity would require a highly acid stable nature (Fig. 4).
3.2 Formate dehydrogenase as a biocatalyst to promote CO2 reduction
FDH is the biological catalyst responsible for the interconversion between formate and CO2. Although always part of an equilibrium, most FDH enzymes evolved to preferentially catalyse the formate oxidation, but some FDHs are reportedly more active in catalysing CO2 reduction.28,32 Kinetic parameters are dependent on the corresponding redox cofactors of the enzyme (oxidants or reductants), the KM values, as well as process parameters that help shifting the equilibrium in a particular direction.33,34FDHs, constitute a very heterogeneous group of enzymes regarding cellular location, functional specialisation and structural organisation.35 For example, as formate can act as an energy source in hydrogenotrophic methanogens (e.g. Methanococcus sp.), cytosolic FDH will facilitate the conversion to CO2via the methanogenic pathway.36 On the other hand, in Gram-negative bacteria, membrane-bound periplasmic FDH serves to funnel electrons to the quinone pool in the periplasm, where the generation of protons powers the proton-motive force that enables energy to be conserved.37,38 For this article, the classification according to structural organisation is the most relevant. Here, there are two separate types of FDH, characterised by the absence or presence of a metal ion in the active site.
Non-metallo FDH are commonly observed in aerobic hosts, and so taken to be oxygen tolerant.39,40 Nicotinamide adenine dinucleotide (NAD+), or in some rare instances nicotinamide adenine dinucleotide phosphate (NADP+),41 is generally used as the cofactor for the redox catalysis reaction. The formate oxidation reaction is typically favoured among non-metallo FDHs,42 and these enzymes have been frequently used in cofactor recycling for cascade enzymatic reactions.43 As a caveat, however, the lower catalytic activity of non-metallo FDH, the prohibitive cost of NAD+ and the high lability of NADH in acids, are obvious disadvantages for practical CO2 reduction applications, and for that reason this class of FDH also tends to be viewed as a less viable option.44,45
By contrast, metallo FDHs exhibit much greater variation based on metal content, subunit composition, quaternary structure, cellular localisation, active-site residues, O2 sensitivity, and physiological function. True to its name, metallo FDH can contain either molybdenum (Mo) or tungsten (W) within its active site.46 Although there is still some debate, it is accepted that the reaction mechanism in metallo FDH can accommodate a range of electron donor–acceptor cofactors, including artificial cofactors.46–48
In the quest to develop novel CO2 reducing biocatalysts, growing research interest has been drawn to the metallo FDHs. In a head-to-head comparison of turnover activity, metallo FDH clearly outperforms the nonmetal-containing enzyme, making it the far better option for potential applications as a CO2 reductase. Still, practical sticking points with the metallo FDHs continue to be their poor and difficult recombinant expression in E. coli, and their short-lived ability to retain full activity due to O2 sensitivity.49–51 In this perspective, we will focus on our work with metallo FDHs.
4. Research implementation and impact
To accomplish our objectives, we structured the scope of the research project into three work phases (enzyme discovery, enzyme performance, and molecular understanding) to foster the transdisciplinary exchange of scientific ideas and knowledge between consortium members. These three phases, although mostly linear in nature, also provided the opportunity for all members to start working in parallel while waiting for more information and results from the other team members. In this way, each team developed their areas of expertise while updating, complementing and informing the other parts of the consortium. Exceptionally high levels of constant communication were crucial for the advancement of the project. It is important to clarify, that whilst the project has now been finished, the authors are still in the process of preparing publications for their results.4.1 Enzyme Discovery
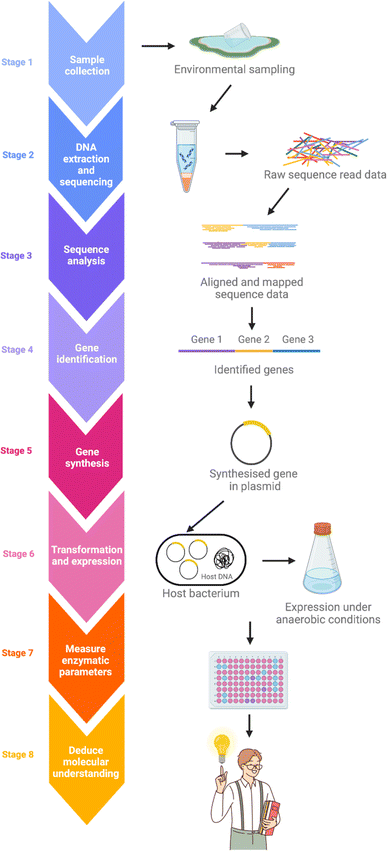
Despite the omnipresence of microorganisms in nature, only a small fraction of these can be readily cultured in the laboratory.52,53 This leaves most of the genetic potential within these natural microbial populations unexplored when traditional culture-based methods are used. Metagenomics is an effective approach for revealing the untapped reservoir of microbial and genomic biodiversity in various environments.54–56 The typical metagenomics workflow begins with collecting samples from the subject environment, followed by isolating the DNA. DNA extraction is much less discriminating than cultivation-dependent approaches, allowing for a more comprehensive representation of the collective genome, or ‘metagenome’, of the microbial community in an environmental sample. After isolation, the metagenome is fragmented and then undergoes high-throughput DNA sequencing. Genes encoding possible novel biological functions are identified from the sequence data by computer algorithms comparing with known genes and proteins (Fig. 5).To discover FDH biocatalysts that are naturally cold adapted and highly acid-tolerant, the researchers at VTT conducted a comprehensive metagenomics survey of the microbial communities in two acidic environments. By selecting an environment that is both sufficiently unique and extreme, and, most importantly, that also reflects the sought-after properties of the enzyme we aimed to significantly expand the overall gene pool and thereby enhance the possibility of discovering FDH enzymes with the desired physical and catalytic properties that were hereto unknown.
The metagenomic approach considerably expanded the available gene pool, yielding to the discovery of novel FDH genes, encoding both metallo and non-metallo FDHs. These newly identified genes represent a wide diversity of microorganisms, as well as enzyme structures and cellular locations, supporting our aim to cover as much functional diversity as possible.
4.2 Protein production and enzymatic performance
In continuation to the metagenomic analysis, these new metal-containing FDHs were explored for their potential to act as stable catalysts that can convert CO2 and H2 to formic acid for industrial applications. Different fdh genes were selected from the novel FDH gene pool; a selection made to represent as wide a protein level and taxonomic level as possible. In this way, it could be established which of the discovered variants could be expressed for activity screening.Overexpression of the enzymes was designed to test different plasmids, vectors, expression conditions and genetic modifications.57–61 Given that the extremophiles in this study were derived from cold and acidic environments, their enzymatic activity was tested at different pHs and temperatures.
To address and circumvent the linearity of the project, parallel work involving other oxygen-sensitive, metal-dependent FDHs was developed to study the enzymatic active site and how its modification affects enzyme performance. At this stage, most of the experimental work was carried out with a formate dehydrogenase from E. coli K12, following Li's protocol.61–65
The procedure is outlined in Fig. 6. A key aspect of the analytical work was to establish the activity of these enzymes with artificial electron donors, such as methyl and benzyl viologens. These compounds afforded an efficient method for measuring enzymatic activities, given their unique colorimetric properties.66,67
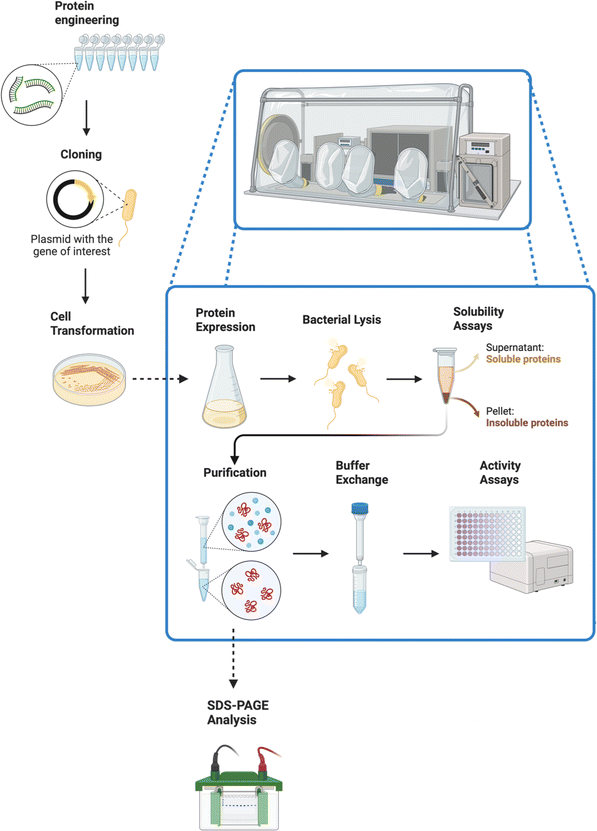 | ||
| Fig. 6 After the metagenomic workflow showed in Fig. 5, a different skill set is needed. This figure displays a protein engineering procedure, starting with a PCR (for protein engineering through DNA mutations) and ending with activity assays. Most of the procedure needs to be carried out inside an anaerobic chamber. The enhanced mutations selected through the activity assays can then be used to expand the synthetic scope, serving as a bridge between the metagenomics field and the organic synthetic chemistry area. Created with https://BioRender.com. | ||
In this project, only modification through rational design was attempted. However, we believe that best results will be obtained through a combination of both directed evolution and rational design. Ideally, if more time was granted, an in-depth study comparing these two parallel lines of work (enzymes from metagenome analysis and available enzymes) would be carried out to extract the precious information. As we stand, those avenues of investigation might have to be saved for future endeavours.
4.3 Molecular understanding and catalytic applications
This phase of the project delivered what might be perceived as the most practical results in terms of CO2 capture and chemical transformation, since it focused on the optimisation of biocatalysis to extend the synthetic product scope.To date, most large-scale efforts have centred around the conversion of CO2 to economically valuable small molecules. Two strong contenders in these processes are methanol and ethanol, which can be easily used as a fuel, as a precursor for more complex fuels or as a building blocks towards other valuable chemicals.68–70 Nowadays, most of the CO2 used as a raw material is converted to fertilisers in the agrochemical industry through urea manufacture.71,72
In chemical terms, carbon dioxide is an electrophilic, but rather stable molecule. Contrary to what might be expected, it is, nonetheless, quick to react with basic compounds;73 or with highly reactive substrates or under harsh conditions.74 Milder chemical methods continue to be developed.75 In terms of biocatalysis, three common enzymes are used in the incorporation of carbon dioxide: carbonic anhydrase for the formation of hydrogen carbonate, RuBisCO in the photosynthetic pathway and formate dehydrogenase to obtain formate.76
It is the latter option that was taken as a stepping-stone to delve into the biocatalysis-synthetic organic chemistry interface. Thus far, the use of FDH to produce compounds other than formate has been practically nonexistent, besides the non-natural reduction of nitrate to nitrite reported by Hartmann et al., and the formic ester cleavage introduced by Frölich.77,78 Given that amines show a strong affinity for electrophiles – including CO2;79,80 the prospect of formylation reactions of amines through FDH was explored. In this way, we hoped to combine disciplines to achieve a wider range of substrates for FDH for the direct synthesis of interesting organic compounds and to provide a milder alternative to conventional carbon dioxide trapping methods (Fig. 7).
The production of C1-derived platform chemicals for the development of new amides would not have been possible – or would have been much slower – if not for the combination of knowledge on enzyme promiscuity, enzyme engineering and synthetic organic chemistry. Once again, the inherent value of the consortium approach and its different skill sets is brought back to the limelight. Through the collaboration, it became possible to not only focus on the discovery and development of new CO2-fixating FDHs (a relevant aim on its own), but we could also delve further into the synthetic value of CO2 in chemistry. At the same time, the linearity of the project did also play a part in the slow deliverance of results from all parts. With more time, the overall benefits of the consortium would easily exceed the downsides, which for this case, were mostly schedule-based.
5. The consortium approach: lessons learned
5.1 The researchers' perspective
In view of our experience, we would like to present this perspective as a case study for the benefits, and drawbacks, of working in the highly collaborative environment that ensued the creation of the project. Besides the success of producing novel FDHs and sustainable new methods for formic acid biosynthesis, the most valuable outcome was the considerable personal and professional growth experienced. Each partner was able to do individual research, and results were enhanced by the overarching input from the network.This multidisciplinary environment fostered the development of transferable skills; such as team working, presentation and communication skills. In this way, monthly meetings and work presentations were supplemented with a well-established knowledge transfer scheme: from bachelor students to principal investigators, everyone was willing to offer their unique perspective on the project, and to share their unique set of skills.
While communication throughout the programme was always present and abundant, the much-needed multidisciplinary approach was, however, demanding. The whole project was much more ambitious than initially expected, and in reality, working together was not always possible. The physical separation constituted a clear disadvantage for building active connections amongst the researchers. Additionally, each round of the project required learning cycles from all its members, along with good doses of willingness and the ability to reach out. The challenging nature of the project marked its fast-learning rhythm: a hectic pace that was only made bearable by the constant help and support from all the network members.
At the same time, the intrinsic nature of ExtremoForm translated into a linear timeline: the results from the metagenomic analysis were needed for the enzyme performance assays, which were needed for the development of the molecular understanding phase. Contingency measures were applied, such as working with readily available enzymes and organisms, but they do not distract from the fact that time was a very important factor and that it was very clearly insufficient.
With regards to time, the most glaringly obvious issue was the 2020 coronavirus pandemic. While perhaps consortia in other fields have a wider choice of workplace (bio)chemistry is very limited on the possibilities for remote work. Unfortunately, the pandemic severely hindered our ability to work in situ, especially during the lockdown stage, meaning that most experiments got a considerable delay from the original plan. Not only that, but working in shifts meant that direct communication with other teams was severed, and we relied mainly on the digital approach – often a lot more convoluted than needs be. The long time that took to get back to in-person meetings was another setback. However, despite all the difficulties, we can say that through collaboration and learning, we have delivered some outstanding research, the results of which we hope will be published separately.
In addition, ExtremoForm also allowed for the development of different branches of work, including two master theses, undergraduate-level participation and bachelor projects. The massive amounts of data produced, not directly related to the original project, can be used in the future, especially focusing on IA development of new methods, closing the cycle and starting a new iteration (Fig. 1).
Clearly, ExtremoForm presented a project that was exponentially more complex than initially thought, mainly because biology is rather less predictable than chemistry. Not only is it not a guarantee that a metagenomic analysis will produce a good match for what is needed, but even if it does, it is a very plausible possibility that it is not feasible to translate the findings into a real laboratory setting. For this reason, we believe that these difficult endeavours should be taken stepwise, from easy to difficult, and taking time to focus on learning cycles, allowing for the ambition to produce its fruit. Despite the hardship, one thing has become apparent: forming a consortium has always been advantageous. In this sense, the implementation of collaborations on a practical level – as opposed to the more theoretical networking approach prevalent in conferences and other sporadic encounters – leads to more grounded science. We now know how each member works, which will no doubt be of benefit in future challenges.
5.2 The consortium managers' perspective
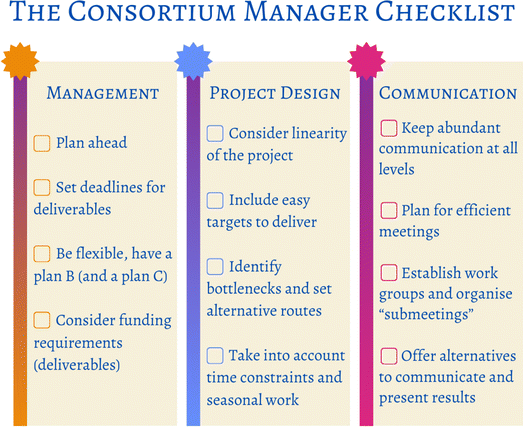
We would like to offer some constructive criticism and some practical advice from the managerial angle, so future consortium leaders can benefit from our experience (Fig. 8). Consortium leaders (management) should plan thoroughly and well-ahead of time. In this stage it is important to consider the big picture and examine how the consortium will address the project and fulfil the research description submitted for the funding application. Be realistic with regards of ambitious research, identify possible bottlenecks and set hard deadlines for specific deliverables; be prepared to continue with an alternative plan in case of negative results, unexpected issues, or other practical inconveniences. Having these “relief” course of actions will result in an efficient and well used time, and possibly to higher quality publications. Flexibility with research will be necessary: the timeline will likely need to be adjusted as the project advances, so this must be reflected upon in the planning stage.Project design is no menial task. On top of the management-related advice in the previous paragraph, here are some specific tips to consider. As we have mentioned all through section 5, the timeline of the project requires attention. In ExtremoForm we worked with a very sequential project that resulted in high linearity, less time efficiency, and a number of lost opportunities. Funded projects have very limited time for often very ambitious research, so make sure your proposal has a carefully studied structure, giving each partner enough room to start early on, and to carry out parallel work. Do not forget to consider external factors, like the weather or growth seasons, which funding bodies do not usually take into account. Here in Finland, for example, the collection of samples and outdoor work in general can be very restrictive during wintertime, translating into delayed lab work. In our case, sample collection needed to be done early on, but could not be carried out during the first few months. We suggest, if you find yourself in the same position, that you plan for alternative work, for example, start with the analysis of previous data so experiments can start as soon as specimens arrive.
Ideally, a consortium should afford high-quality publications. In order to facilitate this, project design should include plenty of accessible branches: smaller divisions of work that will result in fast, easier-to-attain results. If at all possible, including these at the beginning of the research project will also help to ramp up towards the more challenging parts of the project and will generally increase morale, given that results (publications) have already been granted from the start. As stated, ExtremoForm was incredibly ambitious in its entirety and these “side works” were only included towards the end, when the pressure was really high, and researchers were fully immersed in more demanding work. One of the main values of collaborations is having different workers with different skills; therefore, it is possible to have someone working on the low hanging fruits whilst others focus on the most ambitious parts, which also helps with linearity.
The previous subsection highlights the importance of communication. This is a very crucial point for the success of any collaboration: plenty of meetings are needed, but always keeping the same structure can make them overwhelming and inefficient for the participants. We found that, although we had very frequent presentations and discussions, these became draining because everyone needed to be involved all the time. Instead, opt for shorter, more specific gatherings (researchers to discuss practical details with each other and leaders to arrange management separately) and for variety (include retreats, days away or even conference-style poster presentations).
One factor that requires some deliberation is how to implement management itself, and we cannot offer a clear answer. Is micromanaging an asset to science? It will depend on each specific team, and on each individual researcher. It takes time to know your colleagues, and the best approach is to keep an open mind, and again, be ready to be flexible. One thing that we all agree on is that too many restrictions on the work or on the timeline schedule heavily diminish academic creativity, so finding a good management style will determine how the group blossoms.
As a visual aid, Fig. 8 was designed to facilitate the complicated role of the consortium manager, and it is based in everything that was discussed up to this point.
6. Conclusions
All funding agencies, individual researchers and academic institutions are all pointing towards an undeniable reality: an isolated researcher will never accomplish as much as someone who is in touch with different experts, from different fields, promoting an open, and constructive, communication of science. And while perhaps not everyone has the opportunity to form an official bond (i.e., funded consortium) with other fields, collaboration – official or unofficial – should always be encouraged. By merging our different skills, it was possible to create a more complete, and thus realistic, picture of a very complex plan, and has led to high quality-albeit incomplete at this stage-science. Along the way, we have encountered many problems and challenges, and in the hope that others can benefit from our insights, we have prepared this article.Data availability
The manuscript is a critical reflection and discussion based on the bigger picture experience of our researchers rather than an actual detailed account involving primary data and/or concrete experimental/computational results. As such, no data are provided that would require repository access of supporting informations.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research has received financial support from the Research Council of Finland in the form of the ExtremoForm project granted to M. B., S. S. and J. D. (decision no 329510). We thank Dr Heidi Henrickson for helpful discussions. The graphical abstract, as well as Fig. 1, 5 and 6, have been created with BioRender.References
- P. Friedlingstein, M. O’Sullivan, M. W. Jones, R. M. Andrew, D. C. E. Bakker, J. Hauck, P. Landschützer, C. Le Quéré, I. T. Luijkx, G. P. Peters, W. Peters, J. Pongratz, C. Schwingshackl, S. Sitch, J. G. Canadell, P. Ciais, R. B. Jackson, S. R. Alin, P. Anthoni, L. Barbero, N. R. Bates, M. Becker, N. Bellouin, B. Decharme, L. Bopp, I. B. Mandhara Brasika, P. Cadule, M. A. Chamberlain, N. Chandra, T.-T.-T. Chau, F. Chevallier, L. P. Chini, M. Cronin, X. Dou, K. Enyo, W. Evans, S. Falk, R. A. Feely, L. Feng, D. J. Ford, T. Gasser, J. Ghattas, T. Gkritzalis, G. Grassi, L. Gregor, N. Gruber, Ö. Gürses, I. Harris, M. Hefner, J. Heinke, R. A. Houghton, G. C. Hurtt, Y. Iida, T. Ilyina, A. R. Jacobson, A. Jain, T. Jarníková, A. Jersild, F. Jiang, Z. Jin, F. Joos, E. Kato, R. F. Keeling, D. Kennedy, K. Klein Goldewijk, J. Knauer, J. I. Korsbakken, A. Körtzinger, X. Lan, N. Lefèvre, H. Li, J. Liu, Z. Liu, L. Ma, G. Marland, N. Mayot, P. C. McGuire, G. A. McKinley, G. Meyer, E. J. Morgan, D. R. Munro, S.-I. Nakaoka, Y. Niwa, K. M. O’Brien, A. Olsen, A. M. Omar, T. Ono, M. Paulsen, D. Pierrot, K. Pocock, B. Poulter, C. M. Powis, G. Rehder, L. Resplandy, E. Robertson, C. Rödenbeck, T. M. Rosan, J. Schwinger, R. Séférian, T. L. Smallman, S. M. Smith, R. Sospedra-Alfonso, Q. Sun, A. J. Sutton, C. Sweeney, S. Takao, P. P. Tans, H. Tian, B. Tilbrook, H. Tsujino, F. Tubiello, G. R. van der Werf, E. van Ooijen, R. Wanninkhof, M. Watanabe, C. Wimart-Rousseau, D. Yang, X. Yang, W. Yuan, X. Yue, S. Zaehle, J. Zeng and B. Zheng, Global Carbon Budget 2023, Earth Syst. Sci. Data, 2023, 15, 5301 CrossRef.
- K. Miyazaki and K. Bowman, Nat. Commun., 2023, 14, 1604 CrossRef CAS PubMed.
- S. Bierbaumer, M. Nattermann, L. Schulz, R. Zschoche, T. J. Erb, C. K. Winkler, M. Tinzl and S. M. Glueck, Chem. Rev., 2023, 123, 5702 CrossRef CAS PubMed.
- G. Tcherkez and A. M. Limami, New Phytol., 2019, 223(2), 520 CrossRef CAS PubMed.
- D. Archer, M. R. Ashmore, O. Aumont, D. Baker, M. Battle, M. Bender, L. P. Bopp, P. Bousquet, K. Caldeira, P. Ciais, P. M. Cox, W. Cramer, F. Dentener, I. G. Enting, C. B. Field, P. Friedlingstein, E. A. Holland, R. A. Houghton, J. I. House, A. Ishida, A. K. Jain, I. A. Janssens, F. Joos, T. Kaminski, C. D. Keeling, R. F. Keeling, D. W. Kicklighter, K. E. Kohfeld, W. Knorr, R. Law, T. Lenton, K. Lindsay, E. Maier-Reimer, A. C. Manning, R. J. Matear, A. D. McGuire, J. M. Melillo, R. Meyer, M. Mund, J. C. Orr, S. Piper, K. Plattner, P. J. Rayner, S. Sitch, R. Slater, S. Taguchi, P. P. Tans, H. Q. Tian, M. F. Weiring, T. Whorf, A. Yool, G. D. Farquhar, M. J. R. Fasham, M. L. Goulden, M. Heimann, V. J. Jaramillo, H. S. Kheshgi, C. Lé Quéré, R. J. Scholes, D. W. R. Wallace and I. C. Prentice, The Carbon Cycle and Atmospheric Carbon Dioxide, Intergovernmental Panel for Climate Change, https://www.ipcc.ch/site/assets/uploads/2018/02/TAR-03.pdf.
- D. Wei, R. Sang, P. Sponholz, H. Junge and M. Beller, Nat. Energy, 2022, 7, 438 CrossRef CAS.
- M. Z. do Valle Gomes, G. Masdeu, P. Eiring, A. Kuhlemann, M. Sauer, B. Åkerman and A. E. C. Palmqvist, Catal. Sci. Technol., 2021, 11, 6952 RSC.
- A. M. Klibanov, B. N. Alberti and S. E. Zale, Biotechnol. Bioeng., 1982, 24, 25 CrossRef CAS PubMed.
- H. Chen, Y. Huang, C. Sha, J. M. Moradian, Y.-C. Yong and Z. Fang, Renew. Sustain. Energy Rev., 2023, 178, 113270 CrossRef.
- Research Council of Finland, C1 Value Academy Programme https://www.aka.fi/en/research-funding/programmes-and-other-funding-schemes/academy-programmes/alasivut/academy-programmes-under-evaluation/c1-value-20202023/ Search PubMed.
- X. Lan, P. Tans and K. W. Thoning, Trends in globally-averaged CO2 determined from NOAA Global Monitoring Laboratory measurements, Version 2024-02, 2024, DOI:10.15138/9N0H-ZH07, data accessed on 5th March 2024.
- H. McLaughlin, A. A. Littlefield, M. Menefee, A. Kinzer, T. Hull, B. K. Sovacool, M. D. Bazilian, J. Kim and S. Griffiths, Renewable Sustainable Energy Rev., 2023, 177, 113215 CrossRef CAS.
- A. Satanowski and A. Bar-Even, EMBO Rep., 2020, 21, e50273 CrossRef CAS PubMed.
- Q. Hassan, I. D. J. Azzawi, A. Z. Sameen and H. M. Salman, Sustainability, 2023, 15, 11501 CrossRef CAS.
- Y. Kojima, Int. J. Hydrogen Energy, 2019, 44(33), 18179 CrossRef CAS.
- C. Kim, Y. Lee and K. Kim, Energies, 2023, 16(6), 2613 CrossRef CAS.
- J. J. Carroll, J. D. Slupsky and A. E. Mather, J. Phys. Chem. Ref. Data, 1991, 210(6), 1201 CrossRef.
- O. Yishai, S. N. Lindner, J. Gonzalez de la Cruz, H. Tenenboim and A. Bar-Even, Curr. Opin. Chem. Biol., 2016, 35, 1 CrossRef CAS PubMed.
- C. Hao, S. Wang, M. Li, L. Kang and X. Ma, Catal. Today, 2011, 160(1), 184 CrossRef CAS.
- C.-S. He, L. Gong, J. Zhang, P.-P. He and Y. Mu, J. CO2 Util., 2017, 19, 157 CrossRef CAS.
- Y. Himeda, S. Miyazawa and T. Hirose, ChemSusChem, 2011, 4(4), 487 CrossRef CAS PubMed.
- A. Weilhard, M. I. Qadir, V. Sans and J. Dupont, ACS Catal., 2018, 8(3), 1628 CrossRef CAS.
- G. Pietricola, C. Ottone, D. Fino and T. Tommasi, J. CO2 Util., 2020, 42, 101343 CrossRef CAS.
- S. Moret, P. J. Dyson and G. Laurenczy, Nat. Commun., 2014, 5, 4017 CrossRef CAS PubMed.
- J. C. Boyington, V. N. Gladyshev, S. V. Khangulov, T. C. Stadtman and P. D. Sun, Science, 1997, 275(5304), 1305 CrossRef CAS PubMed.
- A. Sharma, Y. Kawarabayasi and T. Satyanarayana, Extremophiles, 2012, 16, 1 CrossRef CAS PubMed.
- L. Calzadiaz-Ramirez and A. S. Meyer, Curr. Opin. Biotechnol., 2022, 73, 95 CrossRef CAS PubMed.
- R. Miyatani and Y. Amao, J. Mol. Catal. B Enzym., 2004, 27(2–3), 121 CrossRef CAS.
- Z. Zhang, T. Vasiliu, F. Li, A. Laaksonen, F. Mocci and X. Ji, J. CO2 Util., 2021, 52, 101679 CrossRef CAS.
- Y. Amao, J. CO2 Util., 2018, 26, 623 CrossRef CAS.
- H. Choe, J. C. Joo, D. H. Cho, M. H. Kim, S. H. Lee, K. D. Jung and Y. H. Kim, PLoS One, 2014, 9(7), e103111 CrossRef PubMed.
- B. R. Crable, C. M. Plugge, M. J. McInerney and A. J. M. Stams, Enzym. Res., 2011, 1, 532536 Search PubMed.
- F. Marpani, Z. Sárossy, M. Pinelo and A. S. Meyer, Biotechnol. Bioeng., 2017, 114, 2762 CrossRef CAS PubMed.
- F. M. Schwarz, K. Schuchmann and V. Müller, Biotechnol. Biofuels, 2018, 11, 237 CrossRef PubMed.
- M. Jormakka, B. Byrne and S. Iwata, Curr. Opin. Struct. Biol., 2003, 13(4), 418 CrossRef CAS PubMed.
- C. F. Nielsen, L. Lange and A. S. Meyer, Biotechnol. Adv., 2019, 37(5), 107408 CrossRef CAS PubMed.
- U. Deppenmeier, Prog. Nucleic Acid Res., 2002, 71, 223 CAS.
- C. E. Price and A. J. Driessen, Biochim. Biophys. Acta, Mol. Cell Res., 2010, 1803(6), 748 CrossRef CAS PubMed.
- A. A. Alekseeva, S. S. Savin and V. I. Tishkov, Acta Nat., 2011, 3(4), 38 CrossRef CAS.
- V. I. Tishkov and V. O. Popov, Biochemistry, 2004, 69(11), 1252 CAS.
- L. Calzadiaz-Ramirez, C. Calvó-Tusell, G. M. M. Stoffel, S. N. Lindner, S. Osuna, T. J. Erb, M. García-Borràs, A. Bar-Even and C. G. Acevedo-Rocha, ACS Catal., 2020, 10(14), 7512 CrossRef CAS.
- M. Takacs, O. V. Makhlynets, P. L. Tolbert and I. V. Korendovych, Protein Eng. Des. Sel., 2017, 30(3), 279 CrossRef.
- V. I. Tishkov and V. O. Popov, Biomol. Eng., 2006, 23, 89 CrossRef CAS PubMed.
- C. A. R. Cotton, C. Edlich-Muth and A. Bar-Even, Curr. Trends Biotechnol., 2018, 49, 49 CAS.
- M. Talvitie, Acid-tolerant enzymes and their biotechnical application in CO2 fixation, Bachelor thesis, Aalto Yliopisto, 2020 Search PubMed.
- H. C. A. Raaijmakers and M. J. Romão, J. Biol. Inorg. Chem., 2006, 11(7), 849 CrossRef CAS PubMed.
- R. Hille, J. Hall and P. Basu, Chem. Rev., 2014, 114(7), 3963 CrossRef CAS PubMed.
- H. Kumar, M. Khosraneh, S. S. M. Bandaru, C. Schulzke and S. Leimkühler, Molecules, 2023, 28, 1537 CrossRef CAS PubMed.
- P. Kaufmann, B. R. Duffus, B. Mitrova, C. Iobbi-Nivol, C. Teutloff, M. Nimtz, L. Jänsch, U. Wollenberger and S. Leimkühler, Biochemistry, 2018, 57(7), 1130 CrossRef CAS PubMed.
- S. Reschke, B. R. Duffus, P. Schrapers, S. Mebs, C. Teutloff, H. Dau, M. Haumann and S. Leimkühler, Biochemistry, 2019, 58(17), 2228 CrossRef CAS PubMed.
- D. Niks, J. Duvvuru, M. Escalona and R. Hille, J. Biol. Chem., 2016, 291(3), 1162 CrossRef CAS PubMed.
- K. Lewis, S. Epstein, A. D'Onofrio and L. L. Ling, J. Antibiot., 2010, 63, 468 CrossRef CAS.
- E. J. Stewart, J. Bacteriol., 2012, 194(16), 4151 CrossRef CAS PubMed.
- N. N. Nam, H. D. K. Do, K. T. L. Trinh and N. Y. Lee, Foods, 2023, 12(11), 2140 CrossRef CAS PubMed.
- E. P. Culligan, R. D. Sleator, J. R. Marchesi and C. Hill, Virulence, 2014, 5(3), 399 CrossRef PubMed.
- R. Li, Y. Wang, H. Hu, Y. Tan and Y. Ma, Nat. Commun., 2022, 13, 7978 CrossRef CAS PubMed.
- C. Pinske, M. Bonn, S. Kruger, U. Lindenstrauss and R. G. Sawers, PLoS One, 2011, 6(8), e22830 CrossRef CAS.
- C. Pinske, Front. Microbiol., 2018, 9, 1238 CrossRef.
- S. Sahin, R. Cai, R. D. Milton, S. Abdellaoui, F. C. Macazo and S. D. Minteer, J. Electrochem. Soc., 2018, 165(3), H109 CrossRef CAS.
- H. Stamatis, Multienzymatic Assemblies: Methods and Protocols (Methods in Molecular Biology), Springer US, Humana, edn. 1st. 2022 Search PubMed.
- F. Li, S. Scheller and M. Lienemann, J. CO2 Util., 2023, 77, 102608 CrossRef CAS.
- R. R. Jameson and A. M. Diamond, RNA, 2004, 10(7), 1142 CrossRef CAS PubMed.
- M. J. Axley, D. A. Grahame and T. C. Stadtman, J. Biol. Chem., 1990, 265(30), 18213 CrossRef CAS.
- T. C. Stadtman, J. N. Davis, W. M. Ching, F. Zinoni and A. Böck, Biofactors, 1991, 3(1), 21 CAS.
- E. Seol, Y. Jang, S. Kim, Y.-K. Oh and S. Park, Int. J. Hydrogen Energy, 2012, 37, 15045 CrossRef CAS.
- S. Ikeyama and Y. Amao, ChemCatChem, 2017, 9(5), 833 CrossRef CAS.
- S. Ikeyama, T. Katagiri and Y. Amao, J. Photochem. Photobiol., A, 2018, 358, 362 CrossRef CAS.
- D. S. Marlin, E. Sarron and Ó. Sigurbjörnsson, Front. Chem., 2018, 6, 446 CrossRef CAS PubMed.
- S. K. Kuk, R. K. Singh, D. H. Nam, R. Singh, J.-K. Lee and C. B. Park, Angew. Chem., Int. Ed., 2017, 56, 3827 CrossRef CAS PubMed.
- X. Wang, P. J. Ramírez, W. Liao, J. A. Rodriguez and P. Liu, J. Am. Chem. Soc., 2021, 143(33), 13103 CrossRef CAS PubMed.
- J. G. Driver, R. E. Owen, T. Makanyire, J. A. Lake, J. McGregor and P. Styring, Front. Energy Res., 2019, 7, 88 CrossRef.
- J. Ding, R. Ye, Y. Fu, Y. He, Y. Wu, Y. Zhang, Q. Zhong, H. H. Kun and M. Fan, Nat. Commun., 2023, 14, 4586 CrossRef CAS PubMed.
- T. Sakakura and K. Kohno, Chem. Commun., 2009, 11, 1312 RSC.
- Q. Liu, L. Wu, R. Jackstell and M. Beller, Nat. Commun., 2015, 6, 5933 CrossRef PubMed.
- M. Franz, T. Stalling, H. Steinert and J. Martens, Org. Biomol. Chem., 2018, 16, 8292 RSC.
- R. Villa, S. Nieto, A. Donaire and P. Lozano, Molecules, 2023, 28, 5520 CrossRef CAS PubMed.
- P. Fröhlich, K. Albert and M. Bertau, Org. Biomol. Chem., 2011, 9, 7941 RSC.
- T. Hartmann, N. Schwanhold and S. Leimkühler, Biochim. Biophys. Acta, Proteins Proteomics, 2015, 1854(9), 1090 CrossRef CAS PubMed.
- J. Vaitla, Y. Guttormsen, J. K. Mannisto, A. Nova, T. Repo, A. Bayer and K. H. Hopmann, ACS Catal., 2017, 7, 7231 CrossRef CAS.
- T. Niemi and T. Repo, Eur. J. Org Chem., 2019, 1180 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |