Open Access Article
Open Access ArticleBioorthogonal probes for L-form conversion visualization and insights into antimicrobial resistance†
Yunzhe
Tao
a,
Yongwei
Feng
ba,
Yu
Peng
a,
Xiang
Wang
a,
Xiangchuan
Meng
a,
Youjun
Xu
 b,
Xiaowan
Han
*a,
Qingyang
Zhang
b,
Xiaowan
Han
*a,
Qingyang
Zhang
 *a and
Hai-Yu
Hu
*a and
Hai-Yu
Hu
 *a
*a
aState Key Laboratory of Bioactive Substance and Function of Natural Medicines, Institute of Materia Medica, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100050, China. E-mail: hanxiaowan@imm.ac.cn; zhangqingyang@imm.ac.cn; haiyu.hu@imm.ac.cn
bSchool of Pharmaceutical Engineering, Key Laboratory of Structure-Based Drug Design & Discovery (Ministry of Education), Shenyang Pharmaceutical University, Shenyang, 110016, China
First published on 31st May 2025
Abstract
Cell wall-deficient bacteria (CWDB) are key contributors to antimicrobial resistance (AMR), enabling persistent infections by evading antibiotics through their transition to L-form states. Therefore, molecular tools for detecting L-form conversion and AMR mechanisms are crucial for developing novel strategies against bacterial infections. Herein, we present the development of small-sized, peptidoglycan-specific fluorogenic probes employing a two-step bioorthogonal strategy that enables real-time visualization of CWDB formation. Tz-FL-S rapidly reacts with the novel D-alanine derivative TCO-D-Ala at a rate of (2.61 ± 0.07) × 103 M−1 s−1, resulting in a 4.9-fold increase in fluorescence intensity. This platform exhibited excellent labeling of peptidoglycan in both Gram-positive and Gram-negative bacteria (signal-to-noise ratio: 15 to 305), effectively capturing the transition from N-form to L-form. Furthermore, we investigated the impact of 14 kinds of antibiotics on L-form conversion and found 13 of them induced CWDB. Besides, we explored the relationship between L-form conversion and AMR. This research enhances our understanding of bacterial adaptations and resistance mechanisms, paving the way for innovative strategies to combat drug-resistant infections.
Introduction
Antimicrobial resistance (AMR) has emerged as a major challenge to global public health.1 Recent studies have identified cell wall-deficient bacteria (CWDB) as one of the key contributors to AMR,2,3 frequently causing chronic and recurrent clinical infections.4,5 Under survival pressures, some bacteria partially or completely lose their cell walls, converting to a cell wall-deficient (L-form) state.6 This adaptation allowed them to evade cell wall-targeting antibiotics, such as β-lactams, making L-form bacteria resistant to many conventional treatments.7 Moreover, research showed that CWDB could even evade phage therapy,8–10 further complicating treatment strategies. A deeper understanding of the biological characteristics of CWDB and the molecular mechanisms underlying the transition from normal (N-form) to cell wall-deficient (L-form) bacteria—including when, why, and how this transition occurred is crucial for developing innovative antibacterial strategies and reversing the rising trend of antimicrobial resistance. Bacterial cell wall defects are typically visualized and analyzed using scanning electron microscopy (SEM) and transmission electron microscopy (TEM).11–13 Besides, phase contrast microscopy has been also employed to observe the L-form conversion.4,10 However, SEM and TEM are limited by complex sample preparation, high equipment costs, and the inability to visualize live bacteria, and phase contrast microscopy lacks the spatial resolution for observing peptidoglycan. In contrast, confocal laser scanning microscopy (CLSM) offers advantages such as high sensitivity and excellent spatial and temporal resolution,14,15 making it a promising tool for monitoring the conversion of L-form bacteria in live cells. Despite the progress of AMR sensing technologies,16–19 the development of effective fluorescent probes capable of visualizing the transition from N-form to L-form bacteria in real time remains a significant challenge. The unique structure of the bacterial cell wall, particularly the outer membrane barrier in Gram-negative bacteria, blocks the penetration of conventional fluorescent probes into live cells.20 Therefore, developing novel probe design strategies that allow for specific and efficient labeling of both Gram-positive and Gram-negative bacterial cell walls is crucial. When combined with modern fluorescence imaging techniques, these probes would enable the real-time visualization of CWDB and the transition from N-form to L-form, providing valuable insights into bacterial physiology and offering potential opportunities for addressing antimicrobial resistance.Peptidoglycan (PGN), a key component of the bacterial cell envelope in both Gram-positive and Gram-negative bacteria, is essential for maintaining cell wall structural integrity.21,22 The use of D-amino acid derivatives for metabolic labeling of bacterial peptidoglycan, as first demonstrated by Prof. Bertozzi and others, is an effective approach.23,24 However, the labeling of peptidoglycan in Gram-negative bacteria remains a challenge because their outer membrane acts as a strong barrier that limits the entry of external compounds, including chemical probes.25 The large size of these probes further reduces their ability to accumulate inside the cells. Consistently, studies have shown that D-alanine derivatives used for metabolic labeling have difficulty penetrating Gram-negative bacterial cells.26,27 In this work, we developed small-sized, peptidoglycan-specific fluorogenic probes to enable real-time visualization of CWDB. Given that small-molecular-weight compounds penetrate bacterial cell walls more effectively,27,28 we employed the two-step bioorthogonal strategy to minimize probe size (Scheme 1A). This approach deconstructed the traditional one-step fluorescent probe into two smaller components: a D-alanine trans-cyclooctene (TCO) derivative as an anchor and a tetrazine-tagged fluorophore. During bacterial biosynthesis, TCO-D-Ala metabolically incorporates at the terminus of the stem peptides in PGN,29 while the tetrazine moiety quenches fluorescence until it undergoes a click reaction with PGN, activating the fluorophore at the bacterial cell wall and enabling no-wash fluorescence imaging. We synthesized clickable probes with various fluorophores, and after screening and optimization, Tz-FL-S exhibited excellent labeling of PGN, successfully capturing the transition from N-form to L-form. Furthermore, we investigated the influence of various antibiotics on L-form conversion and explored the relationship between CWDB and antimicrobial resistance. Notably, by comparing the L-form conversions of drug-susceptible and drug-resistant strains, we found that antimicrobial resistance delayed bacterial L-form conversion, likely due to the increased antibiotic tolerance of drug-resistant strains (Scheme 1B). This research provides valuable insights into bacterial physiological adaptations and resistance mechanisms, contributing to the development of new strategies to combat drug-resistant bacterial infections.
Results and discussion
Design and synthesis of probes
In designing targeted molecules derived from D-amino acids, we employed trans-cyclooctene as the click handle, linking it to the D-alanine structure to obtain TCO-D-Ala. We then synthesized a series of tetrazine-tagged clickable probes with various fluorophores: the Cy7-based probe Tz-CY, the naphthalimide-based probe Tz-NA and the fluorescein-based Tz-FL. Cyanine dye was chosen for its near-infrared (NIR) emission, which allows deeper tissue penetration and reduced background autofluorescence for in vivo imaging.30 Naphthalimide–carbazole fluorophore was selected for its two-photon excitation capability and thermally activated delayed fluorescence (TADF), making it well-suited for both two-photon and time-resolved imaging.31 Fluorescein was chosen for its high quantum yield, good water solubility, and proven biocompatibility.32 We further reduced the molecular weight of Tz-FL, resulting in the development of Tz-FL-S, which optimized its labeling efficiency and ensured robust performance in complex biological environments.The main synthesis is outlined in Scheme S1.† The targeted D-alanine derivative, TCO-D-Ala, was synthesized by reacting the active p-nitrophenyl esters of trans-cyclooctene with modified D-Ala, followed by a deprotection step. Subsequently, we synthesized the tetrazine intermediate, compound 6, according to relevant literature.33 Using this intermediate, we then conjugated it with cyanine, naphthalimide, and fluorescein through 1–2 substitution and coupling reactions to obtain the probes Tz-CY, Tz-NA, and Tz-FL, respectively. Finally, we synthesized a smaller probe, Tz-FL-S, using a simplified tetrazine fragment, compound 15.34 The detailed synthetic procedures and analytical data for the compounds are provided in the ESI.†
The “off–on” fluorescence of Tz-CY, Tz-NA, and Tz-FL
Firstly, we examined the spectral responses of Tz-CY, Tz-NA, and Tz-FL upon reacting with TCO-D-Ala in pH 7.4 phosphate-buffered saline (PBS). As shown in Fig. S1,† the absorption spectra of the three probes exhibited minimal changes after their reaction with TCO-D-Ala, indicating that there was no significant alteration in the chromophore structure. As expected, all three probes demonstrated a pronounced quenching effect of the tetrazine moiety on the chromophore (Fig. 1). Upon the addition of TCO-D-Ala in PBS, the fluorescence intensity of Tz-CY and Tz-FL increased by 7.2-fold at 764 nm and 7.4-fold at 520 nm, respectively (Fig. 1A and D). In contrast, Tz-NA exhibited only a slight fluorescence recovery, which could be attributed to its high lipophilicity potentially inducing aggregation in PBS, thereby diminishing its reactivity with TCO-D-Ala (Fig. 1B). To investigate this hypothesis, we subsequently performed a reaction between Tz-NA and TCO-D-Ala in dimethyl sulfoxide (DMSO). The results indicated a favorable reaction in this solvent, with Tz-NA's fluorescence intensity recovering by up to 21-fold at 590 nm (Fig. 1C). Liquid chromatography-mass spectrometry (LC-MS) analysis further confirmed that the probes Tz-CY, Tz-NA, and Tz-FL could efficiently undergo bioorthogonal reactions with TCO-D-Ala, yielding the corresponding products.Labeling discriminations on Gram-positive and Gram-negative bacteria
Subsequently, we employed S. aureus ATCC 29213, E. coli ATCC 25922, and B. subtilis ATCC 6633 as models to assess the labeling efficacy of our probes for both Gram-positive and Gram-negative bacteria.24 The treated group was treated with 1 mM TCO-D-Ala, whereas the control group received no treatment with TCO-D-Ala. After reaching the logarithmic growth phase, bacteria were subjected to secondary labeling with Tz-CY, Tz-NA, and Tz-FL respectively. As shown in Fig. 1E, in the treated group, Tz-CY demonstrated labeling capability on Gram-positive bacteria, including S. aureus and B. subtilis (Fig. S2†); however, it failed to achieve specific labeling toward TCO-D-Ala, as indicated by fluorescence in the control group. This lack of specificity may be due to the predominantly negatively charged surfaces of bacteria, which attract cations and result in non-specific adsorption that hinders specific labeling of the cell wall.35 Additionally, Tz-CY showed poor labeling of the Gram-negative E. coli, suggesting limited penetration through the outer membrane. Tz-NA displayed no significant labeling on any of the bacterial strains, likely due to its poor solubility (Fig. 2F). In contrast, Tz-FL produced bright labeling in both Gram-positive strains, but showed weak labeling effect on E. coli (Fig. 2G and H). Overall, our results indicate that Tz-CY and Tz-NA did not meet the expected labeling efficacy for TCO-D-Ala in the bacterial PGN, while the better Tz-FL effectively imaged Gram-positive bacteria but struggled with Gram-negative bacteria, likely due to the additional outer membrane that necessitates improved probe penetration.36,37 | ||
| Fig. 2 Probe optimization. (A) Strategy to optimize Tz-FL into Tz-FL-S by removing the benzene ring in the linker and the methyl group from the tetrazine head. (B) Fluorescence spectra of Tz-FL-S before and after reacting with TCO-D-Ala in pH 7.4 PBS, λex = 488 nm. (C) Plot of fluorescence intensity at 520 nm versus time. Tz-FL-S exhibited a faster reaction than Tz-FL. λex = 488 nm. (D) Labeling of S. aureus ATCC 29213, B. subtilis ATCC 6633, and E. coli ATCC 25922 with Tz-FL or Tz-FL-S. Green fluorescence images for each strain were collected and processed under the same conditions. For Hoechst staining images in the Tz-FL-S group, see Fig. S3C.† Scale bar: 5 μm. (E), (F), and (G) mean fluorescence intensity of bacteria labeled by Tz-FL and Tz-FL-S from panel (D) (n ≥ 30). Data are represented as mean ± SD and analyzed using a two-tailed Mann–Whitney U test. ****P < 0.0001. | ||
Optimized Tz-FL-S had a better performance than Tz-FL
To enhance penetration in Gram-negative bacteria, we synthesized a smaller probe, Tz-FL-S, using a simplified tetrazine fragment (Fig. 2A). Spectral response analysis of Tz-FL-S revealed similar absorbance curves before and after incubation with TCO-D-Ala (Fig. S3A†). Notably, the tetrazine moiety maintained a significant quenching effect on fluorescein. Following the reaction with TCO-D-Ala, the fluorescence intensity increased by 4.9-fold at 520 nm (Fig. 2B), and LC-MS analysis confirmed the presence of bioorthogonal products (Fig. S3B†).To investigate the fluorescence quenching mechanism, we conducted density functional theory (DFT) and time-dependent DFT (TD-DFT) calculations on Tz-FL and Tz-FL-S. The results aligned well with the Energy Transfer to a Dark State (ETDS) quenching mechanism (Fig. S4A and B†).38 Our calculations demonstrated that the excited-state energy transferred from the fluorophore to the tetrazine moiety, followed by internal conversion to the low-energy dark state (S1) of tetrazine, ultimately returning to the ground state through non-radiative decay processes. The click reaction with the tetrazine group disrupted ETDS process, restoring fluorescence emission from the fluorophores. Specifically, Tz-FL-S exhibited a low-lying dark state (S1) with a negligible oscillator strength (f = 0.005). After the reaction with TCO-D-Ala, photoexcitation from S0 to S1 was primarily driven by the π → π* transition of the fluorophore moiety, resulting in a substantial increase in oscillator strength (f = 0.520). Fluorescence turn-on efficiency depended on the distance between the fluorophore and tetrazine.38Tz-FL-S had a longer distance between the fluorescein and tetrazine groups than Tz-FL (Fig. S4C†), which likely caused its slightly lower fluorescence recovery.
Furthermore, we determined the reaction kinetics between TCO-D-Ala and either Tz-FL or Tz-FL-S by monitoring the fluorescence increase at 520 nm over time. Upon mixing the tetrazine probes with TCO-D-Ala, a rapid fluorescence activation was observed. The kinetic plots revealed that Tz-FL-S reached its fluorescence plateau within approximately 20 seconds, whereas Tz-FL required around 120 seconds (Fig. 2C). Curve fitting based on a pseudo-first-order reaction model (Fig. S3E and F†) yielded a rate constant of (2.61 ± 0.07) × 103 M−1 s−1 for Tz-FL-S and (5.08 ± 0.04) × 102 M−1 s−1 for Tz-FL. The corresponding half-lives t1/2 were 3.3 ± 0.1 s for Tz-FL-S and 17.0 ± 0.1 s for Tz-FL, respectively. Thus, the reaction rate constant of Tz-FL-S is approximately 5.2 times higher than that of Tz-FL. Studies showed that the H-tetrazine reacted significantly faster than the Me-tetrazine,39–41 and our results are consistent with this trend. More importantly, Tz-FL-S exhibited superior labeling capabilities for both Gram-positive and Gram-negative bacteria compared to Tz-FL (Fig. 2D–G). It showed strong fluorescent signals on S. aureus and B. subtilis, providing specific, brighter, and more uniform labeling for the Gram-negative bacterium E. coli, while the negative control displayed no obvious signal (Fig. S3D†). In Fig. 2G, for the treated E. coli groups, the fluorescence intensity (mean ± SD) for Tz-FL is 6.5 ± 4.3, and for Tz-FL-S, it is 26.0 ± 8.3. Uniformity was assessed using the relative standard deviation (RSD), with Tz-FL-S demonstrating a lower RSD value (0.3 vs. 0.6 for Tz-FL), indicating more consistent labeling. Moreover, we quantitatively analysed TCO-D-Ala uptake in E. coli, B. subtilis, and S. aureus by plotting fluorescence intensity normalized to OD600 against TCO-D-Ala concentration following Tz-FL-S labeling. In E. coli, the uptake profile displayed a saturation curve characteristic of enzyme-catalysed kinetics, suggesting that TCO-D-Ala likely enters the same metabolic pathway as other D-alanine derivatives via transpeptidases.29,42 Fitting the data to the Michaelis–Menten model yielded a Km value of 17 μM for E. coli (Fig. S3G†). The uptake data for B. subtilis and S. aureus exhibited linear relationships within the tested concentration range, suggesting that their Km values exceed the highest concentration tested (1 mM, Fig. S3H and I†). Although exact Km values for B. subtilis and S. aureus could not be determined, the slope of the linear fit is expected to reflect their relative catalytic efficiencies (Vmax/Km). Comparative analysis revealed that TCO-D-Ala incorporation is 4–24 times more efficient in E. coli. Additionally, we assessed the fluorescence stability of the clicked product formed between TCO-D-Ala and Tz-FL-S, and observed no significant decrease in fluorescence intensity after 48 hours at 30 °C (Fig. S3J†). These results indicated that, through molecule optimization, TCO-D-Ala and Tz-FL-S effectively labeled bacterial cell walls and showed strong potential for monitoring bacterial L-form conversion, leading to its selection for further studies on CWDB.
Visualizing bacterial L-form conversion process by fluorescence imaging
During L-form conversion, bacteria undergo significant morphological changes. Although phase contrast microscopy has been employed to monitor this process in some bacterial studies, changes in peptidoglycan molecules remain unclear.10,43,44 To visualize these changes, we used Tz-FL-S with TCO-D-Ala for labeling peptidoglycan in three strains of Gram-positive bacteria (B. subtilis, S. aureus RN 4220, and ATCC 29213) and one strain of Gram-negative bacterium (E. coli ATCC 25922). Hoechst 33342 was also used to label nucleoids. L-form conversion was induced using established agents—penicillin G, lysozyme, lysostaphin, and fosfomycin43,45—and monitored with CLSM (Fig. 3A), with the conversion stages for each strain classified as early, middle, late, and very late.In B. subtilis (Fig. 3B), the early stage exhibited clear labeling of the cell wall structure (green) with Hoechst 33342 staining the nucleoids (blue). As conversion advanced to the middle stage, cracks seemly appeared in the cell wall, leading to bulging, while nucleoids began to extrude. In the late stage, nucleoids exited the peptidoglycan capsule, and the cell wall nearly disappeared. Even in the absence of the cell wall, the cell membrane continued to expand, producing enlarged CWDB in the very late stage, consistent with the literature.46 In S. aureus RN 4220 (Fig. 3C), the conversion process proceeded quickly, with cells displaying bulging during the middle stage before fully detaching from the cell wall to form CWDB. In contrast, S. aureus ATCC 29213 (Fig. 3D) underwent a more gradual reduction in peptidoglycan, evidenced by diminishing cell wall fluorescence and an increase in cell size rather than immediate bulging. For E. coli (Fig. 3E), the process began with cracking in the middle stage, leading to bulging and bending at the cell ends in the late stage. Eventually, nucleoids were squeezed out, resulting in CWDB formation. Interestingly, some late-stage bulges appeared empty, possibly due to vacuole formations known to occur in E. coli.47 We observed that L-form conversion did not occur uniformly among individual bacteria, likely due to variations in bacterial states. Using the fluorogenic bioorthogonal probe Tz-FL-S in combination with TCO-D-Ala, our study effectively captured the temporal and spatial dynamics of key molecular events associated with cell wall loss during bacterial L-form conversion. This approach provided a more accurate and comprehensive view than previous detection methods.
CWDB formed under the induction of different antibiotics
L-form bacteria are capable of evading most cell wall-targeting antibiotics, such as β-lactams, which can contribute to antibiotic resistance. However, their response to other classes of antibiotics remains less understood.2 To thoroughly investigate the impact of antibiotic challenges on bacterial cell walls and to assess the capacity for CWDB formation, we selected 14 antibiotics with distinct antibacterial mechanisms for testing on S. aureus ATCC 29213. These antibiotics target various pathways, including penicillin-binding proteins (PBPs), bacterial protein synthesis, folic acid synthesis, and the synthesis of DNA, RNA, and peptidoglycan.48 We acquired confocal fluorescence images to visualize CWDB formation under 14 different antibiotic treatments, with representative images shown in Fig. 4, and performed quantitative fluorescence analysis (Fig. S6†). Our results demonstrate that all tested antibiotics, except vancomycin, induced CWDB formation. Treatment with fosfomycin at a concentration of 10× MIC continued to induce swollen CWDB (Fig. S5A†). For the antibiotics targeting PBPs (Fig. 4A), CWDB were observed under each treatment. Fluorescent images revealed bright green fusiform peptidoglycan shells or fragments (indicated by white arrows), which may be shed during the transition to CWDB. Upon treatment with ampicillin or penicillin G, almost all bacteria lost their cell walls and transformed into CWDB, while those treated with cefoperazone or meropenem retained visible, weak or strong, green fluorescence. Distinctly swollen cells were observed under ampicillin, cefoperazone, and meropenem treatment (indicated by orange arrows). Antibiotics that inhibit bacterial protein synthesis, such as linezolid, azithromycin, clindamycin, and amikacin (Fig. 4B), ultimately led to bacterial conversion into CWDB. This conversion likely occurred as these antibiotics initially bound to ribosomal subunits, directly disrupting protein synthesis and causing subsequent metabolic disturbances in the cell wall. Similarly, sulfamethoxazole (SMZ) and trimethoprim (TMP), which inhibit folic acid synthesis and thereby block nucleic acid synthesis, also prompted the majority of bacteria to become CWDB (Fig. 4C). Notably, trimethoprim alone produced a comparable effect (Fig. S5B†). Interestingly, we found that levofloxacin, which directly inhibits DNA metabolism, likely affected cell wall synthesis by hindering the transcription process and consequently blocking protein synthesis,49 leading to CWDB formation along with the loss of the cell wall (Fig. 4D). Additionally, rifamycin, which binds to RNA polymerase to interfere with RNA synthesis,50 likely impaired downstream protein synthesis, resulting in the formation of CWDB to some extent, as evidenced by the dim and uneven green fluorescence observed (Fig. 4E). It can be inferred that damage to the robust cell wall is a fundamental mechanism by which antibiotics induce bacterial rupture, either directly or indirectly. In studies involving vancomycin, we observed growth inhibition rather than a significant bactericidal effect, as many bacteria remained brightly labeled on their cell walls (Fig. 4F). This may be due to the extensive modification of D-Ala-D-Ala by the probes, which could hinder vancomycin binding and indirectly contribute to drug resistance.51 As a limitation, probes based on D-alanine derivatives may not be suitable for studies involving vancomycin family drugs that bind to D-Ala-D-Ala. Nonetheless, the lack of L-form conversion observed suggests that bacteria may develop resistance to vancomycin by modifying their D-Ala-D-Ala structure, as previously reported.52 Our findings suggest that CWDB formation can be induced not only by antibiotics that directly target the cell wall but also by those that act on other biological processes.Antimicrobial resistance delayed the bacterial L-form conversion
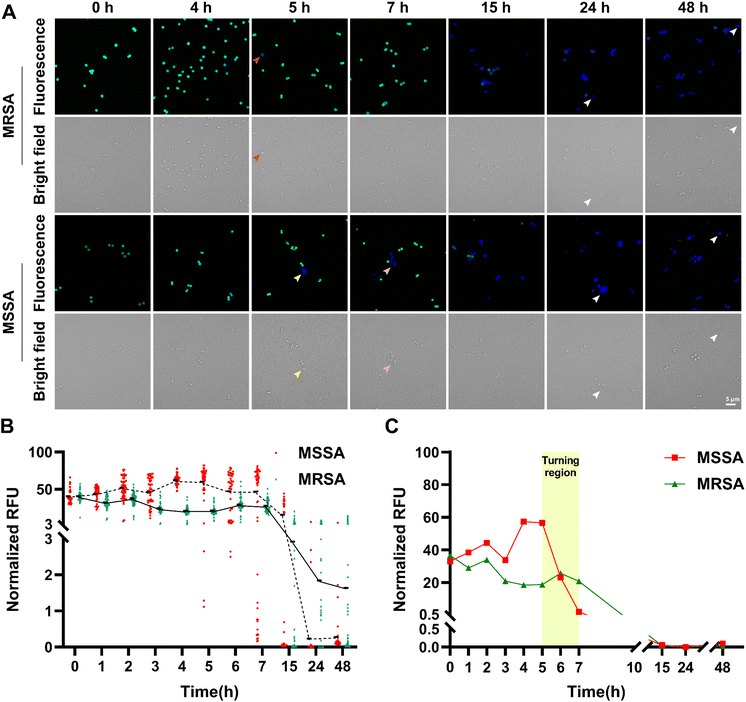
Mutations that reduce drug binding to its target and increased expression of efflux pumps contribute to antibiotic resistance.3 Additionally, tolerance, a mechanism in which non-growing or slow-growing bacteria can survive exposure to antibiotics targeting actively growing cells, highlights the complexity of antibiotic resistance.53 To further explore the relationship between antimicrobial resistance and L-form conversion, we treated methicillin-resistant S. aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) with ampicillin at 10× MIC respectively and monitored bacteria utilizing our probes (Fig. 5 and Table S1†). Although the drug-resistant strain eventually turned into CWDB, it converted more slowly than the drug-susceptible strain. From 0 h to 4 h, no obvious CWDB appeared in either strain. By 5 h, inflated CWDB completely losing their cell wall appeared in the drug-susceptible strain (yellow arrow), while just few cells in the drug-resistant strain exhibited partial loss of cell wall and inflation (orange arrow). At 7 h, the drug-susceptible strain exhibited more CWDB with lost cell wall than at 5 h (pink arrow), while the drug-resistant strain showed few CWDB, retaining a significant amount of peptidoglycan. By 15 h, both strains showed substantial CWDB. Additionally, at 24 h and 48 h, fragile and ruptured CWDB (white arrow) appeared, nearly invisible in the bright field but exhibiting diffuse blue fluorescence. To better illustrate the differences in L-form transition rates between resistant and susceptible strains, we quantified the mean fluorescence intensity of individual bacteria from multiple images over time. Fluorescence intensity was plotted against time (Fig. 5B), and the first quartile (Q1) values at each time point were compared to assess the timing of CWDB formation in MSSA and MRSA (Fig. 5C). The Q1 value represents the fluorescence intensity below which 25% of the bacterial population falls. Notably, a transition phase was observed between 5 h and 7 h, during which MRSA retained higher peptidoglycan levels and exhibited a significantly delayed conversion compared to MSSA. Despite exposure to a high antibiotic concentration, the drug-resistant bacteria took longer to exhibit conversion, suggesting that this delay may be due to antibiotic tolerance. To further investigate this, we applied a higher antibiotic concentration and slightly extended the treatment duration. When treated with a higher concentration of 50× MIC, the drug-resistant strain retained much peptidoglycan at 8 h, but eventually lost its cell wall, forming CWDB (Fig. S7A†). The conversion process can vary under different inducing conditions depending on the strain.43 Therefore, we further induced conversion under a solid medium condition. Under solid medium conditions at 50× MIC, the drug-resistant strain exhibited cell wall loss later than the drug-susceptible strain. At 30 h, the drug-susceptible strain converted into large, expanded CWDB (Fig. S7B†), while the drug-resistant strain only showed weakened cell wall fluorescence (Fig. S7C†). The results indicated that antimicrobial resistance delayed L-form conversion in MRSA. In our results, this delayed conversion may be attributed to the antibiotic tolerance of drug-resistant strains. These strains took longer to lose their cell walls under antibiotic exposure, regardless of the concentration used. Studies have indicated that antibiotic tolerance in E. coli can precede and promote antibiotic resistance.54 However, the interaction between bacterial resistance and tolerance mutations remains underexplored. Our findings suggest potential synergistic interactions between antibiotic resistance and tolerance.55 Nonetheless, the molecular mechanisms underlying antibiotic tolerance, especially in S. aureus, remain unclear. The application of fluorescent probes is anticipated to provide valuable insights into the study of bacterial tolerance.Conclusion
CWDB, formed during the bacterial transition from the N-form to the L-form, plays a crucial role in enhancing bacterial drug resistance and viability. We employed a two-step bioorthogonal labeling strategy to design and synthesize TCO-D-Ala, along with a series of tetrazine-tagged, self-quenched fluorescent probes, ultimately selecting Tz-FL-S as the optimal probe. TCO-D-Ala was metabolically incorporated in bacterial peptidoglycan, and, together with Tz-FL-S, formed the fastest-reacting D-Ala click pair. This rapid reaction enabled bright, uniform labeling of peptidoglycan in both Gram-positive and Gram-negative bacteria, allowing us to capture peptidoglycan dynamics during L-form conversion. Moreover, our bioorthogonal probes served as highly effective sensors, enabling further investigation into bacterial L-form conversion and its relationship with antimicrobial resistance. We discovered that nearly all antibiotics disrupting peptidoglycan homeostasis—whether directly or indirectly—can induce CWDB to varying degrees. Additionally, we observed that antimicrobial resistance delayed bacterial L-form conversion, likely due to the enhanced tolerance of drug-resistant strains. These findings provide valuable insights into the patterns of bacterial L-form conversion and the underlying mechanisms of antimicrobial resistance, advancing global efforts to combat bacterial infections. Together, our probes offer powerful tools for imaging peptidoglycan dynamics, investigating antimicrobial resistance, and studying bacterial adaptation. These tools hold significant potential for advancing microbiological resistance studies, diagnostics, and therapeutic strategies.Abbreviations
| AMR | Antimicrobial Resistance |
| ATCC | American Type Culture Collection |
| B. subtilis | Bacillus subtilis |
| CLSM | Confocal Laser Scanning Microscopy |
| CWDB | Cell Wall-Deficient Bacteria |
| D-Ala | D-Alanine |
| DFT | Density Functional Theory |
| DMSO | Dimethyl Sulfoxide |
| DNA | Deoxyribonucleic Acid |
| E. coli | Escherichia coli |
| ETDS | Energy Transfer to a Dark State |
| FOS | Fosfomycin |
| LC-MS | Liquid Chromatography-Mass Spectrometry |
| MRSA | Methicillin-Resistant Staphylococcus aureus |
| MSM | Magnesium-Sucrose-Maleic Acid |
| MSSA | Methicillin-Susceptible Staphylococcus aureus |
| NB | Nutrient Broth |
| NIR | Near-Infrared |
| NMR | Nuclear Magnetic Resonance |
| OD | Optical Density |
| PBPs | Penicillin-Binding Proteins |
| PBS | Phosphate-Buffered Saline |
| PGN | Peptidoglycan |
| RFU | Relative Fluorescence Intensity |
| RNA | Ribonucleic Acid |
| RSD | Relative Standard Deviation |
| S. aureus | Staphylococcus aureus |
| SEM | Scanning Electron Microscopy |
| SD | Standard Deviation |
| SMZ | Sulfamethoxazole |
| TADF | Thermally Activated Delayed Fluorescence |
| TCO | trans-Cyclooctene |
| TD-DFT | Time-Dependent Density Functional Theory |
| TEM | Transmission Electron Microscopy |
| TMP | Trimethoprim |
Data availability
The data are available in the ESI† or upon request from the authors.Author contributions
Yunzhe Tao: investigation, methodology, formal analysis, writing – original draft, Yongwei Feng: investigation, writing – original draft, Yu Peng: software, writing – original draft, Xiang Wang: software, writing – original draft, Xiangchuan Meng: investigation, writing – original draft, Youjun Xu: writing – review and editing, Xiaowan Han: methodology, writing – review and editing, Qingyang Zhang: methodology, writing – review and editing, Hai-Yu Hu: conceptualization, supervision, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge Dr Yuansheng Sun and Dr Hailin Qiu from ISS for assistance with confocal imaging experiments and data analyses. We also thank Dr Luping Liu, Dr Guang Li, Dr Licheng Yang and Dr Tiantai Zhang from IMM for their insightful discussions and generous support in providing space and equipment. And we thank the IMM Network and Information Center for the assistance of theoretical calculations, especially Dr Yue Zhou and Dr Xueshi Mao. This work was partially supported by the National Natural Science Foundation of China (NSFC) projects (Grant 22477139, 22277143, and 82473886), Beijing Natural Science Foundation (2242025) and CAMS Innovation Fund for Medical Sciences (CIFMS) (Grant 2021-I2M-1-054).References
- C. S. Ho, C. T. H. Wong, T. T. Aung, R. Lakshminarayanan, J. S. Mehta, S. Rauz, A. McNally, B. Kintses, S. J. Peacock, C. D. L. Fuente-Nunez, R. E. W. Hancock and D. S. J. Ting, Antimicrobial Resistance: A Concise Update, Lancet Microbe, 2024, 100947 Search PubMed.
- J. J. Lazenby, E. S. Li and C. B. Whitchurch, Cell Wall Deficiency – an Alternate Bacterial Lifestyle?, Microbiology, 2022, 168(8), 1218 CrossRef CAS PubMed.
- E. M. Darby, E. Trampari, P. Siasat, M. S. Gaya, I. Alav, M. A. Webber and J. M. A. Blair, Molecular Mechanisms of Antibiotic Resistance Revisited, Nat. Rev. Microbiol., 2023, 21(5), 280–295 CrossRef CAS PubMed.
- K. M. Mickiewicz, Y. Kawai, L. Drage, M. C. Gomes, F. Davison, R. Pickard, J. Hall, S. Mostowy, P. D. Aldridge and J. Errington, Possible Role of L-Form Switching in Recurrent Urinary Tract Infection, Nat. Commun., 2019, 10(1), 4379 CrossRef PubMed.
- J. Errington, K. Mickiewicz, Y. Kawai and L. J. Wu, L-Form Bacteria, Chronic Diseases and the Origins of Life, Philos. Trans. R. Soc., B, 2016, 371(1707), 20150494 CrossRef PubMed.
- D. Claessen and J. Errington, Cell Wall Deficiency as a Coping Strategy for Stress, Trends Microbiol., 2019, 27(12), 1025–1033 CrossRef CAS PubMed.
- L. G. Monahan, L. Turnbull, S. R. Osvath, D. Birch, I. G. Charles and C. B. Whitchurch, Rapid Conversion of Pseudomonas Aeruginosa to a Spherical Cell Morphotype Facilitates Tolerance to Carbapenems and Penicillins but Increases Susceptibility to Antimicrobial Peptides, Antimicrob. Agents Chemother., 2014, 58(4), 1956–1962 CrossRef PubMed.
- V. Ongenae, A. S. Mabrouk, M. Crooijmans, D. Rozen, A. Briegel and D. Claessen, Reversible Bacteriophage Resistance by Shedding the Bacterial Cell Wall, Open Biol. J., 2022, 12(6), 210379 CrossRef CAS PubMed.
- A. Petrovic Fabijan, D. Martinez-Martin, C. Venturini, K. Mickiewicz, N. Flores-Rodriguez, J. Errington and J. Iredell, L-Form Switching in Escherichia Coli as a Common β-Lactam Resistance Mechanism, Microbiol. Spectr., 2022, 10(5), e02419–e02422 Search PubMed.
- J. C. Wohlfarth, M. Feldmüller, A. Schneller, S. Kilcher, M. Burkolter, S. Meile, M. Pilhofer, M. Schuppler and M. J. Loessner, L-Form Conversion in Gram-Positive Bacteria Enables Escape from Phage Infection, Nat. Microbiol., 2023, 8(3), 387–399 CAS.
- G. Slavchev, L. Michailova and N. Markova, L-Form Transformation Phenomenon in Mycobacterium Tuberculosis Associated with Drug Tolerance to Ethambutol, Int. J. Microbiol., 2016, 5(4), 454–459 Search PubMed.
- J. J. Jackson and H. Kropp, Differences in Mode of Action of (β-Lactam Antibiotics Influence Morphology), LPS Release and in Vivo Antibiotic Efficacy, J. Endotoxin Res., 1996, 3(3), 201–218 Search PubMed.
- Y. Sumita, M. Fukasawa and T. Okuda, Comparison of Two Carbapenems, SM-7338 and Imipenem. Affinities for Penicillin-Binding Proteins and Morphological Changes, J. Antibiot., 1990, 43(3), 314–320 CrossRef CAS PubMed.
- S. W. Paddock, Principles and Practices of Laser Scanning Confocal Microscopy, Mol. Biotechnol., 2000, 16(2), 127–149 CrossRef CAS PubMed.
- S. W. Paddock, Confocal Laser Scanning Microscopy, BioTechniques, 1999, 27(5), 992–1004 CrossRef CAS PubMed.
- K. J. Fitzpatrick, H. J. Rohlf, T. D. Sutherland, K. M. Koo, S. Beckett, W. O. Okelo, A. L. Keyburn, B. S. Morgan, B. Drigo, M. Trau, E. Donner, S. P. Djordjevic and P. J. De Barro, Progressing Antimicrobial Resistance Sensing Technologies across Human, Animal, and Environmental Health Domains, ACS Sens., 2021, 6(12), 4283–4296 Search PubMed.
- N. Qin, P. Zhao, E. A. Ho, G. Xin and C. L. Ren, Microfluidic Technology for Antibacterial Resistance Study and Antibiotic Susceptibility Testing: Review and Perspective, ACS Sens., 2021, 6(1), 3–21 CrossRef CAS PubMed.
- R. B. P, B. G. Prajapati, R. Rajeshkumar, V. Balasubramaniam and S. Ponnusankar, Revolutionizing Antibacterial Strategies: Lipid Nanoparticles as Game-Changers in Combatting Multi-Drug-Resistant Infections, Curr. Med. Chem., 2024, 32(8), 1488–1506 Search PubMed.
- K. Jani, S. Mehta, R. Patel, B. Prajapati and G. Patel, Focused Insights into Liposomal Nanotherapeutics for Antimicrobial Treatment, Curr. Med. Chem., 2024 DOI:10.2174/0109298673322058241003073312.
- A. M. Stoorza and A. S. Duerfeldt, Guiding the Way: Traditional Medicinal Chemistry Inspiration for Rational Gram-Negative Drug Design, J. Med. Chem., 2024, 67(1), 65–80 Search PubMed.
- W. Vollmer, D. Blanot and M. A. De Pedro, Peptidoglycan Structure and Architecture, FEMS Microbiol. Rev., 2008, 32(2), 149–167 CrossRef CAS PubMed.
- A. J. F. Egan, J. Errington and W. Vollmer, Regulation of Peptidoglycan Synthesis and Remodelling, Nat. Rev. Microbiol., 2020, 18(8), 446–460 Search PubMed.
- M. S. Siegrist, S. Whiteside, J. C. Jewett, A. Aditham, F. Cava and C. R. Bertozzi, D-Amino Acid Chemical Reporters Reveal Peptidoglycan Dynamics of an Intracellular Pathogen, ACS Chem. Biol., 2013, 8(3), 500–505 Search PubMed.
- E. Kuru, H. V. Hughes, P. J. Brown, E. Hall, S. Tekkam, F. Cava, M. A. de Pedro, Y. V. Brun and M. S. VanNieuwenhze, In Situ Probing of Newly Synthesized Peptidoglycan in Live Bacteria with Fluorescent D-Amino Acids, Angew. Chem., Int. Ed., 2012, 51(50), 12519–12523 CrossRef CAS PubMed.
- D. Saxena, R. Maitra, R. Bormon, M. Czekanska, J. Meiers, A. Titz, S. Verma and S. Chopra, Tackling the Outer Membrane: Facilitating Compound Entry into Gram-Negative Bacterial Pathogens, npj Aantimicrob. Resist., 2023, 1(1), 1–22 Search PubMed.
- Y.-P. Hsu, J. Rittichier, E. Kuru, J. Yablonowski, E. Pasciak, S. Tekkam, E. Hall, B. Murphy, T. K. Lee, E. C. Garner, K. Casey Huang, Y. V. Brun and M. S. VanNieuwenhze, Full Color Palette of Fluorescent d -Amino Acids for in Situ Labeling of Bacterial Cell Walls, Chem. Sci., 2017, 8(9), 6313–6321 Search PubMed.
- J. M. Fura, D. Kearns and M. M. Pires, D-Amino Acid Probes for Penicillin Binding Protein-Based Bacterial Surface Labeling*, J. Biol. Chem., 2015, 290(51), 30540–30550 CrossRef CAS PubMed.
- Y. Zheng, X. Zhu, M. Jiang, F. Cao, Q. You and X. Chen, Development and Applications of D-Amino Acid Derivatives-Based Metabolic Labeling of Bacterial Peptidoglycan, Angew. Chem., Int. Ed., 2024, 63(17), e202319400 CrossRef CAS PubMed.
- E. Kuru, A. Radkov, X. Meng, A. Egan, L. Alvarez, A. Dowson, G. Booher, E. Breukink, D. I. Roper, F. Cava, W. Vollmer, Y. Brun and M. S. VanNieuwenhze, Mechanisms of Incorporation for D-Amino Acid Probes That Target Peptidoglycan Biosynthesis, ACS Chem. Biol., 2019, 14(12), 2745–2756 CrossRef CAS PubMed.
- Y. Li, Y. Zhou, X. Yue and Z. Dai, Cyanine Conjugate-based Biomedical Imaging Probes, Adv. Healthcare Mater., 2020, 9(22), 2001327 CrossRef CAS PubMed.
- X. Wang, G. Shi, S. Xu, Y. Sun, H. Qiu, Q. Wang, X. Han, Q. Zhang, T. Zhang and H.-Y. Hu, Unravelling Immune-Inflammatory Responses and Lysosomal Adaptation: Insights from Two-Photon Excited Delayed Fluorescence Imaging, Adv. Healthcare Mater., 2024, 13(15), 2304223 Search PubMed.
- L. D. Lavis, Teaching Old Dyes New Tricks: Biological Probes Built from Fluoresceins and Rhodamines, Annu. Rev. Biochem., 2017, 86, 825–843 Search PubMed.
- A. J. C. Sarris, T. Hansen, M. A. R. de Geus, E. Maurits, W. Doelman, H. S. Overkleeft, J. D. C. Codée, D. V. Filippov and S. I. van Kasteren, Fast and pH-Independent Elimination of Trans -Cyclooctene by Using Aminoethyl-Functionalized Tetrazines, Chem.–Eur. J., 2018, 24(68), 18075–18081 Search PubMed.
- Y. Li, S. Wang, Y. Chen, M. Li, X. Dong, H. C. Hang and T. Peng, Site-Specific Chemical Fatty-Acylation for Gain-of-Function Analysis of Protein S -Palmitoylation in Live Cells, Chem. Commun., 2020, 56(89), 13880–13883 Search PubMed.
- Y. H. An and R. J. Friedman, Concise Review of Mechanisms of Bacterial Adhesion to Biomaterial Surfaces, J. Biomed. Mater. Res., 1998, 43(3), 338–348 Search PubMed.
- A. H. Delcour, Outer Membrane Permeability and Antibiotic Resistance, Biochim. Biophys. Acta, Proteins Proteomics, 2009, 1794(5), 808–816 Search PubMed.
- G. M. Decad and H. Nikaido, Outer Membrane of Gram-Negative Bacteria. XII. Molecular-Sieving Function of Cell Wall, J. Bacteriol., 1976, 128(1), 325 Search PubMed.
- W. Chi, L. Huang, C. Wang, D. Tan, Z. Xu and X. Liu, A Unified Fluorescence Quenching Mechanism of Tetrazine-Based Fluorogenic Dyes: Energy Transfer to a Dark State, Mater. Chem. Front., 2021, 5(18), 7012–7021 Search PubMed.
- G. Beliu, A. J. Kurz, A. C. Kuhlemann, L. Behringer-Pliess, M. Meub, N. Wolf, J. Seibel, Z.-D. Shi, M. Schnermann, J. B. Grimm, L. D. Lavis, S. Doose and M. Sauer, Bioorthogonal Labeling with Tetrazine-Dyes for Super-Resolution Microscopy, Commun. Biol., 2019, 2(1), 1–13 CrossRef CAS PubMed.
- M. R. Karver, R. Weissleder and S. A. Hilderbrand, Synthesis and Evaluation of a Series of 1,2,4,5-Tetrazines for Bioorthogonal Conjugation, Bioconjugate Chem., 2011, 22(11), 2263–2270 CrossRef CAS PubMed.
- J. A. Wagner, D. Mercadante, I. Nikić, E. A. Lemke and F. Gräter, Origin of Orthogonality of Strain-Promoted Click Reactions, Chem.–Eur. J., 2015, 21(35), 12431–12435 CrossRef CAS PubMed.
- A. Bandyopadhyay, S. Cambray and J. Gao, Fast Diazaborine Formation of Semicarbazide Enables Facile Labeling of Bacterial Pathogens, J. Am. Chem. Soc., 2017, 139(2), 871–878 Search PubMed.
- Y. Kawai, K. Mickiewicz and J. Errington, Lysozyme Counteracts β-Lactam Antibiotics by Promoting the Emergence of L-Form Bacteria, Cell, 2018, 172(5), 1038–1049 CrossRef CAS.
- M. Leaver, P. Domínguez-Cuevas, J. M. Coxhead, R. A. Daniel and J. Errington, Life without a Wall or Division Machine in Bacillus Subtilis, Nature, 2009, 457(7231), 849–853 Search PubMed.
- R. Mercier, Y. Kawai and J. Errington, General Principles for the Formation and Proliferation of a Wall-Free (L-Form) State in Bacteria, eLife, 2014, 3, e04629 Search PubMed.
- P. Domínguez-Cuevas, R. Mercier, M. Leaver, Y. Kawai and J. Errington, The Rod to L-Form Transition of Bacillus Subtilis Is Limited by a Requirement for the Protoplast to Escape from the Cell Wall Sacculus, Mol. Microbiol., 2012, 83(1), 52–66 Search PubMed.
- S. Takahashi, M. Mizuma, S. Kami and H. Nishida, Species-Dependent Protoplast Enlargement Involves Different Types of Vacuole Generation in Bacteria, Sci. Rep., 2020, 10(1), 8832 Search PubMed.
- M. A. Kohanski, D. J. Dwyer and J. J. Collins, How Antibiotics Kill Bacteria: From Targets to Networks, Nat. Rev. Microbiol., 2010, 8(6), 423–435 CrossRef CAS PubMed.
- N. G. Bush, I. Diez-Santos, L. R. Abbott and A. Maxwell, Quinolones: Mechanism, Lethality and Their Contributions to Antibiotic Resistance, Molecules, 2020, 25(23), 5662 CrossRef CAS PubMed.
- R. A. Adams, G. Leon, N. M. Miller, S. P. Reyes, C. H. Thantrong, A. M. Thokkadam, A. S. Lemma, D. M. Sivaloganathan, X. Wan and M. P. Brynildsen, Rifamycin Antibiotics and the Mechanisms of Their Failure, J. Antibiot., 2021, 74(11), 786–798 CrossRef CAS PubMed.
- D. H. Williams and B. Bardsley, The Vancomycin Group of Antibiotics and the Fight against Resistant Bacteria, Angew. Chem., Int. Ed., 1999, 38(9), 1172–1193 CrossRef PubMed.
- P. J. Stogios and A. Savchenko, Molecular Mechanisms of Vancomycin Resistance, Protein Sci., 2020, 29(3), 654–669 CrossRef CAS PubMed.
- A. Brauner, O. Fridman, O. Gefen and N. Q. Balaban, Distinguishing between Resistance, Tolerance and Persistence to Antibiotic Treatment, Nat. Rev. Microbiol., 2016, 14(5), 320–330 CrossRef CAS PubMed.
- I. Levin-Reisman, I. Ronin, O. Gefen, I. Braniss, N. Shoresh and N. Q. Balaban, Antibiotic Tolerance Facilitates the Evolution of Resistance, Science, 2017, 355(6327), 826–830 CrossRef CAS PubMed.
- I. Levin-Reisman, A. Brauner, I. Ronin and N. Q. Balaban, Epistasis between Antibiotic Tolerance, Persistence, and Resistance Mutations, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(29), 14734–14739 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details about abbreviations, materials and methods, synthetic procedures, compounds characterization, confocal fluorescence imaging conditions, and theoretical calculations, etc.; UV-vis spectra, mass spectra, additional confocal imaging data, NMR spectra. See DOI: https://doi.org/10.1039/d5sc01586c |
| This journal is © The Royal Society of Chemistry 2025 |