Highly active nanoparticle enhanced rapid adsorption-killing mechanism to combat multidrug-resistant bacteria†
Yunyun
Xue‡
ad,
Zihao
Zhao‡
ad,
Wenbo
Huang‡
b,
Zelin
Qiu
a,
Xiao
Li
b,
Yu
Zhao
a,
Chuyao
Wang
a,
Ronglu
Cui
a,
Shuyang
Shen
a,
Hua
Tian
a,
Lifeng
Fang
 *a,
Rong
Zhou
*bc and
Baoku
Zhu
*a,
Rong
Zhou
*bc and
Baoku
Zhu
 *ad
*ad
aDepartment of Polymer Science and Engineering, ERC of Membrane and Water Treatment (MOE), Key Laboratory of Macromolecular Synthesis and Functionalization (MOE), Zhejiang University, Hangzhou, 310027, China. E-mail: fanglf@zju.edu.cn
bState Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, the First Affiliated Hospital of Guangzhou Medical University, Guangzhou Medical University, Guangzhou 510182, China. E-mail: zhourong@vip.163.com
cGuangzhou Laboratory, Guangzhou 510182, China
dCenter of Healthcare Materials, Shaoxing Institute, Zhejiang University, Shaoxing, 312000, China. E-mail: zhubk@zju.edu.cn
First published on 21st July 2023
Abstract
Contact-killing surfaces with the ability to rapidly adsorb and kill microorganisms are desperately needed since the rapid outbreak of multidrug-resistant (MDR) bacteria poses a serious threat to human health. Therefore, a series of amphiphilic nanoengineered polyquaterniums (ANPQs) were synthesized, and immobilizing ANPQs onto equipment surfaces provided a simple method for preventing microbial infections. The strong charge-positive property of ANPQ offered the possibility of rapid adsorption and efficient killing, such that all bacteria are adsorbed after 10 seconds of contact with ANPQ-treated fabrics, and more than 99.99% of pathogens are killed within 30 seconds. Surprisingly, the adsorption-killing mechanism made it difficult for bacteria to develop resistance to ANPQ coating, even after long-term repeated treatment. Importantly, in a Methicillin-resistant Staphylococcus aureus infection model, ANPQ-treated fabrics exhibited a potent anti-infectious performance while remaining nontoxic. It is envisaged that the strategy of using ANPQ coating undoubtedly provides a promising candidate for fighting MDR strains.
1 Introduction
Owing to the prolonged abuse of antibiotics, the pandemic of antimicrobial resistance has been greatly concerning.1,2 Notably, hospital-acquired infections caused by multidrug-resistant (MDR) strains have endangered patient safety, protracted hospitalization periods, escalated treatment expenses, and resulted in elevated rates of mortality and morbidity.3,4 Contamination of medical implants and equipment surfaces with MDR pathogens has emerged as a severe issue in the healthcare sector.3,4 Urgent measures are imperative to devise tactics that can realize surfaces with rapid adsorption and killing mechanisms. To tackle the aforementioned challenge, endeavors have been undertaken to design diverse strategies, such as coating materials with antimicrobial agents, such as zinc oxide nanoparticles, copper nanoparticles, and silver nanoparticles.5–8 Despite the promising antibacterial efficacy, the leakage of metal ions from metallic nanoparticles may pose a threat to the environment and cause excessive buildup in tissues, which could potentially endanger human health.9,10An alternative option is to employ bactericidal contact-killing surfaces, which execute bacterial death through various mechanisms upon adhesion. Among the most prevalent selections for developing such surfaces are antimicrobial peptides, quaternary ammonium compounds, and polycations.11–14 Benefitting from the advantages of cost-effectiveness, ease of scale-up, and less susceptibility to antibiotic resistance, cationic polymer nanoparticles have recently gained widespread attention.15–20 In contrast to monovalent systems, these nanoparticles have multiple ligands that can bind to several receptors on microbial membranes, leading to stronger interactions.19,21 The higher local cationic charge and hydrophobic bulk in the assembled form enable these nanoparticles to establish improved electrostatic contact with the microbial membrane and confer enhanced antibacterial activity.19,21 However, the majority of synthetic polymer nanoparticles documented in the literature are water-soluble,22 which can lead to uneven release kinetics, reservoir exhaustion concerns, and environmental issues.23–25 Therefore, it is critical to create water-insoluble polymer nanoparticles for bactericidal contact-killing surfaces.
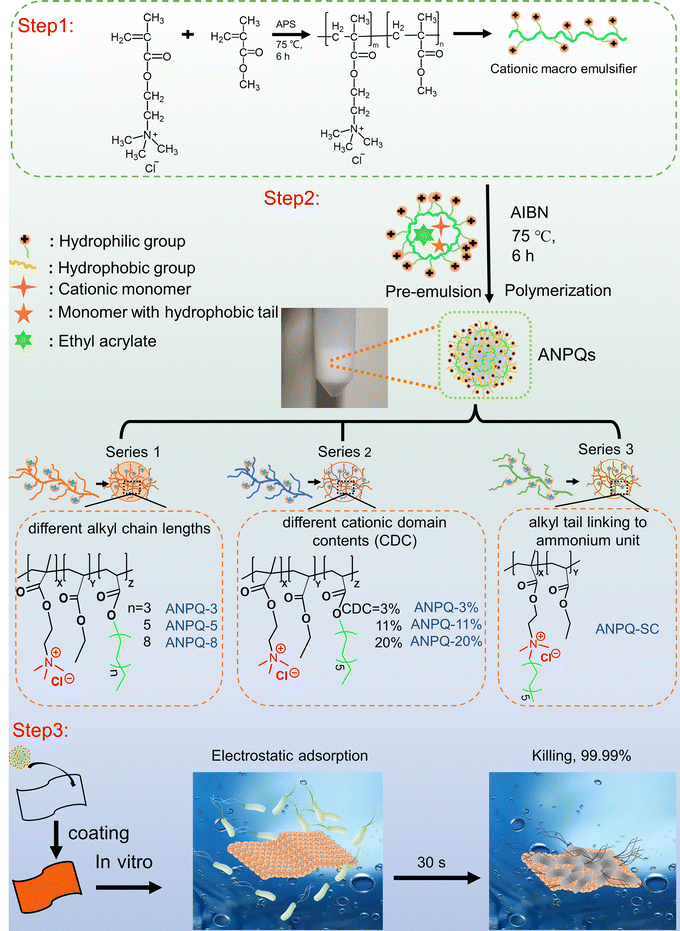
In this study, a series of amphiphilic nanoengineered polyquaterniums (ANPQ) containing cationic groups and hydrophobic alkyl tails were designed (Scheme 1). Escherichia coli (E. coli), Staphylococcus aureus (S. aureus), Pseudomonas aeruginosa (PAO1), and Methicillin-resistant Staphylococcus aureus (MRSA), which are the most common causes of hospital-acquired infections, were used to evaluate in vitro antimicrobial activities of ANPQ. The antibacterial activity of ANPQs was primarily derived from electrostatic adsorption and hydrophobic interaction, severely resulting in membrane translocation and membrane damage. The ability of ANPQ-treated fabrics to promote rapid adsorption and high-efficiency pathogen killing was evaluated. In the presence of a 1/4-fold MBA concentration of ANPQ-11%, E. coli ATCC 8099, S. aureus ATCC 6538, PAO1, and MRSA cells were passed, and MBA was measured at each passage. Importantly, ANPQ-treated fabrics demonstrated fast and high-effective antibacterial activity against MDR bacteria in vitro and MRSA infection in vivo, with negligible toxicity and drug resistance, indicating a high potential for biomedical applications.
2 Experimental section
2.1 Synthesis of amphiphilic nanoengineered polyquaterniums (ANPQs)
In conventional micro-emulsion polymerization, the addition of a significant amount of small molecule emulsifiers necessitated a complex post-processing procedure. Therefore, in this work, ANPQs were synthesized by self-emulsifying polymerization of macromolecular emulsifiers, which eliminated the complex post-processing of small molecule emulsifiers.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5, and the reaction was carried out at 60 °C for 24 hours under magnetic stirring (300 rpm). Finally, the product was precipitated and washed three times with ice ether, followed by vacuum drying to obtain the intermediate monomer (Q-SC).
1.5, and the reaction was carried out at 60 °C for 24 hours under magnetic stirring (300 rpm). Finally, the product was precipitated and washed three times with ice ether, followed by vacuum drying to obtain the intermediate monomer (Q-SC).
| Antibacterial polymers | Alkyl chain component | Cationic domain component | EA (mol%) | ASC (%) | MC (%) | |
|---|---|---|---|---|---|---|
| Note: ASC represents actual solid content. MC represents monomer conversion (%). | ||||||
| ANPQ-6 | Different alkyl chain lengths | 26 mol% C8 | 11 mol% | 63 | 31 | 98.6 |
| ANPQ-10 | 26 mol% C12 | 11 mol% | 63 | 28 | 98.8 | |
| ANPQ-16 | 26 mol% C18 | 11 mol% | 63 | 27 | 87.5 | |
| ANPQ-3% | Different cationic domain | 29 mol% C12 | 3 mol% | 68 | 28 | 99.5 |
| ANPQ-11% | 26 mol% C12 | 11 mol% | 63 | 28 | 98.8 | |
| ANPQ-20% | 23 mol% C12 | 20 mol% | 57 | 26 | 97.8 | |
| ANPQ-SC | Alkyl tail linking to ammonium unit | 43 mol% | 43 mol% | 57 | 29 | 82.5 |
| Q-SC | Q-SC | |||||
Monomer conversion was measured by the weighing method. Briefly, a certain amount of ANPQ emulsions was demulsified using ethanol and cleaned with water and acetone repeatedly to remove unreacted monomers and impurities. The obtained polymer was dried in a vacuum drying oven at 80 °C to constant weight. The monomer conversion (%) was calculated using the formula: MC = (W0 − W1)/W0 × 100%. W0 and W1 are the weights of ANPQ emulsions before and after treatment, respectively.
2.2 Fabrication of antibacterial coatings
In brief, ANPQ emulsion was diluted to 1 wt% using ultrapure water at room temperature. Polypropylene (PP) nonwoven fabrics were then immersed in the emulsion for 30 minutes. Then, the treated fabrics were taken out and dried under 150 °C for 10 min. Afterward, the ANPQ-treated fabrics were ultrasonically cleaned (37 °C, 5 min) to remove the superficially unstable nanoparticles, and finally dried under 150 °C for 10 min. By referring to the previous method,26 the coating amount of ANPQs was calculated according to eqn (1):| Coating amount (g m−2) = (M1 − M0)/S | (1) |
2.3 Antibacterial assays of ANPQs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min). The collected bacterial pellets were fixed with glutaraldehyde solution (2.5%) overnight at 4 °C for further Scanning Electron Microscopy (SEM) (FESEM, S-4800, Japan) and Transmission Electron Microscopy (TEM, Thermo Scientific Talos L120C) observations.
000 rpm for 5 min). The collected bacterial pellets were fixed with glutaraldehyde solution (2.5%) overnight at 4 °C for further Scanning Electron Microscopy (SEM) (FESEM, S-4800, Japan) and Transmission Electron Microscopy (TEM, Thermo Scientific Talos L120C) observations.
2.4 Hemolysis assays of ANPQs
Fresh pig blood (sodium heparin as an anticoagulant) was purchased from Beijing Bersee Science and Technology CO. LTD (Beijing, China) and was used to evaluate the toxicity of ANPQs on normal cells in vitro. Due to the fact that red blood cells (RBCs) are the most fragile cells, the biocompatibility of ANPQs can be directly reflected through hemolytic activity. Firstly, RBCs were collected by centrifugation (3000 rpm, 5 min) and washed until the supernatant was clear and transparent. RBCs were diluted to 2 wt% with sterile saline for use. Equal volume RBCs suspension was incubated with ANPQs emulsions at 37 °C for 60 min to obtain HC50, while 200 μL of RBC suspension was cultured with ANPQs coating at the same condition to calculate HA50%. HC50 and HA50 are defined as the concentration and the coating amount that lyses 50% of the red blood cells and releases hemoglobin, respectively. The experiments were performed in triplicate with two repeats.2.5 Antibacterial assays of ANPQ-fabrics
| Antibacterial ratio (%) = (A0 − A)/A × 100 | (2) |
2.6 Rapid adsorption-killing mechanism
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 5 min). Supernatants were measured for spectrophotometric absorbance readings at 260 nm (corresponding to nucleic acids) and 280 nm (corresponding to proteins)27,28 using a UV-vis spectrophotometer (UV2600, Shimadzu, Japan).
000 rpm, 5 min). Supernatants were measured for spectrophotometric absorbance readings at 260 nm (corresponding to nucleic acids) and 280 nm (corresponding to proteins)27,28 using a UV-vis spectrophotometer (UV2600, Shimadzu, Japan).
2.7 Inhibition zone test
According to the standard GB/T 31713-2015, a zone of inhibition could be used to evaluate the leaching of ANPQ coating. Briefly, test fabric samples were cut into a square shape with a side length of 1.5 cm × 1.5 cm. The bacterial suspension (∼107 CFU mL−1) was uniformly spread on the broth agar using a sterile cotton swab, and the samples were placed. Then, the plates were incubated at 36 °C for 24 hours. After incubation, the inhibition zone was imaged by a digital camera. The antibacterial performance was evaluated by the diameter of the inhibition zone with three bioindependent replicates and two technical replicates each time.2.8 Long-term stability
The long-term stability of antibacterial coatings was examined by leaching tests in 0.9% NaCl (aq). After continuous treatment time (1, 3, 5, 7, 9, and 30 days) under 37 °C and a shaking rate of 200 rpm. Antibacterial ratio, surface zeta potential, ATR-FTIR spectra, and SEM of antibacterial coatings were analyzed, respectively.2.9 Antibacterial resistance test
E. coli ATCC 8099, S. aureus ATCC 6538, PAO1, and MRSA cells were passaged in the presence of 1/4 MBA of ANPQ-11% coating, and MBA was measured at each passage of bacteria. These manipulations were repeated 40 times consecutively to assess the effect of ANPQ-11% on the antibiotic resistance of E. coli ATCC 8099, while 10 times consecutively for the antibiotic resistances of S. aureus ATCC 6538, PAO1, and MRSA. To further investigate the biological mechanism of bacterial response to ANPQ-11%, each group was subjected to transcription sequencing after 40 passages of incubation under 1/4-fold MBA0.2.10 In vivo antibacterial activity
All animal experiments were performed under the Principles of Laboratory Animal Care (NIH Publication No. 86-23, revised 1985) and the guidelines of the Animal Care and Use Committee of Zhejiang University. Female mice (ICR, ∼20 g) were provided by the Zhejiang Academy of Medical Sciences. After anesthetization, the mice's backs were shaved and sterilized. Then, a circular whole-skin wound (8 mm in diameter) was created and all wounds were infected with 50 μL of MRSA suspension (∼108 CFU mL−1). After infection for 24 h, the purulent wounds were treated with medical gauze, PP, and PPANPQ-11%, respectively. Medical gauze served as a control. Treatments were performed every two days and the wound area was recorded by a digital camera. The wound size was calculated using ImageJ-Pro Plus 6.0 software. The wound ratio (%) was calculated by eqn (3):| Wound ratio (%) = (At/A0) × 100% | (3) |
To evaluate the in vivo antibacterial activity, wound exudates were collected using sterile swabs and amplified in liquid LB solution at 37 °C for 4 h and then spread on agar plates for bacterial colony counting. On the 10th day, wound site tissues were collected and stained with hematoxylin and eosin (H&E), Masson's trichrome, and myeloperoxidase (MPO) for evaluating the actual antibacterial activity and wound healing. IL-6 expression in wound tissue from infected wounds was characterized by an ELISA kit.
2.11 In vitro and in vivo biosafety assay
The in vitro cytotoxicity was evaluated by incubating PPANPQ-11% with RBCs or L929 cells. An in vitro irritation test and a guinea pig maximization test were also performed. The experimental details are supplied in the SI. To evaluate the in vivo biosafety of PPANPQ-11%, on the 10th day, blood and major organs (including heart, liver, spleen, lungs, and kidneys) were collected and stained with H&E.3 Results and discussion
3.1 Synthesis and characterization of ANPQ
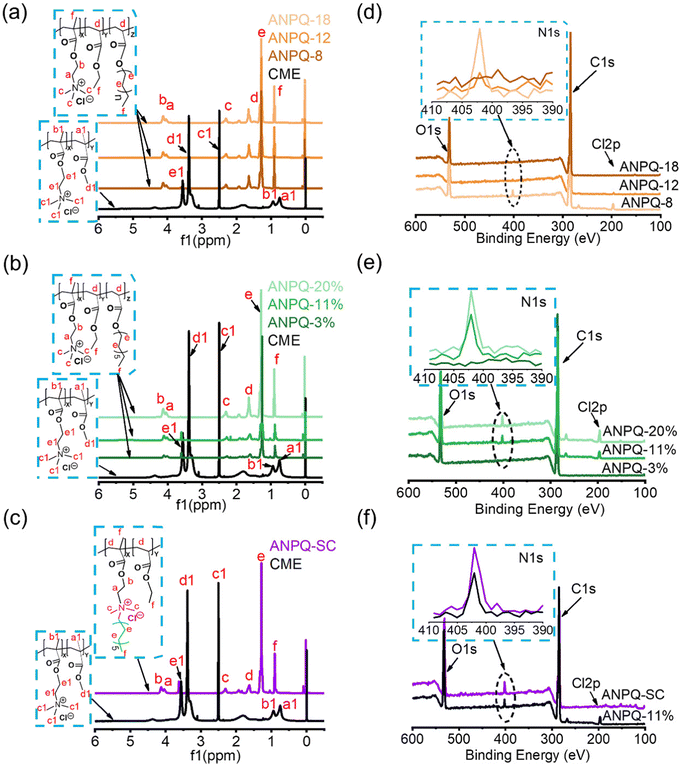
To investigate that ANPQ was synthesized as expected, the structure of ANPQ was characterized using 1H NMR spectra and XPS spectra, and related data are listed in Table 2. As shown in Fig. 1a–c, the 1H NMR spectra of CEM showed peaks at 3.5 ppm for –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2–N+(CH3)3 and –O–C
2–N+(CH3)3 and –O–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2–CH2–N+(CH3)3 groups, 3.4 ppm for –O–C
2–CH2–N+(CH3)3 groups, 3.4 ppm for –O–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3, 2.5 ppm for –N+(C
3, 2.5 ppm for –N+(C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)3, 0.8–0.9 ppm for –C–C
3)3, 0.8–0.9 ppm for –C–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3 group, while in the 1H NMR spectra of ANPQs at varying different alkyl chain lengths and cationic domain contents, there were peaks at 4.0–4.1 ppm for –O–C
3 group, while in the 1H NMR spectra of ANPQs at varying different alkyl chain lengths and cationic domain contents, there were peaks at 4.0–4.1 ppm for –O–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2–N+(CH3)3 and –O–C
2–N+(CH3)3 and –O–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2–CH2–N+(CH3)3 groups, 2.3 ppm for –O–C
2–CH2–N+(CH3)3 groups, 2.3 ppm for –O–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3 group, 1.6 ppm for –CH2–C
3 group, 1.6 ppm for –CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) - group, 1.3 ppm for –C
- group, 1.3 ppm for –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2–C
2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2– group, 0.9 ppm for –C–C
2– group, 0.9 ppm for –C–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3 and –CH2–C
3 and –CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3 groups. These results demonstrated the successful synthesis of ANPQs.
3 groups. These results demonstrated the successful synthesis of ANPQs.
| Name | N atomic (%) | Zeta potential (mV) | MBA (mg cm−2) | MBAm (mg cm−2) | HC50 (mg cm−2) | SI | |||
|---|---|---|---|---|---|---|---|---|---|
| E. coli | S. aureus | PAO1 | MRSA | ||||||
| Note: MBAm was established as the geometric mean value of MBAs; SI: Selectivity index, HA50/MBAm. | |||||||||
| ANPQ-6 | 1.12 | 24.8 ± 1.6 | >2.25 | >2.25 | >2.25 | >2.25 | 2.25 | 9.00 | 3.8 |
| ANPQ-10 | 0.85 | 26.8 ± 1.0 | 0.35 | 0.69 | 0.69 | 0.69 | 0.61 | 9.64 | 15.6 |
| ANPQ-16 | 0.46 | 29.5 ± 1.7 | 0.32 | 0.64 | 1.28 | 0.64 | 0.73 | 3.06 | 4.2 |
| ANPQ-3% | 0.40 | 26.7 ± 1.9 | 0.98 | 1.95 | 1.95 | 1.95 | 1.71 | 10.36 | 6.0 |
| ANPQ-11% | 2.46 | 29.5 ± 1.1 | 0.35 | 0.69 | 0.35 | 0.69 | 0.52 | 9.76 | 18.8 |
| ANPQ-20% | 3.33 | 36.5 ± 2.7 | 0.27 | 0.13 | 0.54 | 0.27 | 0.30 | 3.26 | 10.8 |
| ANPQ-SC | 3.87 | 27.7 ± 2.3 | 2.35 | 1.18 | 2.35 | 1.18 | 1.77 | 1.18 | 0.6 |
While 1H NMR analysis showed the successful synthesis of ANPQs, as depicted in Scheme 1, XPS was needed to further investigate the chemical structures of ANPQs. As shown in Fig. 1d–f and Table 2, the N 1s spectra of ANPQs showed that there was a peak at 401.8 eV, which was reported as the amines in their protonated states (–N+(CH3)3).29 With the increase in the alkyl chain length, the intensity of N 1s spectra was decreased while it increased with the increase in the cationic domain content. As confirmed by XPS data, the N atomic content (%) of ANPQs at varying alkyl chain lengths was 1.1, 0.9, and 0.5, respectively; the N atomic content (%) of ANPQs at varying cationic domain contents was 0.4, 2.5, and 3.3, respectively. The N atomic content (%) of ANPQ-SC was 3.9.
As shown in Table 2, the values of zeta potential were 29.5 ± 1.7, 26.8 ± 1.0, 24.8 ± 1.6, 26.7 ± 1.9, 29.5 ± 1.1, 36.5 ± 2.7, and 27.7 ± 2.3 mV. With the increase in the cationic domain content, the zeta potential value increased. The high zeta potential values proved that the formation of nanoparticles increases the local density of the positive charge. The morphology of ANPQs was observed by SEM, as shown in Fig. S3 (ESI†), the nanoparticles were spherical in nature and had a size range from 250 to 500 nm after self-drying under air. These positively charged nanoparticles may easily interact with the surfaces of microbes by means of electrostatic interaction.
3.2 Antibacterial activity and antibacterial mechanism of ANPQs
The impressive antibacterial activity and biocompatibility observed in the polymers can be attributed to various factors, including the length of the side alkyl chain,30 hydrophilic–hydrophobic balance,31,32 as well as the linkage between functional groups.33 By optimizing these parameters, the polymers can exhibit enhanced performance in both antibacterial activity and biocompatibility. To investigate if ANPQs and ANPQs coating posed efficient killing and good biocompatibility as predicted, the MBC and HC50 values and MBA and HA50 values were determined. As shown in Fig. S4 (ESI†) and Table 2, the antibacterial activity of ANPQs varying chain length indicated that elongation of the chain length resulted in a gradual increase in hemolysis to where hemolysis was stronger than the antimicrobial effect.30 This was probably because the higher surface activities favor the enhancement of antibacterial activities with the increase in the alkyl chain length. However, polymers with a higher hydrophobic content are toxic to all cells and lose selectivity.25,34,35 In Fig. S5 (ESI†) and Table 2, the MBC, MBA, HC50, and HA50 values of ANPQs decreased with the increase of cationic domain content, due to the stronger electrostatic attraction causing lower values of MBC/MBA and higher hemolysis.31,32 ANPQ-SC with cationic groups and alkyl tails in the same center showed lower antimicrobial activity (Fig. S6, ESI†). That may be because the hydrophobic chain on the surface of ANPQ-SC plays a major role and its antimicrobial properties are limited, which was different from that described in Sen's work.33 ANPQ-11% showed the best antibacterial activity and biocompatibility with the highest selectivity index (18.8). This was because the appropriate alkyl chain length and hydrophilic–hydrophobic balance gave excellent antimicrobial properties and biocompatibility to ANPQ-11%. Therefore, ANPQ-11% was selected for the following characterization.To investigate why ANPQs show such great antimicrobial properties, the antibacterial mechanism was directly observed by using SEM and TEM. As shown in Fig. 2b and c, untreated PAO1 showed a complete rod-like morphology, whereas untreated MRSA exhibited a complete spherical structure. However, after incubation with ANPQ-11%, the surface of the bacteria adhered to many particles. Notably, some ANPQ-11% was partially phagocytosed and fused with the bacterial membrane structure, leaving a hemispherical indentation on the bacterial surface (Fig. 2b and c, yellow arrow). Due to the strong positive charge and the long alkyl chains, the electrostatic attraction and hydrophobic interactions between ANPQ-11% and the bacterial surface provided a favorable promotion. As a result, the strong interaction between bacterial membranes and particles caused a phagocytosis and fusion process. Since the spherical structure promoted interfacial interactions, more particles adhered.
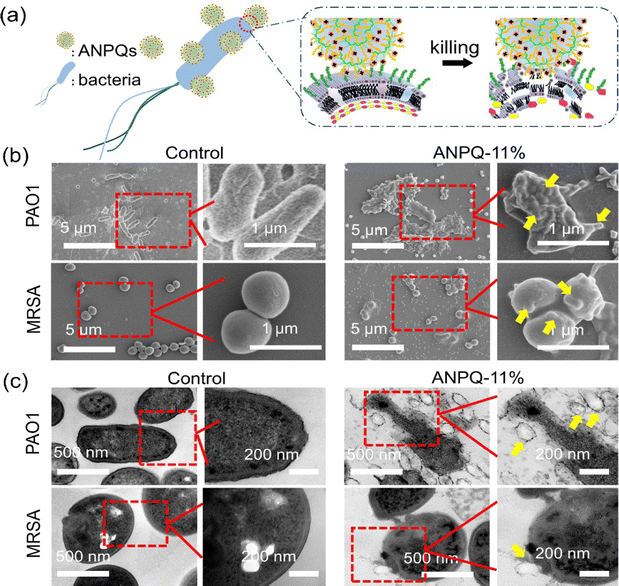 | ||
| Fig. 2 (a) Scheme of ANPQs against bacteria. (b) SEM and (c) TEM of bacteria structure before and after treatment with ANPQ-11%, respectively. | ||
3.3 Antibacterial activity and characterization of ANPQ coating
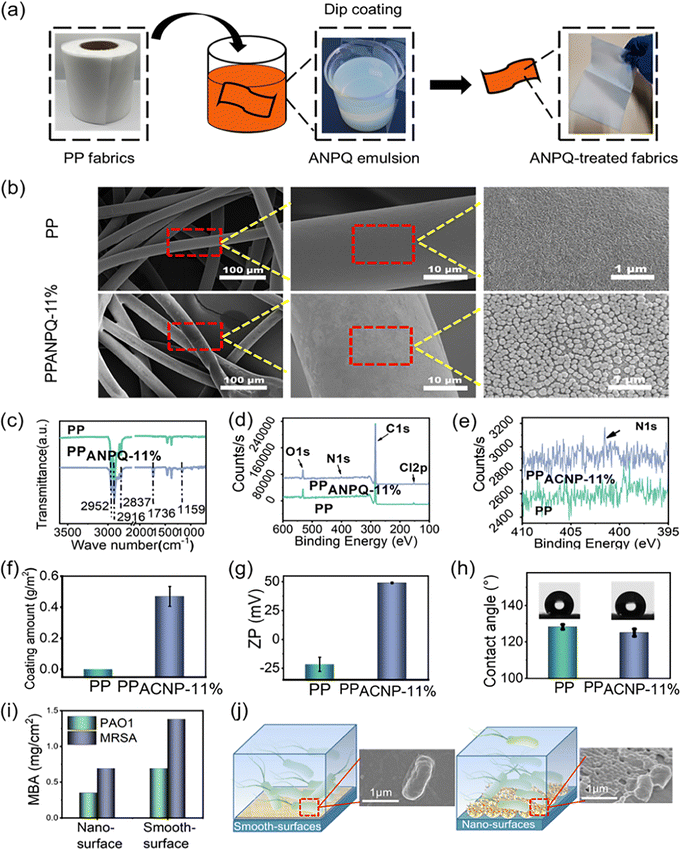
To confirm the utility of these polycations as antimicrobial coating materials for biomedical applications, the ANPQs were immobilized onto the PP nonwoven fabric (Fig. 3a). As shown in Fig. 3b and Fig. S7a, S8a (ESI†), ANPQ-11% nanoparticles were coated on the non-woven fabric uniformly. In ATR-FTIR spectra (Fig. 3c and Fig. S7b, S8b, ESI†), the new absorbance peak at 1727 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) suggested that the ANPQ-11% was successfully coated, which can be confirmed from the XPS results (Fig. 3d and e). The coating amount of ANPQ-11% was 0.47 g m−2 (Fig. 3f) and the surface zeta potential increased to 48.95 mV from −25.61 mV (Fig. 3g and h), which was higher than that of other ANPQs. The results indicated that the strong charge-positive property provided higher killing activity and better biocompatibility than that of other ANPQs (Fig. S7e–g and S8e–g, ESI†), which was consistent with the data from Table 2. More importantly, the water contact angle of PPANPQ-11%(125.08°) was nearly identical to that of pure PP (128.22°), indicating that the coating was water-insoluble.
O) suggested that the ANPQ-11% was successfully coated, which can be confirmed from the XPS results (Fig. 3d and e). The coating amount of ANPQ-11% was 0.47 g m−2 (Fig. 3f) and the surface zeta potential increased to 48.95 mV from −25.61 mV (Fig. 3g and h), which was higher than that of other ANPQs. The results indicated that the strong charge-positive property provided higher killing activity and better biocompatibility than that of other ANPQs (Fig. S7e–g and S8e–g, ESI†), which was consistent with the data from Table 2. More importantly, the water contact angle of PPANPQ-11%(125.08°) was nearly identical to that of pure PP (128.22°), indicating that the coating was water-insoluble.
To further investigate the superiority of the nanoparticle coating method, ANPQ-11% was coated on the wells of a 96-well plate. The coated wells treated with organic solvent were termed smooth-surface, while the coated wells without treatment were named nano-surface. As shown in Fig. 3i, the antimicrobial property of the nano-surface (MBA: 0.35 mg cm−2 for PAO1, 0.69 mg cm−2 for MRSA) was higher than that of the smooth-surface (MBA: 0.69 mg cm−2 for PAO1, 1.38 mg cm−2 for MRSA). To further study this phenomenon, SEM was conducted. After contact with the smooth surface, the cells were only slightly deformed (Fig. 3j). However, the cell structure contact with the nano-surface was significantly deformed and the cell membrane was substantially deformed and torn (Fig. 3j). This result suggested that the surface modified with nanoparticles enhanced the contact area and mechanical tension with bacteria, thus improving the antimicrobial property.36
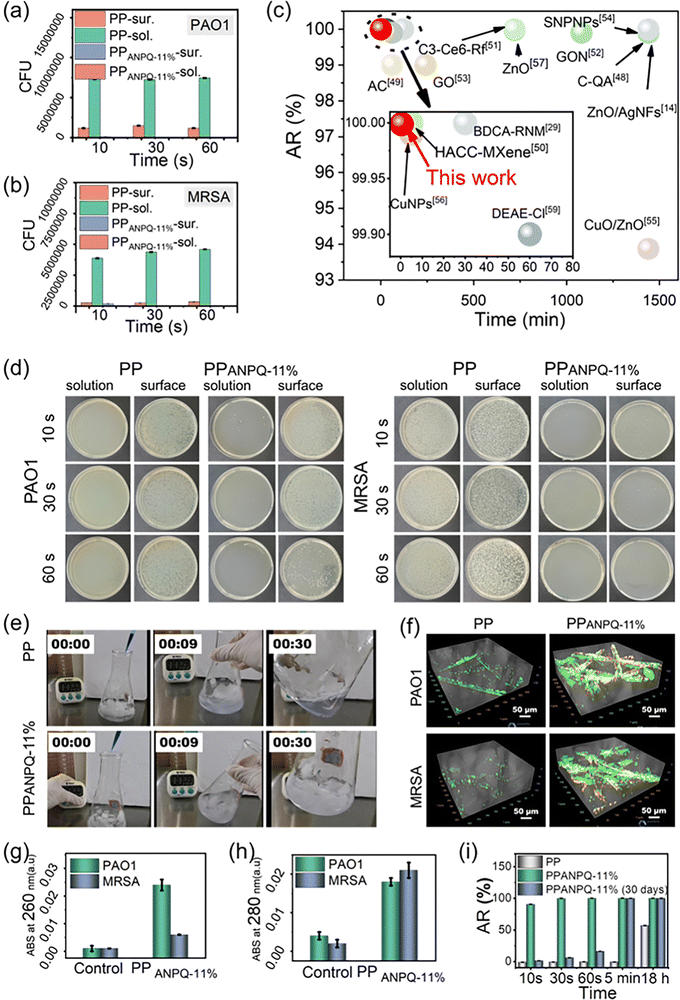
3.5 Microbe adhesion and killing efficiency of ANPQ-11% coating
Medical implants and devices contaminated with MDR pathogens urgently require new coated materials. Hence, it is necessary and critical to investigate whether the ANPQ coating exerted such an ability for enhancing the rapid adsorption-killing mechanism to combat MDR bacteria. As shown in Fig. 4a–c, untreated PP lacked the ability to absorb and kill bacteria. However, after contacting PPANPQ-11%, all the bacteria in the suspension were adsorbed within 10 s. The bacterial killing ratio of PPANPQ-11% reached 99.99% after 30 s and all bacteria were killed within 1 min. ANPQ-11% coating performed noticeably better than other coating materials, such as small molecular organics,27,37 linear quaternary ammonium salts,11–14 inorganic nanoparticles,38,39 metals, and metal oxide nanoparticles.8,40–43To investigate the adsorption-killing mechanism of ANPQ-11% coating, the bacteria were stained with crystal violet dye under Live/Dead staining. For the PPANPQ-11% groups, the solution was completely colorless after 10 s of exposure, while the solution for PP groups was still purple even after 30 s (Fig. 4e and Movies S1, S2, ESI†). As shown in Fig. 4f, only a few bacteria adhered to PP, and the cells were still viable. However, a large number of bacteria were attached to PPANPQ-11%, and the bacteria died due to the destruction of the cell membrane, which was confirmed by SEM images (Fig. S9 and S10, ESI†). Besides, after contact with PP, almost no organic matter was detected. In contrast, significant nucleic acid and protein leakage was observed after contact with PPANPQ-11% (Fig. 4g and h). More importantly, the zone of inhibition test confirmed that ANPQ-11%-coated surfaces killed bacteria by contact rather than through leaching (Fig. S11, ESI†).
As seen in Fig. 4i, after 30 days of continuous treatment, PPANPQ-11% demonstrated a 99.99% bacterial killing rate even after only 30 seconds of contact. The surface zeta potential of PPANPQ-11% only decreased slightly from 48.95 mV to 39.33 mV (Fig. S12, ESI†). From the ATR-FTIR spectra (Fig. S13, ESI†), the absorbance at 1727 cm−1 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) changed little. In addition, the SEM results (Fig. S14, ESI†) further confirmed the good durability of ANPQ-11%. On the one hand, the water-insoluble property demonstrated good durability. On the other hand, ANPQ-11% interacted with PP through hydrophobic interaction, giving it long-lasting antibacterial properties. More impressively, after 2 h of contact with PPANPQ-11%, the virus titer declined by 1.02 log after 2 h (Table S1, ESI†). The antiviral activity of PPANPQ-11% against the H1N1 virus highlighted its strong promise in avoiding the rapid spread of the virus in nosocomial infection.
O) changed little. In addition, the SEM results (Fig. S14, ESI†) further confirmed the good durability of ANPQ-11%. On the one hand, the water-insoluble property demonstrated good durability. On the other hand, ANPQ-11% interacted with PP through hydrophobic interaction, giving it long-lasting antibacterial properties. More impressively, after 2 h of contact with PPANPQ-11%, the virus titer declined by 1.02 log after 2 h (Table S1, ESI†). The antiviral activity of PPANPQ-11% against the H1N1 virus highlighted its strong promise in avoiding the rapid spread of the virus in nosocomial infection.
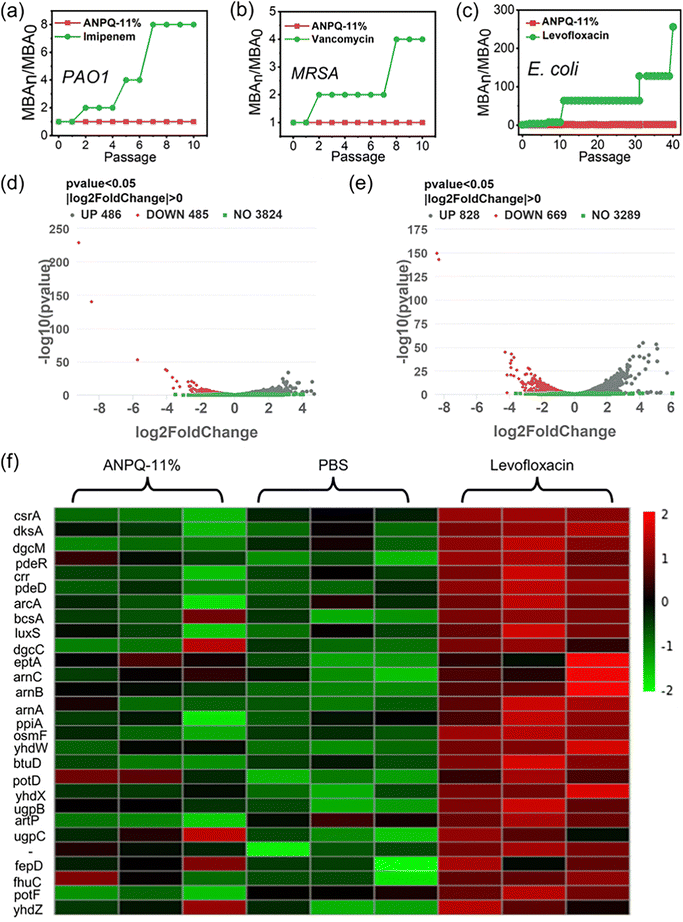
3.6 Bacterial antimicrobial resistance of ANPQ-11% coating
Antibiotic abuse results in the spread of MDR strains associated with nosocomial infection, which has become one of the most severe public health threats.44,45 Therefore, it is necessary to ensure whether incubating repeatedly with the ANPQ-11% coating results in antimicrobial resistance. As shown in Fig. 5a and b and Fig. S15 (ESI†), as compared to the antibiotic imipenem in PAO1, vancomycin in MRSA, and norfloxacin in S. aureus, there was no antimicrobial resistance to ANPQ-11% coating.Surprisingly, as shown in Fig. 5c, following levofloxacin treatment, the MBA value against E. coli increased to 64 times that of MBA0 even after 12 passages and to 256 times that of MBA0 after 40 passages, suggesting significant antimicrobial resistance. In contrast, after treatment with ANPQ-11% for 40 passages, the MBA value increased slightly to 2-fold of MBA0, demonstrating that the antimicrobial resistance was less likely to develop. To go deeper into the biological mechanism of bacterial response to ANPQ-11%, each group was subjected to transcriptome sequencing. As shown in Fig. 5d and e, 971 genes were differentially regulated (p value 0.05; |log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 Fold Chang| > 0) in response to ANPQ-11% treatment, which was significantly lower than levofloxacin (1497 genes changed). Bacterial resistance mechanisms typically include membrane-bound proteases that degrade peptides, efflux pumps, and transport systems that export cationic antimicrobial peptides (CAMPs) from periplasmic and intracellular compartments and regulation of extra-membrane permeability through outer membrane proteins, and so on.46–48 The effect of polymers on bacterial resistance was investigated by analyzing the expression of genes closely associated with antimicrobial resistance.
2 Fold Chang| > 0) in response to ANPQ-11% treatment, which was significantly lower than levofloxacin (1497 genes changed). Bacterial resistance mechanisms typically include membrane-bound proteases that degrade peptides, efflux pumps, and transport systems that export cationic antimicrobial peptides (CAMPs) from periplasmic and intracellular compartments and regulation of extra-membrane permeability through outer membrane proteins, and so on.46–48 The effect of polymers on bacterial resistance was investigated by analyzing the expression of genes closely associated with antimicrobial resistance.
According to Gene Ontology (GO) processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses (Fig. S16–S19, ESI†), genes that are closely associated with drug resistance were selected for detailed analysis (Fig. 5f). CAMP resistance genes, such as arnB, arnC, and arnA, as well as ABC transporters (osmF, yhdW, btuD, and others), were significantly upregulated in the levofloxacin-treated groups. There were fewer distinct differentially regulated genes after treatment with ANPQ-11%, and no significant transcriptional response was observed in any known resistance pathway (KEGG database). A series of major master regulators (including CsrA, dksA, and dgcM) of biofilm formation was significantly inhibited, indicating that the ANPQ-11% coating could inhibit adhesion and biofilm formation. Overall, antimicrobial resistance to ANPQ-11% coating was less likely to develop, which contributed to the potential for effective treatment against antimicrobial resistance.
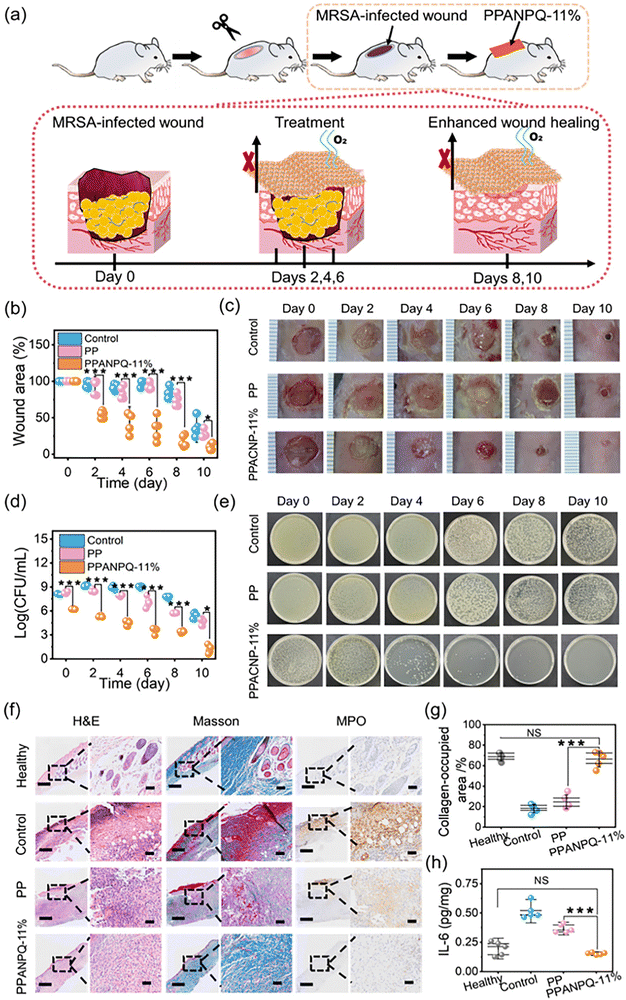
3.7 Improved healing in an MRSA-infected wound model
Infected wound exudate contains large amounts of bacteria, which can delay wound healing and increase bacterial infection.49 To test whether the ANPQ-treated fabrics pose such excellent adsorption-killing properties under pathological settings in vivo, a mouse whole-dermal MARS-infected wound model was established(Fig. 6a). As shown in Fig. 6b–e, on the 6th day, the control groups and the PP groups continued to deteriorate and the area of the wound grew larger with swelling, pus, and blood. Additionally, the bacteria CFUs were comparable to that of the untreated wound on the first day. By contrast, on the fourth day, the PPANPQ-11% treatment group demonstrated obvious improvement. By the eighth day, the wound area was reduced to 25.6% of its initial size (**p < 0.01) and nearly completely healed after ten days of treatment without swelling or scabbing. Significantly, the CFUs were reduced sharply even on the second day.To conduct in-depth research on the wound healing mechanism, wound histology sections were stained via H&E, Masson's trichrome, and MPO stains (Fig. 6f). Compared to the healthy groups, there was infiltration of inflammatory cells and crusting in the control groups without new granulation tissue and blood vessels growing, indicating that the wounds were more severe and difficult to heal. However, after treatment with PPANPQ-11%, the wounds were almost recovered by epithelium cells, the subcutaneous tissues were reconstructed, and the inflammatory cells were reduced significantly. As shown in Fig. 6f and g, a greater amount of nascent collagen was deposited in an orientation in the PPANPQ-11% groups rather than in the control groups and the PP groups, indicating that PPANPQ-11% exerted a potent effect on accelerating wound healing.50,51 MPO and IL-6, two powerful antibacterial proteins secreted by neutrophil granules, are linked to the body's level of inflammation, which can assess wound healing.52,53 In Fig. 6f, MPO expression in the control groups remained high on the last day, indicating a severe wound infection. However, in the PPANPQ-11% groups, MPO tissue area was significantly reduced and expression of IL-6 (0.21 pg mg−1) was obviously lower than that in the control group (0.56 pg mg−1, **p < 0.01), suggesting that the wound tissues had entered the proliferation and remodeling stage of wound healing from the inflammatory stage (Fig. 6h). Overall, ANPQ-treated fabrics showed an excellent antimicrobial effect, and effectively promoted wound healing even in the complex environment in vivo.
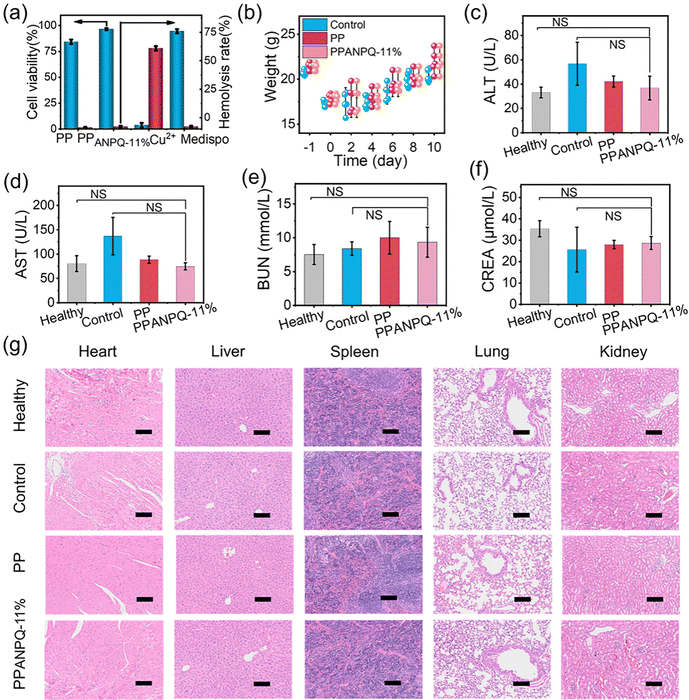
3.8 Biosafety in vitro and in vivo.
In addition to possessing excellent antibacterial properties, the biosafety of PPANPQ-11% is also crucial. Compared with commercial nonwovens coated with copper ions or small molecule quaternary ammonium salts, PPANPQ-11% had no cytotoxicity and hemolysis (Fig. 7a and Fig. S20, ESI†). Besides, PPANPQ-11% also exhibited no skin irritation or allergy (Tables S2–S4, ESI†). During the treatment, the weight of mice in all groups increased from the 4th day (Fig. 7b). At the end of day 10, the blood collected from all groups showed that blood biochemistry results were within the normal range with no significant differences, indicating that PPANPQ-11% had no significant systemic toxicity to liver and kidney functions (Fig. 7c–g). The above in vivo and in vitro results revealed that PPANPQ-11% indicated great promise as a biocompatible antibacterial candidate to fight the spread of MDR strains associated with nosocomial infection.4 Conclusion
In this work, we synthesized a series of amphiphilic nanoengineered polyquaternium (ANPQ) with strong electrostatic attraction and hydrophobic interactions, which showed a favorable promotion, causing bacterial phagocytosis and promoting membrane fusion. Noncovalent immobilization of the ANPQ coating provided a facile method to prevent microbial infections. Due to its water-insoluble properties, ANPQ coating demonstrated strong adhesion to the fabric with long-term durability (30 days). Surprisingly, all the bacteria were absorbed into the ANPQ-treated fabrics within only 10 s of contact, and after 30 s, the bacterial killing rate reached 99.99%. Besides, after 40 times of prolonged use, RNA-sep analysis proved that the bacteria developed resistance to levofloxacin antibiotics, but not to ANPQ-11% coating. More importantly, the ANPQ-treated fabrics exhibited excellent antibacterial properties, suggesting that the ANPQ-treated fabrics could effectively eliminate pathogens in wound exudate and facilitate skin wound healing. Moreover, nearly no cytotoxicity, hemolysis, skin irritation, and allergy were observed for ANPQ-treated fabrics, which undoubtedly made it a promising candidate for combating the spread of MDR strains.Author contributions
Yunyun Xue, Zihao Zhao, Wenbo Huang: conceptualization, methodology, investigation, writing an original draft, data curation. Zelin Qiu, Yu Zhao, Chuyao Wang, Ronglu Cui, Shuyang Shen, Hua Tian: writing – review and editing. Xiao Li, Rong Zhou: conceptualization, methodology, resources, supervision, writing – review and editing. Lifeng Fang, Baoku Zhu: conceptualization, methodology, resources, supervision, project administration, funding acquisition.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships in this paper.Acknowledgements
Funding from Natural Science Foundation of Zhejiang Province, China (No. LD22E030006), New material Platform Project, Ministry of Industry and Information Technology of China (No. TC190H3ZV-1), and Fundamental Research Funds for the Central Universities (Grant No. 226-2022-00020) are highly acknowledged.References
- D. Mukwege, P. Mitangala, A. Byabene, E. Busha, Y. Van Laethem and O. Vandenberg, Lancet, 2022, 399, 2348 CrossRef.
- R. Laxminarayan, Lancet, 2022, 399, 606–607 CrossRef CAS.
- E. Boelee, G. Geerling, B. van der Zaan, A. Blauw and A. D. Vethaak, Acta Trop., 2019, 193, 217–226 CrossRef.
- Water-Associated Infectious Diseases, ed.S. K. Saxena, Springer Singapore, Singapore, 2020 Search PubMed.
- P. Singh and I. Mijakovic, Sci. Rep., 2022, 12, 7902 CrossRef CAS.
- Y. Liu, N. Peng, Y. Yao, X. Zhang, X. Peng, L. Zhao, J. Wang, L. Peng, Z. Wang, K. Mochizuki, M. Yue and S. Yang, Nat. Commun., 2022, 13, 3581 CrossRef CAS.
- Y. Jiang, W. Zheng, K. Tran, E. Kamilar, J. Bariwal, H. Ma and H. Liang, Nat. Commun., 2022, 13, 197 CrossRef CAS.
- S. Karagoz, N. B. Kiremitler, G. Sarp, S. Pekdemir, S. Salem, A. G. Goksu, M. S. Onses, I. Sozdutmaz, E. Sahmetlioglu, E. S. Ozkara, A. Ceylan and E. Yilmaz, ACS Appl. Mater. Interfaces, 2021, 13, 5678–5690 CrossRef CAS.
- T. Ma, X. Zhai, Y. Huang, M. Zhang, X. Zhao, Y. Du and C. Yan, Adv. Healthcare Mater., 2021, 10, 2100033 CrossRef CAS.
- P. Westerhoff, A. Atkinson, J. Fortner, M. S. Wong, J. Zimmerman, J. Gardea-Torresdey, J. Ranville and P. Herckes, Nat. Nanotechnol., 2018, 13, 661–669 CrossRef CAS.
- L. Wang, X. Wen, X. Zhang, S. Yuan, Q. Xu, F. Fu, H. Diao and X. Liu, Cellulose, 2021, 28, 5867–5879 CrossRef CAS.
- Q. Xu, L. Ying, X. Wang, Y. Zhang and P. Wang, Fibers Polym., 2022, 23, 944–953 CrossRef CAS.
- B. Yan, X. Bao, X. Liao, P. Wang, M. Zhou, Y. Yu, J. Yuan, L. Cui and Q. Wang, ACS Appl. Mater. Interfaces, 2022, 14, 2132–2145 CrossRef CAS.
- L. Song, L. Sun, J. Zhao, X. Wang, J. Yin, S. Luan and W. Ming, ACS Appl. Bio Mater., 2019, 2, 2756–2765 CrossRef CAS.
- Z. Guo, S. Li, Y. Qu, J. Lu, W. Xue, X. Yu and Z. Liu, Chem. Eng. J., 2021, 417, 127970 CrossRef CAS.
- S. J. Lam, E. H. H. Wong, C. Boyer and G. G. Qiao, Prog. Polym. Sci., 2018, 76, 40–64 CrossRef CAS.
- L.-Z. Su, Y. Liu, Y.-F. Li, Y.-L. An and L.-Q. Shi, Chin. J. Polym. Sci., 2021, 39, 1376–1391 CrossRef CAS.
- T. Selkälä, T. Suopajärvi, J. A. Sirviö, T. Luukkonen, P. Kinnunen, K. I. Kling, J. B. Wagner and H. Liimatainen, Chem. Eng. J., 2019, 374, 1013–1024 CrossRef.
- F. Nederberg, Y. Zhang, J. P. K. Tan, K. Xu, H. Wang, C. Yang, S. Gao, X. D. Guo, K. Fukushima, L. Li, J. L. Hedrick and Y.-Y. Yang, Nat. Chem., 2011, 3, 409–414 CrossRef CAS.
- J. M. V. Makabenta, A. Nabawy, C.-H. Li, S. Schmidt-Malan, R. Patel and V. M. Rotello, Nat. Rev. Microbiol., 2021, 19, 23–36 CrossRef CAS PubMed.
- S. J. Lam, N. M. O’Brien-Simpson, N. Pantarat, A. Sulistio, E. H. H. Wong, Y.-Y. Chen, J. C. Lenzo, J. A. Holden, A. Blencowe, E. C. Reynolds and G. G. Qiao, Nat. Microbiol., 2016, 1, 16162 CrossRef CAS.
- S. Tian, H. C. van der Mei, Y. Ren, H. J. Busscher and L. Shi, J. Mater. Sci. Technol., 2021, 84, 208–218 CrossRef CAS.
- Y. K. Jo, J. H. Seo, B.-H. Choi, B. J. Kim, H. H. Shin, B. H. Hwang and H. J. Cha, ACS Appl. Mater. Interfaces, 2014, 6, 20242–20253 CrossRef CAS.
- F. Wu, G. Meng, J. He, Y. Wu, F. Wu and Z. Gu, ACS Appl. Mater. Interfaces, 2014, 6, 10005–10013 CrossRef CAS.
- J. Hoque, P. Akkapeddi, V. Yadav, G. B. Manjunath, D. S. S. M. Uppu, M. M. Konai, V. Yarlagadda, K. Sanyal and J. Haldar, ACS Appl. Mater. Interfaces, 2015, 7, 1804–1815 CrossRef CAS.
- J. Lin, X. Chen, C. Chen, J. Hu, C. Zhou, X. Cai, W. Wang, C. Zheng, P. Zhang, J. Cheng, Z. Guo and H. Liu, ACS Appl. Mater. Interfaces, 2018, 10, 6124–6136 CrossRef CAS.
- Y. Si, Z. Zhang, W. Wu, Q. Fu, K. Huang, N. Nitin, B. Ding and G. Sun, Sci. Adv., 2018, 4, eaar5931 CrossRef.
- C. Z. Chen and S. L. Cooper, Biomaterials, 2002, 23, 3359–3368 CrossRef CAS PubMed.
- J. Ederer, P. Janoš, P. Ecorchard, J. Tolasz, V. Štengl, H. Beneš, M. Perchacz and O. Pop-Georgievski, RSC Adv., 2017, 7, 12464–12473 RSC.
- T. N. Siriwardena, M. Stach, R. He, B.-H. Gan, S. Javor, M. Heitz, L. Ma, X. Cai, P. Chen, D. Wei, H. Li, J. Ma, T. Köhler, C. van Delden, T. Darbre and J.-L. Reymond, J. Am. Chem. Soc., 2018, 140, 423–432 CrossRef CAS.
- S. Zhao, W. Huang, C. Wang, Y. Wang, Y. Zhang, Z. Ye, J. Zhang, L. Deng and A. Dong, Biomacromolecules, 2020, 21, 5269–5281 CrossRef CAS PubMed.
- E. F. Palermo, K. Lienkamp, E. R. Gillies and P. J. Ragogna, Angew. Chem., Int. Ed., 2019, 58, 3690–3693 CrossRef CAS PubMed.
- V. Sambhy, B. R. Peterson and A. Sen, Angew. Chem., Int. Ed., 2008, 47, 1250–1254 CrossRef CAS PubMed.
- S. Colak, C. F. Nelson, K. Nüsslein and G. N. Tew, Biomacromolecules, 2009, 10, 353–359 CrossRef CAS.
- E. F. Palermo, I. Sovadinova and K. Kuroda, Biomacromolecules, 2009, 10, 3098–3107 CrossRef CAS.
- C. Q. Lai, Langmuir, 2018, 34, 4059–4070 CrossRef CAS.
- P. Tang, Z. Zhang, A. Y. El-Moghazy, N. Wisuthiphaet, N. Nitin and G. Sun, ACS Appl. Mater. Interfaces, 2020, 12, 49442–49451 CrossRef CAS.
- F. Yaghoubidoust and E. Salimi, Fibers Polym., 2019, 20, 1155–1160 CrossRef CAS.
- J. Zhao, B. Deng, M. Lv, J. Li, Y. Zhang, H. Jiang, C. Peng, J. Li, J. Shi, Q. Huang and C. Fan, Adv. Healthcare Mater., 2013, 2, 1259–1266 CrossRef CAS.
- B. R. Rao, R. Kumar, S. Haque, J. M. Kumar, T. N. Rao, R. V. S. N. Kothapalli and C. R. Patra, ACS Appl. Mater. Interfaces, 2021, 13, 10689–10704 CrossRef CAS PubMed.
- G. Guan, L. Zhang, J. Zhu, H. Wu, W. Li and Q. Sun, J. Hazard. Mater., 2021, 402, 123542 CrossRef CAS PubMed.
- S. Kumar, M. Karmacharya, S. R. Joshi, O. Gulenko, J. Park, G.-H. Kim and Y.-K. Cho, Nano Lett., 2021, 21, 337–343 CrossRef CAS PubMed.
- X. Ni, C. Li, Y. Lei, Y. Shao, Y. Zhu and B. You, ACS Appl. Mater. Interfaces, 2021, 13, 57864–57879 CrossRef CAS PubMed.
- The EuSCAPE Working Group, the ESGEM Study Group, S. David, S. Reuter, S. R. Harris, C. Glasner, T. Feltwell, S. Argimon, K. Abudahab, R. Goater, T. Giani, G. Errico, M. Aspbury, S. Sjunnebo, E. J. Feil, G. M. Rossolini, D. M. Aanensen and H. Grundmann, Nat. Microbiol., 2019, 4, 1919–1929 CrossRef.
- O. O. Ikhimiukor, E. E. Odih, P. Donado-Godoy and I. N. Okeke, Nat. Microbiol., 2022, 7, 757–765 CrossRef CAS.
- Y. Gao, J. Wang, M. Chai, X. Li, Y. Deng, Q. Jin and J. Ji, ACS Nano, 2020, 14, 5686–5699 CrossRef CAS PubMed.
- Y. Yu, J. Dong, Y. Wang and X. Gong, PeerJ, 2021, 9, e11081 CrossRef PubMed.
- W. Chin, G. Zhong, Q. Pu, C. Yang, W. Lou, P. F. De Sessions, B. Periaswamy, A. Lee, Z. C. Liang, X. Ding, S. Gao, C. W. Chu, S. Bianco, C. Bao, Y. W. Tong, W. Fan, M. Wu, J. L. Hedrick and Y. Y. Yang, Nat. Commun., 2018, 9, 917 Search PubMed.
- B. A. Mast and G. S. Schultz, Wound Repair Regener., 1996, 4, 411–420 CrossRef CAS PubMed.
- J. Wu, J. Zhu, C. He, Z. Xiao, J. Ye, Y. Li, A. Chen, H. Zhang, X. Li, L. Lin, Y. Zhao, J. Zheng and J. Xiao, ACS Appl. Mater. Interfaces, 2016, 8, 18710–18721 CrossRef CAS.
- W. Gao, W. Jin, Y. Li, L. Wan, C. Wang, C. Lin, X. Chen, B. Lei and C. Mao, J. Mater. Chem. B, 2017, 5, 7285–7296 RSC.
- S. Kongkiatkamon, L. Terkawi, Y. Guan, V. Adema, M. Hasipek, T. Dombrovski, M. Co, W. Walter, H. Awada, Y. Parker, S. Hutter, S. Pagliuca, C. Gurnari, H. J. Rogers, M. Meggendorfer, D. J. Lindner, T. Haferlach, V. Visconte, T. LaFramboise, B. K. Jha and J. P. Maciejewski, Leukemia, 2022, 36, 2086–2096 CrossRef CAS PubMed.
- T. M. Kistner, B. K. Pedersen and D. E. Lieberman, Nat. Metab., 2022, 4, 170–179 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb01105d |
| ‡ Equally contributing first authors. |
| This journal is © The Royal Society of Chemistry 2023 |